Abstract
Jasmonates are phytohormones that regulate many aspects of plant growth, development, and defense. Within the signaling cascades that are triggered by jasmonates, the JASMONATE-ZIM DOMAIN (JAZ) repressor proteins play a central role. The endogenous bioactive JA-Ile conjugate mediates the binding of JAZ proteins to the F-box protein CORONATINE INSENSITIVE1 (COI1), part of the Skp1/Cullin/F-box SCFCOI1 ubiquitin E3 ligase complex. Upon the subsequent destruction of the JAZ proteins by the 26S proteasome, multiple transcription factors are relieved from JAZ-mediated repression, allowing them to activate their respective downstream responses. However, many questions remain regarding the targets, specificity, function, and regulation of the different JAZ proteins. Here, we review recent studies on the model plant Arabidopsis thaliana that provided essential and novel insights. JAZ proteins have been demonstrated to interact with a broad array of transcription factors that each control specific downstream processes. Recruitment of the corepressor TOPLESS unveiled a mechanism for JAZ-mediated gene repression. Finally, the presence of JAZ proteins was also found to be regulated by alternative splicing and interactions with proteins from other hormonal signaling pathways. Overall, these contemporary findings underscore the value of protein–protein interaction studies to acquire fundamental insight into molecular signaling pathways.
INTRODUCTION
Since their discovery in the 1960s as secondary metabolites in the essential oils of jasmine (Jasminum sp) flowers, jasmonates (JAs) have gradually reached their current undisputable status as plant hormones that are essential for the proper functioning of multiple plant processes, encompassing development, growth, and defense. In the model plant Arabidopsis thaliana, JAs are directly involved in stamen and trichome development, vegetative growth, cell cycle regulation, senescence, and responses to various biotic and abiotic stresses, among others. JAs are ubiquitous in the plant kingdom, from angiosperms to gymnosperms, although there is some degree of species specificity with respect to downstream processes under the direct control of JAs. An example of this principle is the conserved capacity of JAs to elicit plant secondary metabolic pathways, leading to an arsenal of species-specific bioactive metabolites of a wide structural variety and different biochemical origins. Ultimately, through crosstalk with other hormonal pathways, such as of auxins, gibberellins (GAs), abscisic acid (ABA), ethylene (ET), and salicylic acid, JAs are (in)directly involved in the fine-tuning of a myriad of plant cellular processes. This multifaceted role of JAs has been covered by excellent reviews (Wasternack, 2007; Balbi and Devoto, 2008; Kazan and Manners, 2008; Bari and Jones, 2009; Browse, 2009a; Grant and Jones, 2009; Kuppusamy et al., 2009; Pauwels et al., 2009; Reinbothe et al., 2009; Hoffmann et al., 2011; Robert-Seilaniantz et al., 2011).
JA-deficient, insensitive, or hypersensitive mutants substantially contributed to the characterization of JA synthesis and signaling. One of the first and most important signaling mutants identified was coi1-1, insensitive to JAs and defective in the CORONATINE INSENSITIVE1 (COI1) protein, an F-box protein that is part of a Skp1/Cullin/F-box complex (SCFCOI1) (Feys et al., 1994; Xie et al., 1998). SCF complexes act as E3 ubiquitin ligases that target proteins for degradation by the 26S proteasome and in which the F-box protein component provides the specificity for recruitment of the target proteins. Subsequently, the identification of COI1 target proteins became one of the prime quests in the JA field. By means of extensive forward genetic screens for positive and negative effectors of JA signaling, reverse genetics screens with COI1 interactors, COI1-dependent genes or JA-regulated genes, numerous proteins involved in JA signaling have been identified. However, it took nearly a decade after the COI1 locus had been mapped before the family of the JASMONATE ZIM-DOMAIN (JAZ) proteins was discovered, independently and almost simultaneously by several research groups, as the true targets of SCFCOI1 (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Furthermore, the genetic, molecular, and biochemical data established that (1) the JAZ proteins were degraded by the SCFCOI1 complex in response to JA, (2) the JA-Ile conjugate was the active hormone and an essential determinant of the interaction between COI1 and its targets, the JAZ proteins, and (3) the JAZ proteins interact with the positive regulator of JA signaling, MYC2 (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Together, these results, complemented with those from follow-up studies, suggested a model for JA perception and downstream signaling, in which the JAZ proteins block MYC2 activity in the absence of bioactive JAs by recruiting the general corepressors TOPLESS (TPL) and TPL-Related proteins through an interaction with the adaptor protein Novel Interactor of JAZ (NINJA) (Pauwels et al., 2010). MYC2 can be released from this block by JA-Ile–induced and SCFCOI1-mediated degradation of the JAZ proteins. This model and the role of other proteins in JA perception and signaling has been discussed in many reviews (Balbi and Devoto, 2008; Chico et al., 2008; Katsir et al., 2008a; Kazan and Manners, 2008; Staswick, 2008; Browse, 2009a, 2009b; Chini et al., 2009a; Chung et al., 2009; Fonseca et al., 2009a; Memelink, 2009; Gfeller et al., 2010).
