This work demonstrates that low ascorbate triggers abscisic acid-, salicylic acid-, and jasmonate-dependent signaling pathways in leaves that together regulate plant growth and defense responses. It provides insights into how cellular redox state regulates the expression of transcription factors and controls plant growth.
Abstract
Cellular redox homeostasis is a hub for signal integration. Interactions between redox metabolism and the ABSCISIC ACID-INSENSITIVE-4 (ABI4) transcription factor were characterized in the Arabidopsis thaliana vitamin c defective1 (vtc1) and vtc2 mutants, which are defective in ascorbic acid synthesis and show a slow growth phenotype together with enhanced abscisic acid (ABA) levels relative to the wild type (Columbia-0). The 75% decrease in the leaf ascorbate pool in the vtc2 mutants was not sufficient to adversely affect GA metabolism. The transcriptome signatures of the abi4, vtc1, and vtc2 mutants showed significant overlap, with a large number of transcription factors or signaling components similarly repressed or induced. Moreover, lincomycin-dependent changes in LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN 1.1 expression were comparable in these mutants, suggesting overlapping participation in chloroplast to nucleus signaling. The slow growth phenotype of vtc2 was absent in the abi4 vtc2 double mutant, as was the sugar-insensitive phenotype of the abi4 mutant. Octadecanoid derivative-responsive AP2/ERF-domain transcription factor 47 (ORA47) and AP3 (an ABI5 binding factor) transcripts were enhanced in vtc2 but repressed in abi4 vtc2, suggesting that ABI4 and ascorbate modulate growth and defense gene expression through jasmonate signaling. We conclude that low ascorbate triggers ABA- and jasmonate-dependent signaling pathways that together regulate growth through ABI4. Moreover, cellular redox homeostasis exerts a strong influence on sugar-dependent growth regulation.
INTRODUCTION
Ascorbic acid is an essential component of the cellular reduction/oxidation hub that buffers the production of reactive oxygen species (ROS) while allowing essential signaling that regulates plant growth and defense (Gong et al., 2007; Chaouch et al., 2010). Signals from metabolism and the environment as well as biotic and abiotic stress responses converge at the cellular redox hub (Fujita et al., 2006; Foyer and Noctor, 2009). Ascorbate is crucial to continuous ROS processing, and it also acts as a signaling molecule with distinct roles in the regulation of plant growth and defense (Pastori et al., 2003; Foyer and Noctor, 2005; Foyer and Noctor, 2011). Ascorbate and ascorbate oxidase exert a strong influence on plant growth and development (Chinoy, 1984; Pignocchi et al., 2003; Conklin and Barth, 2004; Barth et al., 2006). Arabidopsis thaliana mutants that are completely deficient in ascorbate are embryo lethal (Lukowitz et al., 2001; Dowdle et al., 2007). The vitamin C–defective (vtc) mutants vtc1 and vtc2 are impaired in ascorbate synthesis. vtc1 harbors a point mutation in the ascorbate biosynthetic enzyme GDP-Man pyrophosphorylase (Conklin et al., 1999). vtc2 encodes the enzyme GDP-l-galactose phosphorylase, which catalyzes the conversion of GDP-l-galactose to l-galactose 1-phosphate in the first committed step of the ascorbate synthesis pathway in Arabidopsis leaves (Linster et al., 2007). The vtc1 and vtc2 mutants have a lowered abundance of ascorbate (<30% of the wild-type levels), and they show a slow growth phenotype (Conklin et al., 1999; Veljovic-Jovanovic et al., 2001; Pastori et al., 2003). The leaves of the vtc mutants have smaller cells (Pavet et al., 2005), and they show enhanced basal resistance to biotrophic pathogens (Pavet et al., 2005).
Ascorbate is accumulated to high levels in growing tissues, where it plays a key role in cell growth via regulation of the cell cycle (Potters et al., 2002, 2004), whereas ascorbate depletion is associated with quiescence (Kerk and Feldman, 1995; Potters et al., 2002, 2004). Ascorbate has functions distinct from those of glutathione in the control of cell proliferation (Noctor et al., 2000; Potters et al., 2002; Pellny et al., 2009; Diaz Vivancos et al., 2010a, 2010b). Ascorbate also participates in the synthesis (Sommer-Knudsen et al., 1998) and cross-linking of cell wall components (Smirnoff, 2000; Pellny et al., 2009) and thus participates in the redox regulation of cell expansion (Esaka, 1998; Kato and Esaka, 2000).
The leaves of vtc1 have high abscisic acid (ABA) levels compared with the wild type and show alterations in gene expression patterns that are characteristic of altered ABA signaling (Kiddle et al., 2003; Pastori et al., 2003). ABA and gibberellin (GA) are considered to act antagonistically in plant growth and defense, the ratio of ABA to GA being a fundamental determinant of cell growth or quiescence (Finkelstein and Rock, 2002). The availability of ascorbate can influence growth through effects on GA synthesis because ascorbate is the cofactor involved in the catalysis of 2-oxoacid-dependant dioxygenase reactions (Arrigoni and De Tullio, 2000). Dioxygenases are important in the final stages of GA synthesis, where GA12 is converted to bioactive GAs (Hedden and Kamiya, 1997). The activities of these enzymes are enhanced by the addition of ascorbate in vitro (Graebe, 1987; Hedden, 1992; Lange, 1994; Prescott and John, 1996; Lukacin and Britsch, 1997).
The ABA signaling pathway involves ROS production via the activation of NADPH oxidases (RbohD and RbohF) (Torres et al., 2002; Kwak et al., 2003; Torres and Dangl, 2005). ABA signaling pathways have been implicated in plastid-derived retrograde signaling (Koussevitzky et al., 2007) pathways and in plant–pathogen interactions (Ton and Mauch-Mani, 2004). Ascorbate also fulfils crucial roles in photosynthesis and chloroplast function, and it has been implicated in the control of the expression of genes encoding chloroplast proteins (Kiddle et al., 2003). In the chloroplast, ascorbate protects the photosynthetic machinery by removing ROS (Foyer and Noctor, 2011). It also reduces tocopheroxyl radicals produced in the thylakoid membranes as a result of reactions associated with photosystem II function (Havaux, 2003). Ascorbate in the thylakoid lumen can act as an emergency electron donor to photosystem II when the oxygen-evolving complex is inactivated in stressful conditions (Tóth et al., 2009). The ascorbate pool in the lumen also protects photosystem II because ascorbate is a cofactor for violaxanthin deepoxidase, which is a component of the xanthophyll cycle. The vtc mutants are impaired in nonphotochemical quenching under high light (Müller-Moulé et al., 2003, 2004; Smirnoff, 2000).
The nuclear-localized Apetala 2-type (AP2) transcription factor ABSCISIC ACID-INSENSITIVE-4 (ABI4) is important in ABA signaling during seed development and germination (Finkelstein et al., 1998). In addition, ABI4 fulfils other important roles in the regulation of plant development, such as in Glc responses (Arenas-Huertero et al., 2000), in nitrate responses (Signora et al., 2001), and in chloroplast-to-nucleus retrograde signaling pathways (Kaliff et al., 2007; Koussevitzky et al., 2007) and mitochondria-to-nucleus retrograde signaling pathways (Giraud et al., 2009).
The signaling roles of ROS in regulating plant responses to environmental and metabolic triggers have been extensively studied (Mittler, 2002; Mittler et al., 2004; Mhamdi et al., 2010), but much less information is available concerning how cellular redox buffers, such as ascorbate, regulate plant growth and defense processes. The following experiments were performed to determine the signaling mechanisms that contribute to the low ascorbate-dependent regulation of growth and gene expression. A key goal was to provide new information concerning the complex interplay between redox, Glc, and plastid signaling pathways, particularly in relation to the role of the ABI4 transcription factor. The following analysis of the vtc1, vtc2, abi4, and abi4 vtc2 mutants provides conclusive evidence of strong interactions between ascorbate (redox), sugar, ABA, and jasmonate (JA) signaling pathways in the regulation of plant growth and development.