The partial JA-insensitive phenotype of the myc2 mutant implied that MYC2 was probably not the only transcription factor (TF) controlled by the JAZ proteins. Many intriguing questions remained: which other TFs (or other regulatory proteins) might bind the JAZ proteins, which mechanism would be behind the repressor function of the JAZ, what might be the possible specificity of the different JAZ proteins, and which role might other JA derivatives play? Since the discovery of the JAZ proteins, studies or screens dealing with protein–protein interactions involving the JAZ proteins have provided exciting new insights into the mechanism of JA signaling and its importance in various aspects of plant development, growth, and survival. In this review, we summarize the most recent findings in JA signaling from the perspective of the proteins and protein domains involved and the interactions between them and other (macro)molecules.
JA-ILE MEDIATES BINDING OF JAZ PROTEINS TO COI1
The COI1-JAZ Receptor Complex
When the COI1 locus (At2g39940) was identified and found to encode an F-box protein (Xie et al., 1998), it was predicted to be part of an SCF complex (SCFCOI1) acting as an E3 ubiquitin ligase. Subsequent protein interaction studies identified CULLIN1 (CUL1; At4g02570), RING-BOX1 (At5g20570), the ARABIDOPSIS SKP1 HOMOLOG1 (ASK1; At1g75950), ASK2 (At5g42190), and the COP9 signalosome as interactors and confirmed this hypothesis (Devoto et al., 2002; Xu et al., 2002; Feng et al., 2003). These results and the phenotypes of mutants in the loci of members of the SCFCOI1 complex (Feys et al., 1994; Xie et al., 1998; Xu et al., 2002; Feng et al., 2003; Ren et al., 2005) indicated that protein ubiquitination by SCFCOI1 was essential for the JA response. Unfortunately, these initial yeast two-hybrid (Y2H) screens for COI1-interacting proteins (Devoto et al., 2002; Xu et al., 2002), conducted in the absence of bioactive JAs, failed to identify the SCFCOI1 targets. Indeed, at their discovery, the JAZ proteins were found to interact with COI1 in a manner dependent on the presence of an Ile conjugate of JA, JA-Ile (Thines et al., 2007; Katsir et al., 2008b), later pinpointed as (+)-7-iso-jasmonoyl-l-Ile, which is now accepted as the endogenous bioactive JA (Fonseca et al., 2009b). Interestingly, the phytotoxin coronatine (COR) that is produced by Pseudomonas syringae is a structural mimic of JA-Ile and can substitute for JA-Ile action (Katsir et al., 2008b; Fonseca et al., 2009b).
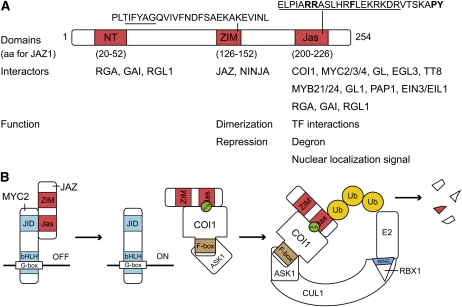
That JAZ proteins were the actual targets of the SCFCOI1 ubiquitin E3 ligase was validated by their rapid and COI1-dependent degradation upon JA perception (Chini et al., 2007; Thines et al., 2007). The C-terminal region containing the JA-associated (Jas) domain was necessary and sufficient for interaction with COI1 and binding of COR to the complex (Katsir et al., 2008b). The Jas domain, first described by Yan et al. (2007), is characterized by an S-L-X(2)-F-X(2)-K-R-X(2)-R core, delimited by a conserved N-terminal Pro and a C-terminal PY sequence (Figure 1A). Two extra N-terminal conserved basic residues (205R and 206R in JAZ1; Figure 1A) within the Jas domain were shown to be essential for COI1 interaction (Melotto et al., 2008). Direct interaction with COI1 has been shown for at least JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ6 (At1g72450), JAZ9 (At1g70700), and JAZ10 (At5g13220) (Table 1) (Thines et al., 2007; Melotto et al., 2008; Chini et al., 2009b; Chung and Howe, 2009; Yan et al., 2009; Sheard et al., 2010). The Jas domain is present in all JAZ proteins, but JAZ7 (At2g34600) and JAZ8 (At1g30135) lack the conserved basic amino acids at positions 205 and 206 in JAZ1 (Figure 1A) essential for COI1 interaction (Chung et al., 2010). To our knowledge, direct binding of JAZ7 and JAZ8 to COI1 remains to be investigated.
Figure 1.
JAZ Domain Structure and JA-Ile–Triggered Degradation by the Ubiquitin-Proteasome Pathway.
(A) Schematic representation of the JAZ1 protein and its conserved domains. Known protein interactors and functions are depicted. The TIFY motif for the ZIM domain and the JAZ degron for the Jas domain are underlined. Important amino acids mentioned in the text are in bold.
(B) In the absence of JA-Ile, JAZ binds MYC2 and represses gene expression. COI1-SKP may form a substrate adaptor that, with JA-Ile and InsP5 as a cofactor, binds the Jas domain of JAZ proteins. Upon formation of the SCFCOI1 E3 ligase complex, JAZ1 is presumably polyubiquitinated, which marks it for degradation by the 26S proteasome.
aa, amino acid; NT, N-terminal domain.
Table 1.
Protein–Protein Interactions with the JAZ Proteins
| The JAZ | JAZ1 | JAZ2 | JAZ3 | JAZ4 | JAZ5 | JAZ6 | JAZ7 | JAZ8 | JAZ9 | JAZ10g | JAZ11 | JAZ12 |
| F-box receptora | ||||||||||||
| COI1 (At2g39940) | X | X | X | X | X | X | ||||||
| bHLH TFsb | ||||||||||||
| MYC2 (At1g32640) | X | X | X | X | X | X | X | X | X | X | X | X |
| MYC3 (At4g17880) | X | X | X | X | X | X | X | X | X | X | X | |
| MYC4 (At5g46760) | X | X | X | X | X | X | X | X | X | X | X | X |
| GL3 (At5g41315) | X | X | X | X | X | X | ||||||
| EGL3 (At1g63650) | X | X | X | X | X | X | ||||||
| TT8 (At4g09820) | X | X | X | X | X | X | ||||||
| R2R3 MYB TFsc | ||||||||||||
| PAP1 (At1g56650) | X | X | X | |||||||||
| GL1 (At3g27920) | X | X | X | X | ||||||||
| MYB21 (At3g27810) | X | X | X | X | ||||||||
| MYB24 (At5g40350) | X | X | X | X | ||||||||
| TFs involved in other hormonal signaling pathwaysd | ||||||||||||
| EIN3 (At3g20770) | X | X | X | |||||||||
| EIL1 (At2g27050) | X | X | X | |||||||||
| GAI (At1g14920) | X | |||||||||||
| RGA (At2g01570) | X | X | X | |||||||||
| RGL1 (At1g66350) | X | |||||||||||
| Corepressor proteinse | ||||||||||||
| NINJA (At4g28910) | X | X | X | X | X | X | X | X | X | X | ||
| TOPLESS (At1g15750) | X | X | ||||||||||
| HDA6 (At5g63110) | X | X | X | |||||||||
| JAZ proteinsf | ||||||||||||
| JAZ1 (At1g19180) | X | X | X | X | X | X | X | X | X | X | X | |
| JAZ2 (At1g74950) | X | X | X | X | X | X | X | X | X | X | ||
| JAZ3 (At3g17860) | X | X | X | X | X | X | X | X | ||||
| JAZ4 (At1g48500) | X | X | X | X | X | X | X | |||||
| JAZ5 (At1g17380) | X | X | X | X | X | X | ||||||
| JAZ6 (At1g72450) | X | X | X | X | X | X | X | |||||
| JAZ7 (At2g34600) | ||||||||||||
| JAZ8 (At1g30135) | X | X | X | X | X | X | X | X | X | |||
| JAZ9 (At1g70700) | X | X | X | X | X | X | X | X | ||||
| JAZ10g (At5g13220) | X | X | X | X | X | X | X | X | X | X | ||
| JAZ11 (At3g43440) | X | X | X | X | X | X | ||||||
| JAZ12 (At5g20900) | X | X | X | X | X | X | ||||||
Crosses indicate direct protein–protein interactions with a particular JAZ protein demonstrated with Y2H.
Y2H data from Thines et al. (2007), Melotto et al. (2008), Chini et al. (2009b), Chung and Howe (2009), and Sheard et al. (2010).
Y2H data from Chini et al. (2009b), Cheng et al. (2011), Fernández-Calvo et al. (2011), Niu et al. (2011), and Qi et al. (2011).
Y2H data from Qi et al. (2011) and Song et al. (2011).
Y2H data from Hou et al. (2010) and Zhu et al. (2011).
Y2H data from Pauwels et al. (2010), Zhu et al. (2011), and Arabidopsis Interactome Mapping Consortium (2011).
Y2H data from Chini et al (2009b) and Chung and Howe (2009).
Only Y2H with the clone corresponding to the JAZ10.1 splicing variant is shown.
The minimal amino acid sequence sufficient for COR binding (also called the JAZ degron) was mapped to an N-terminal part of the Jas domain extended with two nonconserved amino acids (Figure 1A) (Sheard et al., 2010), creating a bipartite structure consisting of a loop and an amphiphatic α-helix. The loop, formed by the sequence ELPIARR in JAZ1 (Figure 1A), acts as a lid of the JA-Ile binding pocket and binds both JA-Ile and COI1. The α-helix of the JAZ degron binds COI1 near the JA-Ile binding site, and this interaction is necessary for coreceptor function (Sheard et al., 2010).
The hormone is probably perceived by a coreceptor complex consisting of a JAZ protein and COI1 (Figure 1B). Both the structural data and the observation that COR binding to COI1-JAZ was more than 50-fold higher than to COI1 alone support this hypothesis (Sheard et al., 2010). Nonetheless, it appears that COI1 alone can bind JA-Ile to some extent (Yan et al., 2009; Sheard et al., 2010), suggesting a model in which COI1 might initially bind to JA-Ile and subsequently recruit JAZ proteins, which would in turn significantly enhance the interaction (Yan et al., 2009).
F-box proteins may form substrate adaptors together with the Skp1 proteins that, only when bound to their target, are recruited by cullins to complete the SCF E3 ubiquitin ligase complex. It is assumed that this maintains an uncommitted pool of cullin-RING ligases in the cell (Hua and Vierstra, 2011). It is widely accepted that binding with SCFCOI1 is followed by JAZ polyubiquitination and degradation by the proteasome (Figure 1B). The dependence of JA-mediated JAZ degradation by the 26S proteasome has been confirmed (Chini et al., 2007; Thines et al., 2007). Nevertheless, information on JAZ (poly-)ubiquitination is lacking and only JAZ6 has been reported to be modified by ubiquitin (Saracco et al., 2009).