RESULTS
Characteristics of the abi4, vtc1, and vtc2 Mutants
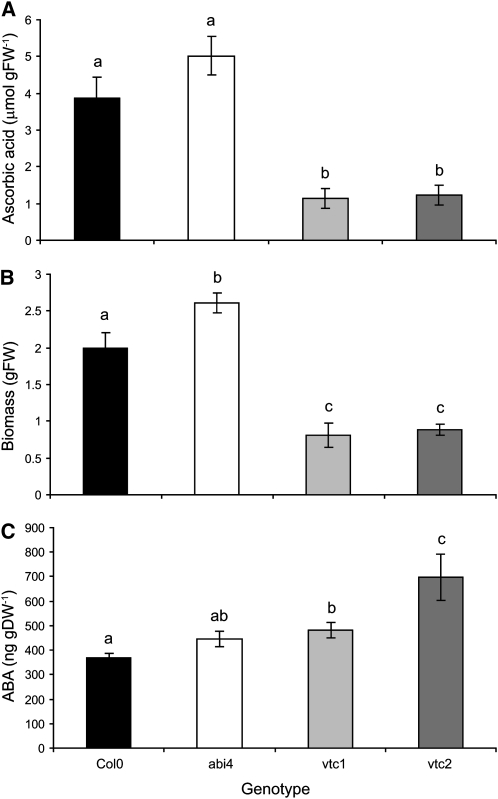
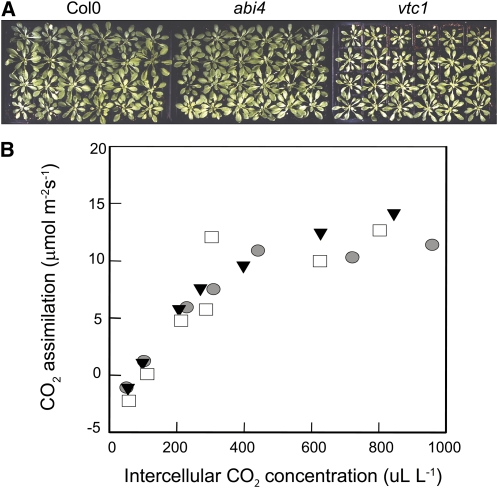
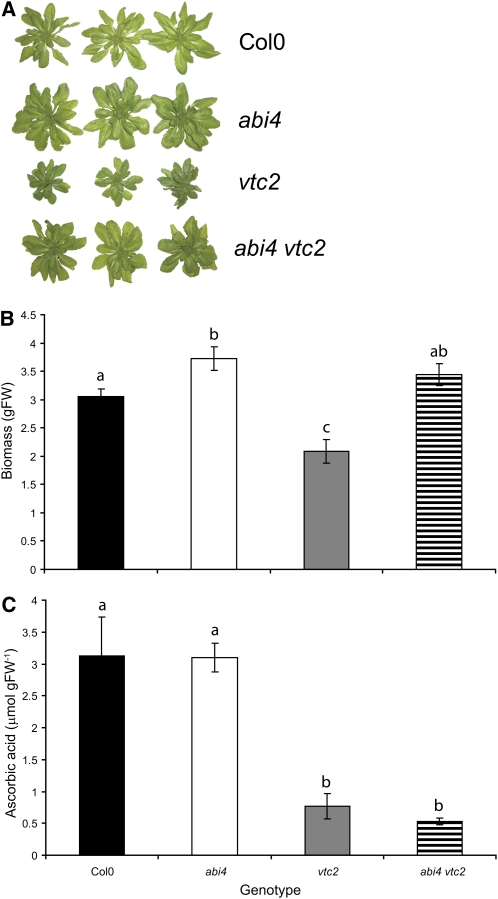
The ascorbate-defective vtc1 and vtc2 mutants had significantly lower levels of ascorbate than Columbia-0 (Col-0; Figure 1A), as previously reported (Conklin et al., 1999; Veljovic-Jovanovic et al., 2001; Pavet et al., 2005). The vtc1 and vtc2 mutants also showed a slow growth phenotype relative to Col-0 (Figures 1B and 2A), as reported previously (Pavet et al., 2005). By contrast, the abi4 mutants had similar ascorbate levels (Figure 1A), a slightly increased growth phenotype (Figures 1B and 2A), and similar ABA levels (Figure 1C) to Col-0. The leaves of the vtc1 and vtc2 mutants had higher ABA levels than the Col-0 leaves (P = 0.027 and 0.026 respectively; Figure 1C). The photosynthetic CO2 assimilation rates measured in rosette leaves at 6 weeks were similar in all genotypes (Figure 2).
Figure 1.
Characterization of Col-0, abi4, vtc1, and vtc2.
(A) Rosette ascorbic acid content in 4-week-old plants (n = 3).
(B) Rosette fresh weight (FW) in 4-week-old plants (n = 6).
(C) Rosette ABA content in 6-week-old plants (n = 3). DW, dry weight.
Plants were germinated and grown in soil in controlled environment chambers as described in the text. Values are represented as mean ± se, and those that were significantly different from one another according to Fisher’s protected LSD test are indicated by different letters (P < 0.05).
Figure 2.
Characterization of Photosynthesis in Col-0, abi4, and vtc2.
Plants were germinated and grown for 6 weeks as described in the text.
(A) Images of genotypes following 6 weeks of growth.
(B) CO2 assimilation plotted as a function of intercellular CO2 concentration. Filled triangles, Col-0; gray circles, abi4; filled squares, vtc1.
[See online article for color version of this figure.]
Chloroplast-to-Nucleus Retrograde Signaling in the abi4, vtc1, and vtc2 Mutants
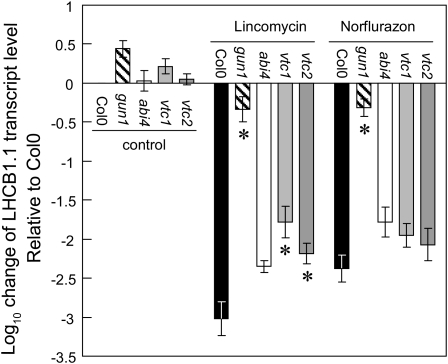
The nucleus has a preeminent role in regulating chloroplast development, the nuclear genome encoding >95% of all chloroplast-located proteins. Nevertheless, chloroplasts retain some control over the expression of many of these nuclear genes via chloroplast-to-nucleus retrograde signaling pathways. The following experiments were conducted to determine whether chloroplast-to-nucleus retrograde signaling was altered in the vtc1 and vtc2 mutants, using norflurazon and lincomycin to induce the retrograde signaling pathways. The addition of norflurazon (a carotenoid biosynthesis inhibitor) or lincomycin (an inhibitor of translation on 70S chloroplast ribosomes) to Col-0 decreased the accumulation of transcripts of nuclear genes encoding photosynthesis-related proteins, such as LHCB1.1 (Figure 3). By contrast, the repression of LHCB1 transcripts by these inhibitors was largely absent in the genomes uncoupled1 (gun1) mutant, which is defective in retrograde plastid-to-nucleus signaling (Figure 3). Under the growth irradiances (100 μmol m−2 s−1) used in this study, the abundance of LHCB1.1 transcripts in the abi4 mutants was similar to that of Col-0 in the presence of norflurazon or lincomycin. However, the repression caused by lincomycin treatments was significantly less marked in the vtc1 and vtc2 mutants than that observed in Col-0 and abi4 (Figure 3). With the exception of the gun1 mutant, the abundance of LHCB1.1 transcripts was similar in all genotypes in the presence of norflurazon (Figure 3).
Figure 3.
Impact of Lincomycin and Norflurazon Treatment on LHCB1.1 Transcript Levels in Col-0, gun1, abi4, vtc1, and vtc2.
LHCB1.1 transcript levels were quantified following lincomycin and norflurazon treatments, and data are presented as mean transcript expression ± se (n = 3). Values significantly different from those of Col-0 estimated by performing the Student’s t test on individual Δ cycle threshold values as suggested by Yuan et al. (2006) are indicated by asterisks (P < 0.05).
Characteristics of the abi4 vtc2 Double Mutants
Despite extensive efforts, we were unable to isolate abi4 vtc1 double mutants (see Supplemental Figure 1 online). In these studies, double mutants were selected either first for insensitivity to ABA and then screened for low ascorbate (see Supplemental Figure 1A online) or on the basis of low ascorbate with subsequent sequencing to identify the abi4 mutation (see Supplemental Figure 1B online). This failure might be explained by the proximity of the sequences encoding GDP-mannose pyrophosphorylase (Conklin et al., 1999) and ABI4 on Arabidopsis chromosome 2. However, according to The Arabidopsis Information Resource (TAIR), these sequences are ~200 kb apart. Another possibility, given the embryo lethality of severe vtc1 mutants (CYTOKINESIS DEFECTIVE1) and the semidominance of the allele used in this study, is that the double mutation is also lethal. This possibility is indicated by the developmental failure observed in ~25% of the seeds (see Supplemental Figure 1C online, top seedpod). The vtc1 mutation resides in an early step in the pathway of ascorbate synthesis, which also affects cell wall metabolism (Veljovic-Jovanovic et al., 2001). By contrast, the vtc2 mutation occurs in the Gal pathway, which is specific to ascorbate synthesis (Dowdle et al., 2007; Linster et al., 2007). We therefore crossed the abi4 and vtc2 mutants and verified the homozygous abi4 vtc2 double mutants by sequencing, insensitivity of germination to ABA, and ascorbate contents.
Glc Signaling
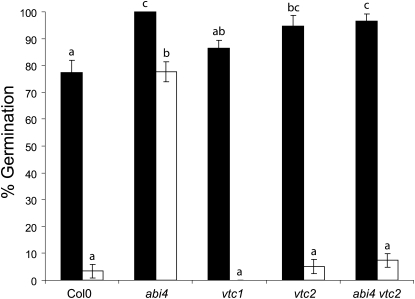
The following experiments were undertaken to determine the effects of low ascorbate on Glc signaling, which regulates a wide range of metabolic and developmental processes in plants. Literature evidence, concisely summarized by Bossi et al. (2009), indicates that abi4 plays an important role in sugar signaling, as well as ABA responses. The abi4 seeds were able to germinate on media containing a high level of Glc, whereas Col-0 seeds showed a greatly decreased ability to germinate under these conditions (see Supplemental Figure 2 online). The germination of the vtc1 and vtc2 mutant seeds showed a similar inhibition by high Glc to that observed in Col-0 (see Supplemental Figure 2 online). Like vtc2, the abi4 vtc2 double mutants exhibited a Glc-sensitive phenotype (see Supplemental Figure 2 online). A quantitative comparison of seed germination rates (Figure 4) revealed that all genotypes showed high germination (>75%) on Murashige and Skoog (MS) media. The germination of Col-0 seeds was decreased to <5% when the MS media was supplemented with 6% Glc (Figure 4). However, the germination rate of the abi4 seeds remained high (>75%) on high Glc-containing media (Figure 4). The germination rates of the vtc1, vtc2, and the abi4 vtc2 double mutants were similar to Col-0 under the high Glc growth conditions (Figure 4). Thus, the Glc-insensitive phenotype of the abi4 seeds was repressed in the low ascorbate (vtc2) background.