Inositol Pentakisphosphate Is a Cofactor of the JA Receptor
The similarity between the JA and auxin signaling pathways in regulation of JAZ and auxin/indole-3-acetic acid (Aux/IAA) protein stability, respectively, is striking. COI1 is a close homolog of the auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1; At3g62980), and the binding of the latter to the Aux/IAA proteins is mediated by the auxin IAA. Because COI1 and TIR1 share the core cullin-RING ligase subunits, such as CUL1, and the CUL1 regulation by rubylation, many mutants in these common components are compromised in both auxin and JA signaling (Hoffmann et al., 2011).
In the structure of the TIR1-Aux/IAA coreceptor, inositol hexakisphosphate (InsP6) was identified as a cofactor (Tan et al., 2007). Intriguingly, it is not InsP6, but presumably inositol pentakisphosphate (InsP5) that acts as a cofactor for the COI1-JAZ coreceptor (Sheard et al., 2010). This corresponds with the hypersensitivity to JA of the Arabidopsis ipk1-1 mutant (Mosblech et al., 2011), which is defective in the conversion of InsP5 to InsP6 by the INOSITOL POLYPHOSPHATE KINASE1 (IPK1) and accumulates InsP4 and InsP5 (Stevenson-Paulik et al., 2005). Accordingly, in Y2H assays, deletion of the yeast IPK1 gene enhanced the COR-mediated COI1 interaction with JAZ proteins. This interaction depended on the amino acid residues in COI1 that coordinate the inclusion of InsP5 near the JA-Ile binding pocket (Mosblech et al., 2011). Finally, InsP5 is essential for COR binding to the COI1-JAZ coreceptor in vitro (Sheard et al., 2010). Whether and how InsP5 levels are controlled by environmental stimuli, such as wounding, remains to be determined, but these results suggest that InsP5 has the potential to play a role as a signaling molecule in plants by fine-tuning of the COI1-JAZ coreceptor formation.
Alternative Splicing Leads to Dominant JAZ Repressors
The involvement of JAZ proteins in JA signaling has been proven by the expression of dominant JAZ variants resulting in JA-insensitive plants. These dominant variants all lacked (part of) the C-terminal Jas domain, either through a mutation in a splice acceptor site in JAZ3 (Chini et al., 2007), overexpression of a splice variant of JAZ10 (Yan et al., 2007), or overexpression of a truncated JAZ1 sequence lacking the Jas domain (Thines et al., 2007).
Several JAZ pre-mRNAs are subject to alternative splicing, which, at least for JAZ10, results in truncated JAZ proteins with a reduced COI1 binding potential, thereby causing dominant JA-insensitive phenotypes (Yan et al., 2007; Chung et al., 2010). The alternative splicing event provokes retention of an intron with the loss of the C-terminal X5PY (PY) motif in the Jas domain and the expression of ΔPY JAZ proteins as a consequence. Notably, the conserved PY sequence itself is not part of the JAZ degron (Sheard et al., 2010), and its replacement by two Ala residues does not affect COI1 binding (Chung et al., 2010). Stable ΔPY JAZ proteins might be produced to terminate the activated JA pathway (Chung et al., 2010). This hypothesis raises a new question as to which mechanism is responsible for the clearing of the ΔPY JAZ proteins that would be necessary to reset the JA pathway when required.
A possible role for the conserved PY sequence might reside in the subcellular localization of JAZ proteins. Whereas full-length JAZ1 is exclusively located in the nucleus, deletion of the Jas domain leads to the accumulation of the mutant JAZ1 proteins in the cytoplasm, at least in transformed tobacco (Nicotiana tabacum) suspension cells. A green fluorescent protein–tagged version of the Jas domain alone was localized specifically to the nucleus only when the conserved PY sequence was included (Grunewald et al., 2009). Hence, JAZ proteins might contain a type of nuclear localization signal (PY-NLS) that is recognized by karyopherin β in yeast and humans (Lee et al., 2006; Bai et al., 2011). Nevertheless, the splice variants JAZ10.3 and JAZ10.4 that lack the PY motif and the entire Jas domain, respectively, are still localized in the nucleus, arguing for an alternative mode of nuclear localization, at least for the JAZ10 proteins (Chung and Howe, 2009).
AN EVER-EXTENDING ARRAY OF JAZ-CONTROLLED TFs
JAZ Proteins Control the MYC-Type Basic Helix-Loop-Helix Activators
The first TF described to be regulated by the JAZ proteins was MYC2 (At1g32640) (Chini et al., 2007). The importance of this basic helix-loop-helix (bHLH) protein for JA signaling initially was discovered through a forward genetics approach (Berger et al., 1996; Lorenzo et al., 2004). MYC2 acts both as an activator and repressor of downstream JA responses. For example, it is a positive regulator of JA-mediated inhibition of primary root growth, anthocyanin biosynthesis, and oxidative stress tolerance but a negative regulator of JA-mediated resistance to necrotrophic fungi and biosynthesis of Trp and indole-glucosinolates (Lorenzo et al., 2004; Dombrecht et al., 2007). Nevertheless, MYC2 functions as a transcriptional activator when targeted to a heterologous promoter (Pauwels and Goossens, 2008).
After its discovery by forward genetics, JAZ3 was found to interact with MYC2, indicating that JAZ proteins might function as regulators of JA responses by binding and inhibiting the function of TFs, such as MYC2 (Figure 2A) (Chini et al., 2007). MYC2 binds most JAZ proteins, and, conversely, most JAZ proteins interact with its closest homologs, MYC3 (At4g17880) and MYC4 (At5g46760) (Table 1) (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). Although these three bHLH proteins have similar DNA binding preferences, they seem to regulate specific subsets of JA responses (Figure 2A). For example, in contrast with MYC2, MYC3 and MYC4 play only a weak role in regulating JA-mediated inhibition of primary root growth, whereas MYC3 and MYC4 are important for JA-mediated resistance to the herbivore Spodoptora littoralis. These differences between MYC2 and MYC3/MYC4 are postulated to correlate with their preferential production in root and shoot tissues, respectively (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011).
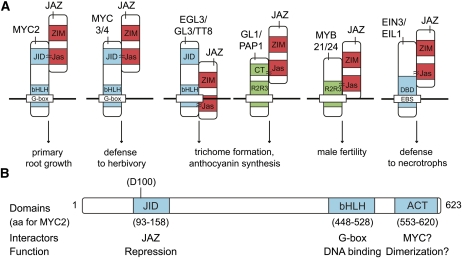
Figure 2.
Inhibition of JA Responses by Interaction of JAZ Proteins with a Wide Array of TFs.
(A) Model depicting different JAZ-TF combinations and the protein domains involved. The most important known JA responses controlled by the TFs are shown. Approximation symbols connect domains or approximate regions involved in physical interactions.
(B) Schematic representation of the MYC2 protein and its conserved interaction domains. Known protein interactors and functions are depicted. The location of the Asp-100 mutation in the JID domain is shown.
aa, amino acid; CT, C-terminal domain; DBD, DNA binding domain; EBS, EIN3 binding sites.
All three MYC proteins belong to group IIIe of the bHLH family (Heim et al., 2003), in which at least five different protein domains have been identified (Figure 2B). Besides the DNA binding bHLH domain, MYC2/MYC3/MYC4 and other bHLH proteins contain a conserved ACT-like domain at their C terminus that is necessary and essential for homo- and heterodimerization of the related bHLH proteins GLABRA3 (GL3; At5g41315) and ENHANCER OF GL3 (EGL3; At1g63650) (Zhang et al., 2003). Therefore, the ACT-like domain in MYC2/MYC3/MYC4 might have a similar dimerization function. At their N terminus, they contain three adjacent conserved domains of which the second domain is essential for interaction with JAZ proteins and designated the JAZ-Interacting Domain (JID) (Fernández-Calvo et al., 2011). A mutation in a conserved Asp (Asp94Asn) in the JID of MYC3 (Asp100 in MYC2; Figure 2B) resulted in a dominant phenotype (Smolen et al., 2002), which might correspond to a loss of interaction with and repression by JAZ proteins (Fernández-Calvo et al., 2011).
JAZ Regulation of WD40/bHLH/MYB Complexes Involved in Anthocyanin and Trichome Development
The JID domain is also found in several other bHLH proteins, suggesting that JAZ proteins might target more bHLH proteins than MYC2/MYC3/MYC4 (Fernández-Calvo et al., 2011). For instance, the JID domain is present in GL3, EGL3, and TRANSPARENT TESTA8 (TT8; At4g09820), all of which belong to group IIIf of the bHLH family (Heim et al., 2003) and interact with eight different JAZ proteins (Table 1) (Qi et al., 2011). Remarkably, however, this interaction has been observed only with the C-terminal part of TT8 and EGL3, which lacks the JID domain. Nonetheless, the Jas domain of JAZ1 and JAZ8 is as essential for interaction with these bHLH proteins as for the MYC2/MYC3/MYC4 proteins (Figure 2A) (Qi et al., 2011).
GL3, EGL3, and TT8 function in complexes in which they interconnect directly with the WD40 protein TRANSPARANT TESTA GLABRA1 (At5g24520), on the one hand, and different R2R3 MYB proteins, on the other hand. The GL3/EGL3/TT8 complexes regulate multiple processes, including development of trichomes and root hairs, biosynthesis of flavonoids (anthocyanins and proanthocyanidins), stomata patterning on hypocotyls, and seed coat mucilage production (Zhang et al., 2003; Zimmermann et al., 2004; Broun, 2005; Serna and Martin, 2006; Gonzalez et al., 2008; Ishida et al., 2008; Balkunde et al., 2010; Dubos et al., 2010). Anthocyanin biosynthesis and trichome initiation are both inducible by JAs (Feys et al., 1994; Traw and Bergelson, 2003; Maes et al., 2008). This induction requires both the JA receptor component COI1 and the GL3/EGL3/TT8-type bHLH proteins (Maes et al., 2008; Yoshida et al., 2009; Qi et al., 2011). Interestingly, the MYC2/MYC3/MYC4 complex is also involved in the JA-mediated induction of anthocyanin biosynthesis (Lorenzo et al., 2004; Dombrecht et al., 2007; Niu et al., 2011), but trichome induction seems to be independent of the MYC2 presence (Yoshida et al., 2009). How and whether MYC2/3/4 on the one hand and GL3/EGL3/TT8 on the other hand interact to regulate anthocyanin biosynthesis remains to be resolved.