Figure 4.
Germination of Col-0, abi4, vtc1, vtc2, and abi4 vtc2 in the Presence of Glc.
Mature, stratified seeds were plated onto MS media (black bars) or MS supplemented with 6% Glc (white bars). Plates were incubated as described for 10 d and the percentage germination estimated visually. Data are presented as mean ± se (n = 3), and values that were significantly different within a treatment group according to Fisher’s protected LSD test are indicated by different letters.
Growth Repression and Restoration
As shown in Figures 1B and 2A, the vtc2 mutants have a marked slow growth phenotype relative to Col-0. However, the abi4 vtc2 double mutants showed a similar growth phenotype (Figure 5A) and rosette biomass accumulation to Col-0 (Figure 5B), despite having similar ascorbate levels to the vtc2 mutants (Figure 5C).
Figure 5.
Characterization of abi4 vtc2 in Comparison with Col-0, abi4, and vtc2.
(A) Visual phenotype of Col-0, abi4, vtc2, and abi4 vtc2 following 6 weeks of growth under controlled environment conditions as described in the text.
(B) Rosette fresh weight (FW) following 4 weeks of growth (n = 6).
(C) Rosette ascorbic acid content following 4 weeks of growth (n = 4). Data in (B) and (C) are represented as mean ± se, and values that were significantly different according to Fisher’s protected LSD test are indicated by different letters (P < 0.05).
[See online article for color version of this figure.]
Comparisons of the abi4, vtc1, vtc2, and abi4 vtc2 Leaf Transcriptomes
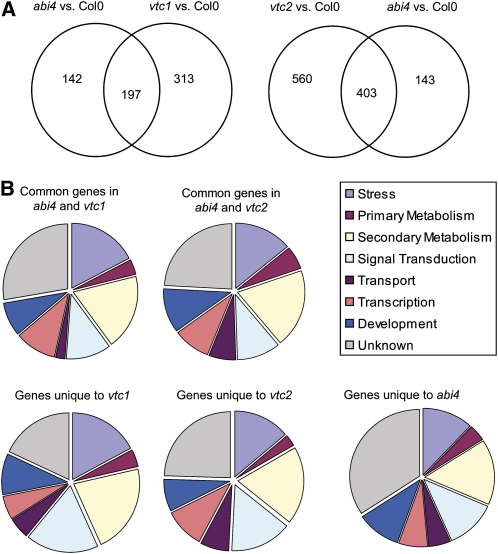
Comparisons of the leaf transcriptomes of Col-0, abi4, vtc1, vtc2, and abi4 vtc2 double mutants were made at the 6-week stage (Figure 6). The abi4 leaf transcriptome was analyzed relative to Col-0 in separate series of experiments. In one set of comparisons, the abi4 and vtc1 transcriptomes were analyzed relative to Col-0 (see Supplemental Data Set 1 online), and in the second set of comparisons the abi4, vtc2, and abi4 vtc2 double mutant transcriptomes were analyzed relative to Col-0 (see Supplemental Data Set 2 online). A total of 339 transcripts were differentially expressed in abi4 relative to Col-0 with 249 of these increased relative to Col-0 and 90 decreased. Many of the transcripts that were enhanced in abi4 relative to Col-0 encode proteins that are associated with defense, such as PATHOGENESIS RELATED1 (PR1), PR4, β-glucanase, glutaredoxin, heat shock protein 17, and harpin-induced protein 1 (see Supplemental Data Set 3 online).
Figure 6.
Differential Expression of Transcripts in abi4, vtc1, and vtc2 relative to Col-0.
(A) Comparison of transcripts differentially expressed in abi4, vtc1, and vtc2 relative to Col-0. Venn diagrams indicate differentially expressed transcripts that were common or separate in abi4 and vtc1 or abi4 and vtc2.
(B) Functional categorization of transcripts uniquely differentially expressed in abi4, vtc1, and vtc2 or commonly differentially expressed in abi4 and vtc1 or abi4 and vtc2.
Analysis of the vtc1 transcriptome revealed that 510 transcripts were differentially expressed relative to Col-0 (see Supplemental Data Set 4 online), while 963 transcripts were differentially expressed in vtc2 (see Supplemental Data Set 5 online). Of the transcripts that were differentially expressed in vtc1, vtc2, and abi4 relative to Col-0 (Figure 6A), a large number were identical in all mutants. The nature of the genes that were either common or differentially expressed in the mutants and double mutant relative to Col-0 were broadly comparable in terms of functional categories (Figure 6B).
Of the transcripts that were repressed or induced in a similar manner in all three mutants (abi4, vtc1, and vtc2; see Supplemental Data Sets 3 to 5 online), a large number are either transcription factors or involved in signaling. This includes several WRKY transcription factors (40, 47, and 53), NAC transcription factors (anac036 and anac090), and ZAT 10, which are significantly overexpressed, along with a number of other transcription factors in all three mutants compared with Col-0 (Table 1). Moreover, several ethylene-responsive transcription factors, such as ERF104, were also highly expressed in the abi4, vtc1, and vtc2 mutants relative to Col-0 (Table 1; see Supplemental Data Sets 3 to 5 online). A large number of transcripts associated with salicylic acid (SA)–dependent and SA-independent defense responses were also more highly expressed in the abi4, vtc1, and vtc2 mutants compared with Col-0 (see Supplemental Data Sets 3 to 5 online).
Table 1.
Transcripts Encoding Transcription Factors Commonly Expressed in vtc1, vtc2, and abi4 Relative to Col-0
| Expression Ratio Relative to Col-0b | ||||
| AGIa | vtc1 | vtc2 | abi4 | TAIR Annotationc |
| At4g23810 | +5.13 | +5.96 | +4.92 | ATWRKY53__WRKY53; DNA binding/protein binding/transcription activator/transcription factor |
| At2g17040 | +3.43 | +3.91 | +2.73 | anac036 (Arabidopsis NAC domain containing protein 36); transcription factor |
| At1g80840 | +3.29 | +2.81 | +3.04 | ATWRKY40__WRKY40; transcription factor |
| At1g27730 | +3.05 | +3.83 | +3.28 | ZAT10__STZ (salt tolerance zinc finger); nucleic acid binding/transcription factor/transcription repressor/zinc ion binding |
| At5g22380 | +3.00 | +2.84 | +3.19 | anac090 (Arabidopsis NAC domain containing protein 90); transcription factor |
| At4g01720 | +3.00 | +2.20 | +2.37 | AtWRKY47__WRKY47; transcription factor |
| At1g74930 | +2.71 | +3.01 | +2.78 | ORA47; DNA binding/transcription factor |
| At5g61600 | +2.20 | +2.15 | +2.23 | ERF104__ethylene-responsive element-binding family protein |
| At5g62165 | −2.81 | −2.86 | −2.69 | AGL42; transcription factor |
Arabidopsis Genome Initiative number.
+, Transcript abundance was enhanced compared to Col-0; −, transcript abundance was repressed compared to Col-0. All ratios are expressed on a linear scale.
Database annotation of the protein product.
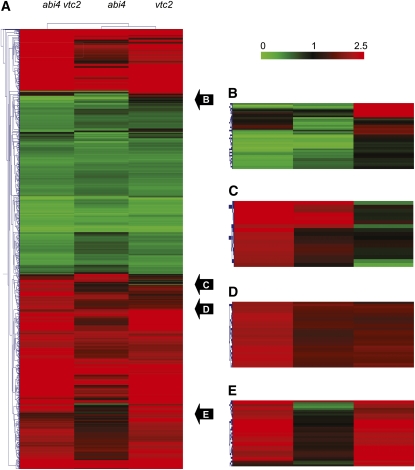
Comparisons of the leaf transcriptome profile patterns also revealed transcripts that were uniquely expressed in either vtc1, vtc2, or abi4 relative to Col-0 (Figure 7). The comparisons illustrated in Figure 7 demonstrate that many transcripts were expressed in a similar manner in vtc2 and abi4 vtc2 relative to Col-0. Of the transcripts that were differentially expressed in vtc2 relative to Col-0 (see Supplemental Data Set 5 online), a small number showed a reversal in expression in the abi4 vtc2 double mutant (Table 2). Many of these transcripts encode proteins involved in pathogen responses and defense (Table 2). Relatively few mRNAs in this group encode transcription factors or proteins that are involved in signaling. However, the AP2/EREBP transcription factor ORA47 (At1g74930; an AP2-type transcription factor of the DREB family), which was enhanced in the abi4 and vtc2 single mutants relative to Col-0 (Table 1), was strongly repressed in abi4 vtc2 double mutants (Table 2). Moreover, a gene encoding histone H3 (At3g27360) was strongly repressed in the abi4 vtc2 double mutant and yet enhanced in vtc2 relative to Col-0 (Table 2). Similarly, the gene encoding ABI5 binding protein 3 (AFP3; At3g29575) was strongly repressed in the abi4 vtc2 double mutant but enhanced in vtc2 relative to Col-0 (Table 2).
Figure 7.
Cluster Analysis Comparison of the abi4, vtc2, and abi4 vtc2 Transcriptomes Relative to Col-0.
(A) Whole transcript profile.
(B) Examples of transcripts uniquely expressed in vtc2 relative to Col-0.
(C) Examples of transcripts whose expression is reversed in vtc2 relative to abi4 vtc2.
(D) Examples of transcripts that are expressed in a similar manner in abi4 and vtc2 relative to Col-0.