Within the GL3/EGL3/TT8 complexes, the R2R3 MYB proteins provide the specificity for the downstream effects (Zhang et al., 2003; Broun, 2005; Serna and Martin, 2006; Gonzalez et al., 2008; Ishida et al., 2008; Balkunde et al., 2010; Dubos et al., 2010). Several JAZ proteins interact directly with the R2R3 MYB proteins PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1; At1g56650) and GL1 (At3g27920), which have specific roles in anthocyanin synthesis and trichome initiation, respectively (Table 1). These connections are mediated by the Jas domain of the JAZ proteins and the C-terminal domain of PAP1/GL1, respectively (Figure 2A). Moreover, overexpression of JAZ1 could disturb the bHLH–MYB interaction between PAP1-TT8 and GL1-GL3 (Qi et al., 2011). Correspondingly, in plants producing stabilized JAZ proteins, such as the coi1-2 knockout mutant plant, the levels of anthocyanin production and trichome initiation are lower than those of wild-type plants, either in the absence or presence of JA elicitation. Overexpression of PAP1, GL3, and EGL3 suppresses the inhibition of these processes by the JAZ proteins (Qi et al., 2011). To date, no R2R3 MYB proteins have been identified that interact with MYC2/MYC3/MYC4.
More bHLH-type interactors of JAZ proteins may exist; hence, additional cellular processes under yet unrevealed control of JA signaling might be discovered. For instance, JID occurs in bHLH013 (At1g01260), which was found to interact with JAZ1, JAZ8, and JAZ10 in systematic Y2H screens (Arabidopsis Interactome Mapping Consortium, 2011).
JAZ Regulation of Male Fertility via MYB21/MYB24
JA signaling is essential for male fertility in Arabidopsis. Mutants defective in JA biosynthesis or JA-insensitive mutants, such as coi1-1, are male sterile because of a combination of defective anther dehiscence, insufficient filament elongation, and severely reduced pollen viability (Browse, 2009b). Transcriptome analysis of JA-treated stamens identified two R2R3 MYB proteins, MYB21 (At3g27810) and MYB24 (At5g40350), as key regulators of the stamen maturation processes triggered by JA (Mandaokar et al., 2006). Overexpression of MYB21 in the coi1-1 or oxophytodienoate reductase3 background could partially restore male fertility (Cheng et al., 2009; Song et al., 2011), whereas the myb21-1 knockout mutant had strongly reduced fertility that could not be rescued by exogenous JA (Mandaokar et al., 2006).
A select set of JAZ proteins (i.e., JAZ1, JAZ8, and JAZ11 [At3g43440]) interact directly with MYB21 and MYB24 (Figure 2A, Table 1) (Song et al., 2011), revealing a model in which developmentally regulated JA biosynthesis triggers COI1-dependent JAZ degradation to control MYB21 and MYB24 levels and thereby stamen development. Like PAP1 and GL1, MYB21 and MYB24 bind the Jas domain of the JAZ proteins, but in contrast with PAP1 and GL1, the connection between JAZ proteins and MYB21/MYB24 is mediated through the N-terminal R2R3 domain (Song et al., 2011).
Ectopic expression of JAZ1ΔJas (Thines et al., 2007) and JAZ10.4 (Chung and Howe, 2009), both of which lack the full Jas domain, results in male sterility, whereas the JAZ3 splice acceptor mutant jai3-1, which expresses JAZ3 without the Jas domain, is still fertile (Chini et al., 2007). Overall, this suggests that production of truncated JAZ proteins under the control of their own promoter might help reveal the specific roles of the different JAZ proteins in plants. In a related approach, cell type–specific production of a nondegradable DELLA protein in the endodermis established that GA-mediated regulation of cell proliferation occurs in a subset of root cells rather than in all root cells (Ubeda-Tomás et al., 2009).
Repression of EIN3/EIL1 Mediates JA/ET Crosstalk
Crosstalk between ET and JA signaling occurs in many developmental and defense-related processes (Bari and Jones, 2009; Grant and Jones, 2009; Kuppusamy et al., 2009; Pauwels et al., 2009; Van der Ent et al., 2009; Robert-Seilaniantz et al., 2011). Part of this crosstalk apparently is mediated through the connection of JAZ proteins with ETHYLENE INSENSITIVE3 (EIN3; At3g20770) and EIN3-LIKE1 (EIL1; At2g27050), two TFs that are the central positive regulators of the ET response (Table 1) (Zhu et al., 2011). At least JAZ1, JAZ3, and JAZ9 can bind EIN3 and EIL1. The Jas domain–harboring C terminus of JAZ1 is necessary and sufficient for interaction with a fragment of EIN3 (comprising amino acids 200 to 500) that overlapped with its DNA binding domain (amino acids 59 to 359) (Figure 2A). Thereby, the JAZ proteins can repress the function of EIN3/EIL1, possibly by suppressing the DNA binding of EIN3 (Zhu et al., 2011). The emerging model, in which ET is needed for EIN3/EIL1 stabilization and JA for EIN3/EIL1 release from the JAZ protein repression, might provide a plausible explanation for the synergy in many ET/JA-regulated processes (Zhu et al., 2011).