(E) Examples of transcripts that are expressed in vtc2 and abi4 vtc2 in a similar manner relative to Col-0.
Table 2.
Transcripts with Reversed Expression in vtc2 abi4 Relative to vtc2
| Expression Ratio Relative to Col-0b | |||
| AGIa | vtc2 | abi4 vtc2 | TAIR Annotationc |
| At1g76960 | +4.23 | −20.26 | Unknown protein |
| At3g48650 | +3.55 | −1.57 | Pseudogene, At14a-related protein, similar to At14a |
| At1g74930 | +3.01 | −1.23 | ORA47, DREB subfamily A-5 of ERF, AP2 transcription factor family |
| At1g66970 | +1.75 | −2.19 | SHV3-like 2 (SVL2) |
| At3g26230 | +1.70 | −3.71 | Putative cytochrome P450 |
| At3g16420/ At3g16430d | +1.43 | −1.19 | Jacalin-related lectin 30 (JAL30), PYK10-binding protein 1 (PBP1)/Jacalin-related lectin 31 (JAL31) |
| At1g80960 | +1.36 | −2.64 | F-box protein-related |
| At3g43740 | +1.24 | −8.57 | Leucine-rich repeat family protein |
| At5g44580 | +1.24 | −2.95 | Unknown protein |
| At3g47250 | +1.23 | −19.02 | Unknown protein |
| At3g29575 | +1.94 | −1.61 | ABI5 binding protein 3 (AFP3) |
| At5g39030 | +1.18 | −3.16 | Protein kinase family protein |
| At5g23010 | +1.17 | −2.29 | 2-Isopropylmalate synthase 3 (IMS3), Methylthioalkylmalate synthase 1 (MAM1) |
| At1g08105/ | +1.16 | −2.10 | Hypothetical protein similar to putative transposase of transposable element Ac GB:CAA25635 (Zea mays) |
| At1g62766/ | |||
| At2g29120/ | |||
| At5g39060d | |||
| At1g28290 | +1.15 | −3.56 | AGP31, Arabinogalactan-protein 31 (AGP31) |
| At3g27360 | +1.12 | −3.21 | Histone H3 |
| At3g46980 | +1.10 | −2.48 | Phosphate transporter 4.3 |
| At3g46530 | +1.07 | −2.61 | Recognition of Peronospora parasitica 13 (RPP13) |
| At3g26290 | +1.04 | −4.09 | CYP71B26, Cytochrome P450, family 71, subfamily B, polypeptide 26 (CYP71B26), putative cytochrome P450 |
| At1g68050/ At5g53410d | +1.04 | −3.37 | Flavin binding kelch repeat F box 1 (FKF1)/unknown protein |
| At5g28500 | +1.03 | −2.90 | Unknown protein |
| At5g24240 | −1.03 | +4.54 | Phosphatidylinositol 3- and 4-kinase family protein, ubiquitin family protein |
| At3g52390 | −1.05 | +1.75 | TatD-related deoxyribonuclease family protein |
| At5g38260 | −1.08 | +2.05 | Ser/Thr protein kinase, putative |
| At3g49580 | −1.16 | +2.86 | Response to low sulfur 1 (LSU1) |
| At3g14990 | −1.29 | +1.18 | 4-Methyl-5(β-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative |
| At5g48850 | −2.10 | +2.02 | Homologous to the wheat sulfate deficiency-induced gene sdi1 |
| At5g65080 | −3.23 | +1.04 | AGL68, MADS affecting flowering 5 (MAF5) |
Arabidopsis Genome Initiative number.
+, Transcript abundance was enhanced compared to Col-0; −, transcript abundance was repressed compared to Col-0. All ratios are expressed on a linear scale.
Database annotation of the protein product.
Probe did not return a unique AGI number.
Glutathione and Glutathione-Associated Transcripts
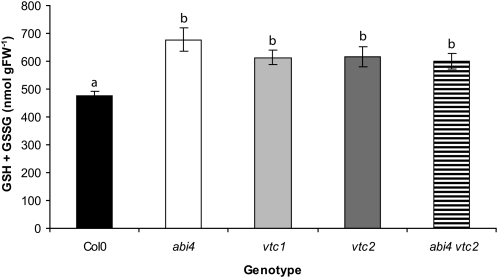
Like ascorbate, the tripeptide thiol antioxidant glutathione is a modulator of abiotic and biotic stress signaling pathways in plants (Noctor et al., 2011). The following experiments were therefore performed to determine whether low ascorbate affects glutathione accumulation and signaling pathways. The leaf glutathione pool was significantly increased in the vtc1, vtc2, abi4, and abi4 vtc2 mutants relative to Col-0 (Figure 8). Two transcripts encoding tau-type glutathione peroxidases (GSTtau4 and GSTtau10) and two encoding glutaredoxins (GRX10 and GRX480) were uniformly increased in the vtc1, vtc2, abi4, and abi4 vtc2 double mutants relative to Col-0 (Table 3). GSTtau8 transcripts were also increased only in abi4 but not in the vtc mutants or the abi4 vtc2 double mutants (Table 3). The expression of more genes encoding glutathione S-transferases (GSTs), glutathione peroxidases, and glutaredoxins was changed relative to Col-0 in the vtc2 mutant than in abi4 (Table 3). Moreover, the altered expression of these genes was retained in the abi4 vtc2 double mutants (Table 3).
Figure 8.
Glutathione Content of Col-0, abi4, vtc1, vtc2, and abi4 vtc2 Rosettes.
Plants were germinated and grown in controlled environments as described in the text. Whole rosettes were harvested and glutathione quantified as described in the text. Data are presented as mean ± se (n = 8), and values that were significantly different according to Fisher’s protected LSD test are indicated by different letters (P < 0.05). FW, fresh weight.
Table 3.
Differentially Expressed Transcripts in abi4, vtc1, vtc2, and abi4 vtc2 Mutants Encoding GSTs, GRXs, and Glutathione Peroxidases
| Expression Ratio Relative to Col-0b | |||||
| AGIa | abi4 | vtc1 | vtc2 | abi4 vtc2 | TAIR Annotationc |
| At2g02390 | nsd | ns | +2.0 | +2.2 | GSTzeta 1 |
| At2g29460 | +2.8 | +2.0 | +2.4 | +3.7 | GSTtau4 |
| At2g29450 | −2.2 | ns | −2.1 | −2.3 | GSTtau5 |
| At2g29440 | ns | ns | ns | −2.4 | GSTtau6 |
| At3g09270 | +3.8 | +2.8 | ns | ns | GSTtau8 |
| At1g74590 | +2.9 | +3.5 | +3.9 | +6.5 | GSTtau10 |
| At1g69930 | ns | ns | +2.0 | +2.3 | GSTtau11 |
| At1g10370 | −2.5 | ns | −5.1 | −4.9 | GSTtau17 |
| At4g02520 | +3.7 | +2.1 | +4.9 | +5.7 | GSTphi2 |
| At1g02930 | ns | +2.5 | +4.2 | +4.8 | GSTphi3 |
| At2g47730 | ns | ns | −2.0 | ns | GSTphi8 |
| At1g03850 | +2.3 | +2.4 | +5.4 | +9.8 | Glutaredoxin family protein |
| At5g11930 | ns | +2.1 | ns | ns | Glutaredoxin family protein |
| At1g28480 | +2.0 | +2.1 | +2.6 | +3.4 | GRX480 |
| At4g11600 | ns | ns | +2.0 | +2.4 | Glutathione peroxidase 6 |
| At4g31870 | ns | −2.6 | ns | −2.1 | Glutathione peroxidase 7 |
| At1g63460 | ns | ns | +2.1 | +2.0 | Glutathione peroxidase 8 |
Arabidopsis Genome Initiative number.
+, Transcript abundance was enhanced compared to Col-0; −, transcript abundance was repressed compared to Col-0. All ratios are expressed on a linear scale.
Database annotation of the protein product.
Gene expression not significantly different from Col-0.
Responsiveness to SA
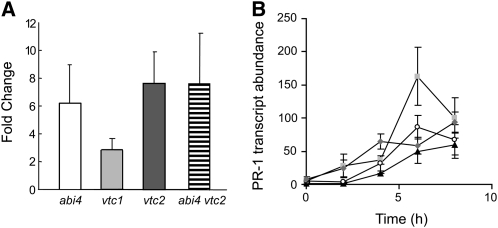
Glutathione and thioredoxins participate in the regulation of the SA-dependent NONEXPRESSOR OF PATHOGENESIS RELATED GENES1 (NPR1) pathway (as discussed by Noctor et al. (2011). To determine the extent to which SA-dependent signaling pathways are modified in the vtc1, vtc2, abi4, and the abi4 vtc2 double mutants, we examined the abundance of transcripts encoding PR1 protein, together with the induction of PR1 transcripts by SA. The leaves of vtc1, vtc2, abi4, and vtc2 abi4 genotypes had higher basal levels of PR1 mRNAs than Col-0 (Figure 9). The expression of PR1 was rapidly induced in all genotypes following SA treatment (Figure 9). Whereas the greatest level of stimulation was observed in Col-0, SA spraying resulted in the comparable absolute levels of PR1 transcripts in all lines at the end of the experiment (Figure 9).
Figure 9.
Expression of PR-1 Transcripts in Col-0, abi4, vtc1, vtc2, and abi4 vtc2.
All plants were grown under controlled environments for 6 weeks prior to analysis of gene expression.
(A) Basal level of PR-1 transcript abundance relative to Col-0.
(B) Impact of SA treatment on PR-1 transcript abundance relative to untreated Col-0 (triangles, Col-0; circles, abi4; squares, vtc2; diamonds, abi4vtc2). Relative transcript abundance is represented as mean values ± se (n = 4).
GA Status of the abi4, vtc1, vtc2, and abi4 vtc2 Mutants
Extensive literature evidence supports the view that ABA acts antagonistically to GA in the control of organ development, with, for example, ABA catabolism and GA biosynthesis required for processes such as seed germination (Liu et al., 2010). We therefore conducted a series of experiments to determine whether GA content was altered in the ascorbic acid–deficient mutants relative to Col-0. First, the vtc1 and Col-0 plants were sprayed every week after sowing either with water or with GA3 for 6 weeks. At 6 weeks, GA spraying increased the shoot biomass of both genotypes, such that they were visibly larger than the respective water-treated controls (Figure 10). To explore the interactions between ascorbate and GA synthesis in more detail, the abundance of the bioactive GAs, GA1 and GA4, and some of their precursors and inactivation products was compared in the vtc1, vtc2, abi4, and vtc2 abi4 mutants (Table 4). However, the abundance of these GA forms was similar in the rosettes of all genotypes (Table 4).
Figure 10.
Impact of GA3 Treatment on Growth Phenotype of Col-0 and vtc1.
Plants were germinated and placed under controlled environmental conditions as described in the text. Following transfer, rosettes were sprayed with either water or GA3 weekly for 6 weeks.
[See online article for color version of this figure.]
Table 4.
A Comparison of Rosette GA Concentrations (ng g Dry Weight−1)
| Genotype | GA1 | GA19 | GA44 | GA53 | GA4 | GA34 | GA9 | GA51 | GA24 |
| Col-0 | 0.3 (0.1) | 2.9 (0.0) | 0.7 (0.4) | 9.1 (0.5) | 2.5 (0.8) | 3.1 (0.7) | 1.2 (0.8)a | 1.5 (1.6) | 20.6 (6.0) |
| abi4 | 0.4 (0.1) | 2.6 (0.6) | 0.5 (0.3) | 9.4 (1.9) | 2.2 (1.8) | 2.9 (0.8) | 2.7 (0.4)a | 1.5 (1.1) | 24.3 (4.7) |
| vtc1 | 0.5 (0.0) | 2.3 (0.1) | 0.4 (0.1) | 5.3 (0.2) | 1.3 (1.2) | 3.6 (0.4) | 0.0b | 2.9 (0.2) | 21.5 (0.9) |
| vtc2 | 0.5 (0.2) | 2.1 (0.2) | 0.5 (0.2) | 5.4 (1.0) | 2.9 (1.9) | 2.4 (0.6) | 0.5 (0.1) | 2.1 (1.2) | 20.4 (2.7) |
| abi4 vtc2 | 0.4 (0.2) | 3.1 (0.5) | 1.0 (0.2) | 10.1 (0.6) | 4.4 (2.5) | 3.0 (0.7) | 1.3 (1.4) | 1.2 (1.1) | 24.0 (3.5) |
Values are means of three to five biological replicates (sd), except where indicated.
Mean of two biological replicates.
Not detected. GAs A8, A20, and A29 were also analyzed but were below the level of detection in all samples.
DISCUSSION
The vtc1 and vtc2 mutants, which are defective in ascorbate synthesis, have constitutively low levels of ascorbate compared with the wild-type plants (Veljovic-Jovanovic et al., 2001; Colville and Smirnoff, 2008). However, they have similar levels of oxidants, and under optimal growth conditions they do not show symptoms of oxidative stress (Veljovic-Jovanovic et al., 2001; Olmos et al., 2006). In an earlier study, we presented the hypothesis that ABA synthesis and signaling were important factors in the slow growth phenotype of the vtc mutants (Pastori et al., 2003). The in-depth analysis of the vtc1, vtc2, abi4, and abi4 vtc2 mutants reported here demonstrates unequivocally that the ascorbate-dependent regulation of plant growth requires the ABI4 transcription factor. In the absence of a functional ABI4 transcription factor, the ascorbate-dependent slow growth phenotype is not expressed. Thus, like other ABA signaling components, such as ABI1 and ABI2, which have long been known to function in stress signaling cascades involving ROS as second messengers (Allen et al., 1999), the data presented here also implicate ABI4 in redox signaling.
Interactions among ascorbate, ABA, and GA signaling pathways have been described in processes such as floral induction (Barth et al., 2006). Moreover, redox regulation of the balance between ABA and GA signaling pathways has been suggested to affect the dormancy and germination of Arabidopsis seeds (Liu et al., 2010). Seed germination rates were not adversely affected by the higher ABA contents of the vtc mutants compared with Col-0. In agreement with the recent observation that ABA did not affect GA biosynthesis (Ross et al., 2011), the amount and composition of GAs was similar in vtc, abi4, and abi4 vtc2 to that observed in Col-0. These results demonstrate that a 75% decrease in the leaf ascorbate pool (Figure 1) is not sufficient to adversely affect GA metabolism or alter the abundance of bioactive GAs (GA1 and GA4; Table 4).
The ABI4 transcription factor is required for ABA signaling during seed development and germination (Finkelstein et al., 1998). Moreover, the abi4 mutants are able to germinate in the presence of high levels of Glc that suppress germination in the wild-type plants (Arenas-Huertero et al., 2000). The data presented here demonstrate that the expression of the sugar-insensitive phenotype of the abi4 mutant is dependent on cellular redox homeostasis because it is repressed by low ascorbate. Thus, ascorbate plays a role in sugar signaling pathways. High sugar levels inhibit photosynthetic gene expression (Van Oosten et al., 1997) and limit the extent of ascorbate accumulation in leaves (Yabuta et al., 2007), as well as stimulating the expression of defense genes (Price et al., 2004; Sulmon et al., 2004; Thibaud et al., 2004; Loreti et al., 2005; Nishikawa et al., 2005; Couée et al., 2006). Whereas Suc regulates the expression of genes encoding enzymes of the ascorbate synthesis pathway, the negative effect of Suc on ascorbate accumulation is abolished in the abi4 mutant (Yabuta et al., 2007), as is the Suc-dependent repression of photosynthetic gene expression (Huijser et al., 2000; Oswald et al., 2001; Yabuta et al., 2007). The finding that the abi4 vtc2 mutant can no longer grow in the presence of high levels of Glc provides further evidence of a strong interaction between the redox and sugar signaling pathways in the regulation of plant growth, particularly through the ABI4 pathway.
The ABI4 transcription factor is also considered to be important in the transmission of signals from the chloroplasts to the nucleus (Koussevitzky et al., 2007). We therefore examined the possibility that low ascorbate might also participate in the multiple signal transduction pathways that transmit information between the chloroplast and nucleus. The data presented here show that the low ascorbate levels in the vtc1 and vtc2 mutants exert an influence over plastid-derived retrograde signaling pathways because LHCB1.1 expression was much less repressed in the vtc1 and vtc2 mutants than in Col-0 in the presence of lincomycin. While not as marked as in the gun1 mutants, the level of repression observed in the vtc1 and vtc2 mutants was more pronounced than in abi4. This finding that ascorbate affects plastid-derived retrograde signaling pathways supports the results of earlier studies, which showed that the expression of nuclear genes encoding photosynthetic proteins was modulated by cellular ascorbate levels (Kiddle et al., 2003). Differences in the results presented here regarding the effects of the abi4 mutation on gene expression from those reported previously (Koussevitzky et al., 2007) are probably related to the light levels used for plant growth. In this study, where plants were grown under more physiological light levels (100 μmol m−2 s−1) for Arabidopsis, which are ~10-fold higher than those used by Koussevitzky et al. (2007), the abi4 mutant did not display the marked gun phenotype with regard to LHCB1.1 expression (Figure 3).
The high level of similarity among the vtc1, vtc2, and abi4 mutant transcriptomes relative to Col-0 provides evidence of a significant overlap in the signal transduction pathways triggered by these mutations. The ethylene response factor ERF104, which is involved in the regulation of innate immunity and mitogen-activated protein kinase signaling cascades (Bethke et al., 2009), was more highly expressed in the vtc1, vtc2, and abi4 mutant leaves than in Col-0. ABA is known to elicit H2O2, which acts as a second messenger, in ABA and mitogen-activated protein kinase signaling cascades (Schroeder et al., 2001; Zhang et al., 2007). Transcripts encoding ZAT10 and several WRKY transcription factors were also expressed to higher levels in the vtc1, vtc2, and abi4 mutants relative to the wild type. These transcription factors are involved in the control of stress tolerance, particularly the expression of genes that enhance resistance to oxidative stress (Mittler et al., 2006). Enhanced expression of ZAT10 is also associated with the systemic acquired acclimation responses of leaves to high light levels that cause an inhibition of photosynthesis (Rossel et al., 2007). The high levels of WRKY53 transcripts in the vtc1, vtc2, and abi4 mutant leaves are consistent with alterations in plant development (Miao and Zentgraf, 2007) as is the enhanced expression of the NAC transcription factor ABSCISIC ACID-RESPONSIVE NAC036 (ANAC036) (Kato et al., 2010). Overexpression of ANAC036 resulted in a dwarf phenotype in Arabidopsis (Kato et al., 2010). The dwarf phenotype and accelerated leaf senescence are phenotypic characteristics of the vtc1 and vtc2 mutants, but these traits are not observed in abi4. The MADS box gene AGAMOUS-LIKE42 (AGL42), which is a marker for the Arabidopsis root quiescent center, was uniformly repressed in the vtc1, vtc2, and abi4 mutant leaves compared with Col-0.