EXPECTED AND UNEXPECTED FRIENDS
The DELLA Proteins Modulate JA Responses through JAZ Interaction
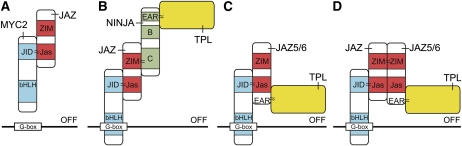
A mechanism similar to that of auxin and JA has also been revealed with respect to GA signaling. The GA-degradable DELLA proteins are well-known central repressors of GA signaling, and their target TFs from the bHLH family are recruited to regulate different GA-dependent developmental pathways (Davière et al., 2008; Schwechheimer and Willige, 2009). A remarkable finding has been that the DELLA proteins GIBBERELLIC ACID INSENSITIVE (GAI; At1g14920), REPRESSOR OF GA (RGA; At2g01570), and RGA-LIKE1 (RGL1; At1g66350) could also interact with JAZ proteins and compete with MYC2 for JAZ binding (Hou et al., 2010). Both the N-terminal and the Jas domains, but not the ZIM domain of JAZ1, are required for this interaction (Figure 1A). When DELLA levels decrease by GA treatment or increase by blocking of GA biosynthesis, binding of MYC2 to G-box–containing promoter fragments decreases or enhances, respectively. These findings provided a molecular mechanism for the observed crosstalk between the two hormone signaling pathways (Navarro et al., 2008) and led to the hypothesis that in the absence of GA, the DELLA proteins remove JAZ proteins from MYC2, allowing the TF to bind its targets and activate JA responses (Hou et al., 2010). Moreover, this model implies that JAZ inhibits MYC2 activity not only by recruiting corepressors (see below) but also by inhibiting the binding of MYC2 to the promoter DNA (Figure 3A).
Figure 3.
Hypothetical Models of Repression by JAZ Proteins.
(A) In the absence of JA-Ile, JAZ proteins bind and inhibit MYC2 DNA binding activity.
(B) to (D) In the absence of JA-Ile, the corepressor TPL is recruited to the MYC2 complex through EAR motif–containing proteins.
(B) The adaptor protein NINJA links TPL to the MYC2-bound JAZ proteins.
(C) and (D) Some JAZ proteins (such as JAZ5 and JAZ6) contain endogenous EAR motifs and might link TPL to MYC2 by direct binding (C) or through heterodimerization with other JAZ proteins (D).
Homo- and Heterodimerization of JAZ Proteins
Most of the 12 Arabidopsis JAZ proteins form homo- and heterodimers (Table 1) (Chini et al., 2009b; Chung and Howe, 2009). Although some JAZ proteins are more prone to dimerization than others, the exact combinations of JAZ pairs that can form in planta remain elusive, due to possible false negative results in the techniques employed, or, conversely, false positive interactions by forcing dimerization under artificial conditions. For example, no interaction of JAZ7 with other JAZ proteins has been observed, in contrast with its close homolog JAZ8. Dimerization is mediated by the conserved ZIM domain that possesses a conserved TIFY motif (Figure 1A) (Vanholme et al., 2007) in which point mutations abolish dimerization (Chung and Howe, 2009). Interestingly, by removal of the Jas domain, the range of JAZ proteins able to heterodimerize with the truncated protein is higher than that of the full-length JAZ proteins (Chini et al., 2009b; Chung and Howe, 2009).
Currently, the in vivo function of JAZ dimerization remains unclear. Extensive dimerization by JAZ, MYC-type bHLH proteins, and other components of the JAZ complexes might account for the occurrence of JAZ proteins in very high molecular weight complexes in plant cells (Figure 3) (Geerinck et al., 2010). Also, JAZ dimerization can account for JA insensitivity resulting from the production of JAZΔJas proteins. As the Jas domain is essential for both interaction with COI1 and MYC2, nondegradable JAZΔJas proteins might still be able to bind MYC2 through heterodimerization with full-length JAZ proteins, thereby dominantly repressing MYC2 function (Chung et al., 2009). However, Jas-independent binding of some JAZ proteins, such as JAZ1 and JAZ10, to MYC2 has been observed as well (Zhang, 2008; Chung and Howe, 2009), providing another model not requiring dimerization (Memelink, 2009).
EAR Motifs Recruit the Corepressor TPL to the Complex
The protein NINJA (At4g28910) binds directly to most JAZ proteins (Table 1) through their ZIM domain, which is also responsible for JAZ homo- and heterodimerization (Figure 1A). NINJA overexpression and knockdown lines have an attenuated JA response, corresponding to a role for NINJA as a negative regulator of JA signaling (Pauwels et al., 2010). The ABI5 BINDING PROTEINS (AFPs) that are involved in the control of ABA responses are homologs of NINJA. However, none of the AFPs interacts with JAZ proteins; conversely, NINJA does not interact with ABA INSENSITIVE5 (At2g36270). Both NINJA and AFP proteins contain an ETHYLENE RESPONSIVE FACTOR–associated amphiphilic repression (EAR) motif in their N-terminal domain by which they interact with the corepressor TPL (At1g15750) and TPL-related proteins (Pauwels et al., 2010). NINJA has been proposed as an adaptor protein linking the TF-bound JAZ proteins to TPL complexes (Figure 3B) (Pauwels et al., 2010).