A comparison of transcripts whose expression was reversed in the abi4 vtc2 double mutant compared with vtc2 (Table 2) provides new insights into the signaling pathways by which low ascorbate represses the growth of the vtc2 mutant. Of these, the ORA47 transcription factor, the ABI5 binding protein, AFP3, and the histone H3 are likely to be involved in the ascorbate-dependent regulation of growth in the vtc2 mutants. These transcripts were enhanced in vtc2 relative to Col-0, and they were strongly repressed in the abi4 vtc2 double mutant. The ability to execute an ABA-mediated growth arrest depends on ABI5. The stability of ABI5 is regulated by ABA through ubiquitin-related events (Lopez-Molina et al., 2003). It has recently been shown that a member of the ABI5 binding protein family called Novel Interactor of JAZ (NINJA) functions as an adaptor protein that interacts with the ZIM domain of most JA ZIM domain (JAZ) proteins that act as substrates of the E3 ubiquitin ligase SCF complex in the repression of JA response genes (Pauwels et al., 2008). Moreover, NINJA contains an ERF-associated amphiphilic repression (EAR) motif that recruits the corepressor TOPLESS, which interacts with an EAR motif on auxin/indole-3-acetic acid substrates of TRANSPORT INHIBITOR RESPONSE1 to repress auxin responses (Pauwels et al., 2010).
Like ORA47, ZAT10 is highly expressed in vtc2 and abi4 mutant leaves. ZAT10 and ORA47 are regulators of JA biosynthesis whose action depends on the F-box protein CORONATINE INSENSITIVE1 (COI1), which plays a central and conserved role in JA signaling pathways. ORA47 function is downstream of MYC2 (Pauwels and Goossens, 2008). Of the nine transcription factors that are expressed in a similar manner in the vtc and abi4 single mutants, ORA47 is the only one whose expression is reversed in the abi4 vtc2 leaves compared with vtc2 rosettes. It is possible that the overexpression of ORA47 in vtc2 confers a JA-sensitive phenotype and contributes to the inhibition of growth. This situation would also favor the induction of a subset of MYC2-regulated genes in the JA pathway (Pauwels and Goossens, 2008). The repression of ORA47 together with the ABI5 binding protein AFP3 in the abi4 vtc2 leaves is consistent with the restoration of the wild-type growth phenotype in abi4 vtc2. Transcripts encoding ATERF2, an ERF/AP2-type transcription factor of the ERF subfamily B, which is a positive regulator of JA-responsive genes, were higher in vtc2 than Col-0. The PROTODERMAL FACTOR1.2a (PDF1.2a) and PDF1.2b transcripts were also higher in the abi4 mutants than Col-0. By contrast, ARABINOGALACTAN PROTEIN31 transcripts whose expression is repressed by methyl jasmonate were increased in vtc2 but repressed in the abi4 vtc2 leaves. Similarly, JASMONIC ACID CARBOXYL METHYLTRANSFERASE transcripts were decreased in all mutant genotypes relative to Col-0. Taken together, these seemingly conflicting findings reveal a complex interplay between ABA, JA, and redox signaling pathways.
JA stimulates synthesis of the two major low molecular weight antioxidants of plant cells, ascorbate and glutathione, by enhancing the transcription of component enzymes (Xiang and Oliver, 1998; Wolucka et al., 2005). The leaves of vtc1, vtc2, and abi4 mutants have significantly more glutathione than those of the wild type. Similarly, many of the genes that are altered in expression in the mutant genotypes relative to Col-0 encode defense proteins that can be classified as related to glutathione-associated gene expression (Mhamdi et al., 2010). The significant overlap in gene expression patterns among abi4, vtc1, and vtc2 relative to Col-0 implies a strong link between low ascorbate signaling and signaling pathways that are triggered when ABI4 function is impaired. The observed changes in glutathione and in glutathione-associated gene expression patterns in the abi4, vtc1, vtc2, and abi4 vtc2 mutants might indicate something regarding the nature of this common signaling pathway. Accumulating evidence suggests that glutathione is a modulator of SA and JA signaling (for discussion, see Noctor et al., 2011). For example, a suite of JA genes are repressed in gr1 mutants that lack the cytosolic/peroxisomal form of glutathione reductase (GR), whereas gr1 cat2 double mutants that lack both GR and the major leaf form of catalase show H2O2-induced expression of these and other JA-associated genes (Mhamdi et al., 2010).
Among the many components that could link JA signaling and glutathione are GRXs and GSTs. In particular, transcripts encoding the SA-inducible protein GRX480, which represses upregulation of JA-induced genes (Ndamukong et al., 2007), were increased in expression in all the mutant genotypes relative to Col-0. Similarly, increased expression of GSTtau4, GSTtau10, and At1g03850, which is identical to the monothiol glutaredoxin, S13 (GrxS13), was observed in all mutant genotypes. The expression of GSTU4 and GrxS13 has been linked to expression of JA/COI1 signaling pathways (Tamaoki et al., 2008; Armengaud et al., 2010). Taken together, these data suggest that low ascorbate and defective ABI4 signaling drive gene expression through common glutathione/SA/JA-mediated signaling pathways to regulate defense gene expression. The higher levels of glutathione observed in all the mutant genotypes would serve to repress JA signaling through SA-dependent induction of NPR1 and GRX480, both of which interact with TGA transcription factors.
A large number of transcripts associated with SA-dependent defense responses were more highly expressed in all the mutant lines relative to Col-0. For example, all the mutant lines displayed higher basal levels of PR1 transcripts than did Col-0, suggesting that the SA signaling is primed in all the mutant genotypes. However, treatment with SA induced a comparable absolute level of PR1 transcript abundance in all lines. Moreover, the SA-dependent signaling pathway in the vtc2 mutants was not altered by the absence of ABI4, at least in terms of the induction of PR gene expression, showing that this SA-dependent defense response does not require this transcription factor.
Taken together, these results demonstrate that low ascorbate triggers ABA-, SA-, and JA-dependent signaling pathways in leaves that together regulate plant growth and defense responses. The absence of the slow growth phenotype in the abi4 vtc2 double mutant, together with the reversal of gene expression patterns relative to the vtc2 mutant, demonstrates that JA signaling contributes to growth regulation but that these pathways are dependent on ABI4. Further evidence in support of strong interactions between ascorbate and sugar signaling in the regulation of plant growth and development is provided by the demonstration of a requirement for ascorbate in the execution of the sugar insensitivity phenotype of the abi4 mutant. These results provide significant new insights into how cellular redox state and particularly the abundance of ascorbate regulate plant growth.
METHODS
In the following experiments, we used wild-type Arabidopsis thaliana accession Col-0, vtc1 (Conklin et al., 1999), vtc2 (Jander et al., 2002), abi4 (Laby et al., 2000), gun1-1 (Susek et al., 1993), and vtc2 abi4 double mutants that we produced as described below.
Growth on Soil
Plants were grown in controlled environment cabinets under a light intensity of 250 μmol m−2 s−1, with a 12-h photoperiod at a constant temperature of 20°C and a relative humidity of 70%.
Selection of abi4 vtc2 Double Mutants
F2 generation seeds from the abi4 × vtc2 crosses were surface sterilized with 15% (v/v) household bleach for 15 min, immersed in ethanol 70% (v/v) for 1 min, and then rinsed three times with sterile distilled water. Seeds were germinated on plates supplemented with 7 μM ABA as described below, and germinated seedlings were transferred to moist compost. The homozygous abi4 vtc2 double mutant plants were validated by sequencing. DNA was extracted from leaf discs according to Edwards et al. (1991), and two PCR reactions were performed using ReadyMix Taq PCR reaction mix (Sigma-Aldrich) to amplify the regions where mutations were localized. Primers used for amplification of the region containing the vtc2 mutation were 5′-CCTTTTGCTTGCAGTTCACA-3′ and 5′-TGAAGGCAAACACAGCAGTC-3′, while those used for amplification of the abi4 mutation were 5′-GATTCCACCACCGACTCATC-3′ and 5′-CTATACGGGTCACGTGCTCA-3′. PCR products were sequenced on Applied Biosystems 3730xl DNA analyzer (Genevision), and DNA sequences were analyzed using FinchTV software. The validated homozygous abi4 vtc2 plants were grown under the conditions described below and seeds collected from individual plants.
Glc Sensitivity Experiments
Col0, vtc1, vtc2, abi4, and abi4 vtc2 were germinated on 0.8% agar containing full-strength MS medium alone or MS medium supplemented with 6% Glc. The seedlings were grown for up to 10 d in controlled environment chambers under a light intensity of 30 μmol m−2 s−1, with a 12-h photoperiod.
Treatment of Arabidopsis Rosettes with GA
After germination, the rosettes of soil-grown vtc1 and Col-0 seedlings were exposed to either water or 100 μM GA3 every 7 d.
Photosynthesis
Photosynthetic gas exchange measurements and stomatal function were determined essentially as described by Soares et al. (2008).
Chloroplast-to-Nucleus Retrograde Signaling
Norflurazon and lincomycin treatments were conducted according to Koussevitzky et al. (2007). Seeds of Col-0, gun1, abi4, vtc1, and vtc2 were placed on media containing half-strength MS, 0.5 g/L MES, 2% Suc as a control, or with the addition of either 5 μM norflurazon or 500 μM lincomycin, both supplied by Sigma-Aldrich. Seeds were incubated at 4°C for 48 h in the dark and then transferred to a 16/8-h light/dark cycle at 100 μmol m−2 s−1 and 22°C. After 5 d, seedlings in the cotyledon state were harvested in the middle of the photoperiod, snap frozen in liquid nitrogen, and total RNA extracted using the RNeasy kit (Qiagen). cDNA synthesis and quantitative RT-PCR were performed as described by Pellny et al. (2008) using primers for LHCB1.1 (Ankele et al., 2007) and PP2D and SAND as reference genes (Czechowski et al., 2005). Data for three biological replicates were analyzed using the SDS 1.3 software (Applied Biosystems) with the untreated Col-0 as the control sample. Results for each reference gene were averaged.
Metabolite Quantification
Ascorbate was extracted and determined as described by Veljovic-Jovanovic et al. (2001). Glutathione was extracted and quantified according to the method of Queval and Noctor (2007).
GAs were analyzed in freeze-dried plant material as described by Griffiths et al. (2006). Ground tissue samples (500 mg) were extracted with 80% methanol-water (100 mL) spiked with 3H- (833 Bq) and 2H-GA (5 ng) internal standards. The methanol extracts were purified, fractionated by HPLC, and analyzed by gas chromatography–mass spectrometry (GC-MS) as described previously.
ABA was extracted from freeze-dried plant material (50 mg per sample) and determined using the radio-immunoassay described by Le Page-Degivry et al. (1997) using an ABA-specific monoclonal antibody (AFRC MAC252), which was prepared and characterized by Quarrie and Galfre (1985). Three biological replicates per genotype were analyzed by analysis of variance and significant differences between genotypes determined using Fisher’s protected LSD test.
Microarray Analysis
Between three and six biological replicates were used for each of the genotypes Col-0, abi4, vtc1, vtc2, and abi4 vtc2. Each replicate consisted of a whole rosette harvested at 6 weeks. Gene expression profiles were analyzed using commercial oligonucleotide microarrays (Genechip Arabidopsis ATH1 genome arrays; Affymetrix). Affymetrix standard protocols were followed throughout. The data were analyzed using GeneSpring GX 11.00, and P values were calculated by asymptotic unpaired t test and subjected to multiple testing correction (Benjamini Hochberg false discover rate). A cutoff with P value <0.05 and log2 expression ratio ±1 was adopted. The probe targets were as defined by Affymetrix (https://www.affymetrix.com/analysis/netaffx/index.affx), and these targets were used to guide annotation of the genes through the use of BLAST against the TAIR database (http://www.Arabidopsis.org/).
Real-Time PCR
Real-time quantitative PCR was performed as described previously (Pellny et al., 2009). Total RNA was extracted from samples using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s protocol. RNA reverse transcription and quantitative PCR was performed on an Eppendorf Realplex2 real-time PCR system by one-step RT-PCR using the Quantifast SYBR Green RT-PCR kit (Quiagen) following the manufacturer’s instructions. The expression of the genes of interest was normalized with two endogenous controls, PDF and SAND. Arabidopsis accessions and primer sequences used are as follows: PR1 (AT2G14610) sense 5′-TGCTCTTGTTCTTCCCTCGAA-3′ and antisense 5′-TGCCTGGTTGTGAACCCTTAG; SAND (AT2G28390; 5′-AACTCTATGCAGCATTTGATCCACT-3′; 5′-TGATTGCATATCTTTATCGCCATC-3′); PDF2 (AT1G13320; 5′-TAACGTGGCCAAAATGATGC-3′; 5′-GTTCTCCACAACCGCTTGGT-3′).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ABI4 (At2g40220), VTC1 (At2g39770), VTC2 (At4g26850), LHCB1.1 (At1g29920), ORA47 (At1g74930), AFP3 (At3g29575), PR1 (At2g14610), WRKY53 (At4g23810), NAC036 (At2g17040), WRKY40 (At1g80840), ZAT10 (At1g27730), NAC090 (At5g22380), WRKY47 (At4g01720), ERF104 (At5g61600), AGL42 (At5g62165), GSTZ1 (At2g02390), GSTU4 (At2g29460), GSTU5 (At2g29450), GSTU6 (At2g29440), GSTU8 (At3g09270), GSTU10 (At1g74590), GSTU11 (At1g69930), ERD9 (At1g10370), GSTF2 (At4g02520), ERD11 (At1g02930), GSTF8 (At2g47730), F21M11.22 (At1g03850), F14F18.100 (At5g11930), GRX480 (At1g28480), GPX6 (At4g11600), GPX7 (At4g31870), and GPX8 (At1g63460). All microarray data from this article are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE23331.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genetic and Phenotypic Characterization of Putative abi4 vtc1 Mutants.
Supplemental Figure 2. Representative Plates Illustrating Germination of Col-0, abi4, vtc1, vtc2, and abi4 vtc2 in the Presence of Glc.
Supplemental Data Set 1. Common Transcripts Differentially Expressed in abi4 and vtc1 Relative to Col-0.
Supplemental Data Set 2. Common Transcripts Differentially Expressed in abi4 and vtc2 Relative to Col-0.
Supplemental Data Set 3. Transcripts Differentially Expressed in abi4 Relative to Col-0.
Supplemental Data Set 4. Transcripts Differentially Expressed in vtc1 Relative to Col-0.
Supplemental Data Set 5. Transcripts Differentially Expressed in vtc2 Relative to Col-0.
Acknowledgments
P.I.K. thanks the James Hutton Institute and University of Leeds for funding via the James Hutton Institute joint PhD fellowship scheme. P.D.V. thanks the Fundación Séneca (Spain) for a postdoctoral research fellowship. Part of this work was funded by Biotechnology and Biological Sciences Research Council Grant BB/C51508X/1 and by EU Initial Training Project, PITN-GA-2008-215174: Chloroplast Signals. Rothamsted Research receives strategic funding from the UK Biotechnology and Biological Sciences Research Council. We thank Ron Mittler and Shai Koussevitzky for their help regarding setting up the assay conditions for the GUN retrograde signaling pathways and Tatjana Kleine and Dario Leister for providing seeds of the gun1 mutant. We also thank Agnès Reyss for help with the ABA analysis, Fan Gong for assistance with the GA analysis, and Susan Verrall for assistance in statistical analysis.
AUTHOR CONTRIBUTIONS
P.I.K. performed the experimental work, except where attributed to other authors. T.K.P. generated the abi4 vtc2 double mutants, assisted with the characterization of the mutants, and produced the data shown in Figures 1C and 3. P.D.V. undertook the isolation and molecular characterization of the abi4 vtc2 double mutants. G.K. assisted with the characterization of the mutants and generated the data shown in Figure 10 and Supplemental Figure 1 online, characterized the abi4 vtc1 crossed lines in search for double mutants, and undertook some of the transcriptome comparisons of the abi4 and vtc1 mutants relative to Col-0 shown in Table 3, Figure 6, and the Supplemental Data Sets online. P.H. generated the data shown in Table 4. S.D. generated the data shown in Figure 2B. H.V. generated the initial crosses for the abi4 vtc1 double mutants. P.V. performed all the initial bioinformatics analysis of the microarray data. R.D.H. jointly supervised the experimental work of P.I.K. with C.H.F. and produced all the final versions of the figures and the Supplemental Figures and Supplemental Data Sets online. C.H.F directed the work overall and cowrote the article with R.D.H.
References
- Allen G.J., Kuchitsu K., Chu S.P., Murata Y., Schroeder J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankele E., Kindgren P., Pesquet E., Strand A. (2007). In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F., Arroyo A., Zhou L., Sheen J., León P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Armengaud P., Breitling R., Amtmann A. (2010). Coronatine-insensitive 1 (COI1) mediates transcriptional responses of Arabidopsis thaliana to external potassium supply. Mol. Plant 3: 390–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni O., De Tullio M.C. (2000). The role of ascorbic acid in cell metabolism: Between gene-directed functions and unpredictable chemical reactions. J. Plant Physiol. 157: 481–488 [Google Scholar]
- Barth C., De Tullio M., Conklin P.L. (2006). The role of ascorbic acid in the control of flowering time and the onset of senescence. J. Exp. Bot. 57: 1657–1665 [DOI] [PubMed] [Google Scholar]
- Bethke G., Scheel D., Lee J. (2009). Sometimes new results raise new questions: The question marks between mitogen-activated protein kinase and ethylene signaling. Plant Signal. Behav. 4: 672–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F., Cordoba E., Dupré P., Mendoza M.S., Román C.S., León P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., Van Breusegem F., Saindrenan P., Noctor G. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinoy N.J. (1984). Ascorbic acid in plant growth and development. In The Role of Ascorbic Acid in Growth, Differentiation and Metabolism of Plants, Martinus Nijhoff W., (The Hague, The Netherlands: Junk Publishers; ), pp. 68–195 [Google Scholar]
- Colville L., Smirnoff N. (2008). Antioxidant status, peroxidase activity, and PR protein transcript levels in ascorbate-deficient Arabidopsis thaliana vtc mutants. J. Exp. Bot. 59: 3857–3868 [DOI] [PubMed] [Google Scholar]
- Conklin P.L., Barth C. (2004). Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 17: 959–970 [Google Scholar]
- Conklin P.L., Norris S.R., Wheeler G.L., Williams E.H., Smirnoff N., Last R.L. (1999). Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc. Natl. Acad. Sci. USA 96: 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couée I., Sulmon C., Gouesbet G., El Amrani A. (2006). Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57: 449–459 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Vivancos P., Dong Y.-P., Ziegler K., Markovic J., Pallardó F., Pellny T.K., Verrier P., Foyer C.H. (2010a). Recruitment of glutathione into the nucleus during cell proliferation adjusts whole cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 64: 825–838 [DOI] [PubMed] [Google Scholar]
- Diaz Vivancos P., Wolff T., Markovic J., Pallardó F.V., Foyer C.H. (2010b). A nuclear glutathione cycle within the cell cycle. Biochem. J. 431: 169–178 [DOI] [PubMed] [Google Scholar]
- Dowdle J., Ishikawa T., Gatzek S., Rolinski S., Smirnoff N. (2007). Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 52: 673–689 [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaka M. (1998). Gene expression and function of ascorbate oxidase in higher plants. Recent Res. Dev. Phytochem. 2: 315–326 [Google Scholar]
- Finkelstein R.R., Rock C.D. (2002). Abscisic acid biosynthesis and response. In The Arabidopsis Book 1: e0058 doi/10.1199/tab.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Wang M.L., Lynch T.J., Rao S., Goodman H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2005). Oxidant and antioxidant signaling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 29: 1056–1071 [Google Scholar]
- Foyer C.H., Noctor G. (2009). Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M., Hay S., Marshall K.R., Munro A.W., Scrutton N.S. (2007). DNA binding suppresses human AIF-M2 activity and provides a connection between redox chemistry, reactive oxygen species, and apoptosis. J. Biol. Chem. 282: 30331–30340 [DOI] [PubMed] [Google Scholar]
- Graebe J.E. (1987). Gibberellin biosyntheis and control. Annu. Rev. Plant Physiol. 38: 419–465 [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M. (2003). Spontaneous and thermoinduced photon emission: New methods to detect and quantify oxidative stress in plants. Trends Plant Sci. 8: 409–413 [DOI] [PubMed] [Google Scholar]
- Hedden P. (1992). 2-Oxoglutarate-dependent dioxygenases in plants: Mechanism and function. Biochem. Soc. Trans. 20: 373–377 [DOI] [PubMed] [Google Scholar]
- Hedden P., Kamiya Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Huijser C., Kortstee A., Pego J., Weisbeek P., Wisman E., Smeekens S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliff M., Staal J., Myrenås M., Dixelius C. (2007). ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant Microbe Interact. 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Kato N., Esaka M. (2000). Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid than that of wild-type protoplasts. Planta 210: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Kato H., Motomura T., Komeda Y., Saito T., Kato A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167: 571–577 [DOI] [PubMed] [Google Scholar]
- Kerk N.M., Feldman L.J. (1995). A biochemical model for the initiation and maintenance of the quiescent center – Implications for organization of root meristems. Development 121: 2825–2833 [Google Scholar]
- Kiddle G., Pastori G.M., Bernard S., Pignocchi C., Antoniw J., Verrier P.J., Foyer C.H. (2003). Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana. Antioxid. Redox Signal. 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockley T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D.G., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby R.J., Kincaid M.S., Kim D., Gibson S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lange T. (1994). Purification and partial amino-acid sequence of gibberellin 20-oxidase from Cucurbita maxima L. endosperm. Planta 195: 108–115 [DOI] [PubMed] [Google Scholar]
- Le Page-Degivry M.T., Garello G., Barthe P. (1997). Changes in abscisic acid biosynthesis and catabolism during dormancy breaking in Fagus sylvatica embryo. J. Plant Growth Regul. 16: 57–61 [Google Scholar]
- Linster C.L., Gomez T.A., Christensen K.C., Adler L.N., Young B.D., Brenner C., Clarke S.G. (2007). Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J. Biol. Chem. 282: 18879–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ye N., Liu R., Chen M., Zhang J. (2010). H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 61: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Poggi A., Novi G., Alpi A., Perata P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacin R., Britsch L. (1997). Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3beta-hydroxylase. Eur. J. Biochem. 249: 748–757 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Nickle T.C., Meinke D.W., Last R.L., Conklin P.L., Somerville C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., Saindrenan P., Gouia H., Issakidis-Bourguet E., Renou J.-P., Noctor G. (2010). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Müller-Moulé P., Golan T., Niyogi K.K. (2004). Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 134: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P., Havaux M., Niyogi K.K. (2003). Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol. 133: 748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Nishikawa F., Iwama T., Kato M., Hyodo H., Ikoma Y., Yano M. (2005). Effects of sugars on ethylene synthesis and responsiveness in harvested broccoli florets. Postharvest Biol. Technol. 36: 157–165 [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C.H. (2011). Glutathione. In The Arabidopsis Book 9: e0142, doi/10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Veljovic-Jovanovic S., Foyer C.H. (2000). Peroxide processing in photosynthesis: Antioxidant coupling and redox signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos E., Kiddle G., Pellny T.K., Kumar S., Foyer Ch. (2006). Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana. J. Exp. Bot. 57: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Oswald O., Martin T., Dominy P.J., Graham I.A. (2001). Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98: 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori G.M., Kiddle G., Antoniw J., Bernard S., Veljovic-Jovanovic S., Verrier P.J., Noctor G., Foyer C.H. (2003). Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V., Olmos E., Kiddle G., Mowla S., Kumar S., Antoniw J., Alvarez M.E., Foyer C.H. (2005). Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 139: 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2008). Fine-tuning of early events in the jasmonate response. Plant Signal. Behav. 3: 846–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellny T.K., Locato V., Vivancos P.D., Markovic J., De Gara L., Pallardó F.V., Foyer C.H. (2009). Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol. Plant 2: 442–456 [DOI] [PubMed] [Google Scholar]
- Pellny T.K., Van Aken O., Dutilleul C., Wolff T., Groten K., Bor M., De Paepe R., Reyss A., Van Breusegem F., Noctor G., Foyer C.H. (2008). Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris. Plant J. 54: 976–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C., Fletcher J.M., Barnes J., Foyer C.H. (2003). The function of ascorbate oxidase in tobacco. Plant Physiol. 132: 1631–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G., De Gara L., Asard H., Horemans N. (2002). Ascorbate and glutathione: Guardians of the cell cycle, partners in crime? Plant Physiol. Biochem. 40: 537–548 [Google Scholar]
- Potters G., Horemans N., Bellone S., Caubergs R.J., Trost P., Guisez Y., Asard H. (2004). Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiol. 134: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott A.G., John P. (1996). Dioxygenases: Molecular structure and role in plant metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 245–271 [DOI] [PubMed] [Google Scholar]
- Price J., Laxmi A., St Martin S.K., Jang J.-C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie S.A., Galfre G. (1985). Use of different hapten-protein conjugates immobilized on nitrocellulose to screen monoclonal antibodies to abscisic acid. Anal. Biochem. 151: 389–399 [DOI] [PubMed] [Google Scholar]
- Queval G., Noctor G. (2007). A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Ross J.J., Weston D.E., Davidson S.E., Reid J.B. (2011). Plant hormone interactions: How complex are they? Physiol. Plant. 141: 299–309 [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J.I., Allen G.J., Hugouvieux V., Kwak J.M., Waner D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Signora L., De Smet I., Foyer C.H., Zhang H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Soares A.S., Driscoll S.P., Olmos E., Harbinson J., Arrabaça M.C., Foyer C.H. (2008). Adaxial/abaxial specification in the regulation of photosynthesis and stomatal opening with respect to light orientation and growth with CO2 enrichment in the C4 species Paspalum dilatatum. New Phytol. 177: 186–198 [DOI] [PubMed] [Google Scholar]
- Smirnoff N. (2000). Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 3: 229–235 [PubMed] [Google Scholar]
- Sommer-Knudsen J., Bacic A., Clarke A.E. (1998). Hydroxyproline-rich plant glycoproteins. Phytochemistry 47: 483–497 [Google Scholar]
- Sulmon C., Gouesbet G., Couée I., El Amrani A. (2004). Sugar-induced tolerance to atrazine in Arabidopsis seedlings: Interesting effects of atrazine and soluble sugars on psbA mRNA and D1 protein levels. Plant Sci. 167: 913–923 [Google Scholar]
- Susek R.E., Ausubel F.M., Chory J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tamaoki M., Freeman J.L., Pilon-Smits E.A.H. (2008). Cooperative ethylene and jasmonic acid signaling regulates selenite resistance in Arabidopsis. Plant Physiol. 146: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud M.-C., Gineste S., Nussaume L., Robaglia C. (2004). Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signaling pathway. Plant Physiol. Biochem. 42: 81–88 [DOI] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). β-Amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth S.Z., Puthur J.T., Nagy V., Garab G. (2009). Experimental evidence for ascorbate-dependent electron transport in leaves with inactive oxygen-evolving complexes. Plant Physiol. 149: 1568–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosten J.J.M., Gerbaud A., Huijser C., Dijkwel P.P., Chua N.H., Smeekens S.C.M. (1997). An Arabidopsis mutant showing reduced feedback inhibition of photosynthesis. Plant J. 12: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S.D., Pignocchi C., Noctor G., Foyer C.H. (2001). Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 127: 426–435 [PMC free article] [PubMed] [Google Scholar]
- Wolucka B.A., Goossens A., Inzé D. (2005). Methyl jasmonate stimulates the de novo biosynthesis of vitamin C in plant cell suspensions. J. Exp. Bot. 56: 2527–2538 [DOI] [PubMed] [Google Scholar]
- Xiang C., Oliver D.J. (1998). Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10: 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y., Mieda T., Rapolu M., Nakamura A., Motoki T., Maruta T., Yoshimura K., Ishikawa T., Shigeoka S. (2007). Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. J. Exp. Bot. 58: 2661–2671 [DOI] [PubMed] [Google Scholar]
- Yuan J.S., Reed A., Chen F., Stewart C.N., Jr (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Ding H., Xu S., Hu X., Tan M. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175: 36–50 [DOI] [PubMed] [Google Scholar]