Four out of the 12 JAZ proteins (i.e., JAZ5 [At1g17380], JAZ6, JAZ7, and JAZ8) contain EAR motifs themselves (Kagale et al., 2010). Correspondingly, direct interaction of JAZ5 and JAZ8 with TPL has been confirmed in a plant interactome project (Arabidopsis Interactome Mapping Consortium, 2011), but no direct interactions of other JAZ proteins with TPL have been described to date. Interestingly, JAZ7 and JAZ8 are the only JAZ proteins for which no indication of a NINJA interaction has been found with Y2H (Pauwels et al., 2010). Although this is based solely on Y2H data, which need confirmation by alternative methods, it seems that the TPL corepressor proteins can be recruited to the JAZ-regulated TFs by multiple means (Figure 3). In addition to NINJA, EAR motif–containing JAZ proteins might also link TPL to MYC2, for instance by direct binding (Figure 3C) or through heterodimerization with other JAZ proteins (Figure 3D). This interactor redundancy might account for the subtle JA-related phenotypes that can be observed in NINJA knockdown lines (Pauwels et al., 2010).
The Non-JAZ TIFY Proteins
Besides the 12 JAZ proteins, the Arabidopsis genome harbors six other proteins that contain a ZIM domain. Collectively, these 18 proteins are known as the TIFY proteins (Vanholme et al., 2007; Bai et al., 2011). The six additional TIFY proteins are ZIM (At4g24470), ZIM-LIKE1 (ZML1; At3g21175), and ZML2 (At1g51600); the PEAPOD (PPD) proteins PPD1 (At4g14713) and PPD2 (At4g14720); and TIFY8 (At4g32570). The first three are classified as group I TIFY proteins due to their divergent ZIM domain. All three are capable of homo- and heterodimerization (Chung and Howe, 2009), but heterodimerization of group-I TIFY proteins with JAZ proteins or interaction with NINJA has not been reported. They are also characterized by the presence of a CO, CO-like, and TOC1 (CCT) protein–protein interaction domain and a C2C2-GATA zinc-finger domain (Vanholme et al., 2007). The N-terminal part of the CCT domain has similarity to the Jas domain but has no known function in the ZIM proteins (Chung et al., 2009). The GATA domain is a DNA binding domain, suggesting that group I TIFY proteins might act as genuine TFs. Accordingly, and in contrast with JAZ1 (Pauwels et al., 2010), the ZIM protein functions as a transcriptional activator when targeted to a heterologous promoter (Shikata et al., 2003). Arabidopsis plants that overexpress ZIM show elongated petioles and hypocotyls (Shikata et al., 2004), but no JA-related phenotypes have been studied in plants with altered levels of group I TIFY proteins.
The PPD proteins PPD1 and PPD2 have a domain architecture resembling that of JAZ proteins more closely than the group I TIFY proteins. Besides the ZIM domain and a modified Jas domain, they also contain a PPD domain at their N terminus (Bai et al., 2011). The PPD proteins can bind NINJA (Pauwels et al., 2010), but based on the primary amino acid sequence, it is difficult to predict whether the modified Jas domain of PPD proteins allows interaction with COI1 or the TFs listed above. The PPD Jas-like domain contains several amino acid residues important for COI1 binding but has a Tyr instead of a conserved Phe in the α-helix, which has been shown to be essential for coreceptor formation (Sheard et al., 2010). PPD2 is used as a host factor by begomoviruses and is able to bind directly to DNA elements in the viral coat protein promoter (Lacatus and Sunter, 2009). Hence, it is tempting to speculate that the PPD proteins are true TFs and that the specific PPD domain is involved in the observed protein–DNA interaction. PPD proteins have been shown to additively repress the proliferation of dispersed meristematic cells (DMCs) in leaves (White, 2006), but this process has not been related to JAs. In DMCs, which give rise to the stomatal lineage and pavement cells and are usually scattered over the developing leaf, the cell cycle remains active after the cells from the primordium are arrested. Loss of PPD function results in an increased number of DMCs and dome-shaped leaves (White, 2006).
TIFY8 is an outlier in the TIFY family because it has only the ZIM domain in common with the other TIFY proteins. Nevertheless, this domain is functional because TIFY8 interacts at least with NINJA (Pauwels et al., 2010). To date, nothing is known about the function of TIFY8, but interactome data suggest that it is capable of interacting with a large number of TFs (Arabidopsis Interactome Mapping Consortium, 2011).
CONCLUSIONS AND FUTURE PROSPECTS
JAZ Function
The identification of NINJA as an adaptor for the recruitment of TPL proteins to complexes with JAZ proteins has given novel insight into JAZ function and has revealed how multiple signaling pathways can use similar mechanisms and the same proteins to repress gene expression (Pauwels et al., 2010). The exact mechanism by which TPL restrains gene expression remains unclear. Some data point to a link with the histone deacetylase HDA19 (At4g38130; Long et al., 2006; Zhu et al., 2010), and a direct interaction has been reported between TPL and Jumonji8, a histone demethylase (Macrae and Long, 2011). Furthermore, a direct link between JAZ1 and HDA6 (At5g63110) has recently been established, although the JAZ domain necessary for this interaction has not been mapped precisely (Zhu et al., 2011). Accordingly, it has been postulated that JAZ proteins might use different repression mechanisms for different TF sets, an idea that was further supported by the observation that the histone deacetylase inhibitor trichostatin A affects EIN3-dependent but not MYC2-dependent gene expression (Zhu et al., 2011).
A possible complementary action of JAZ repression of their target TFs is prevention of DNA binding. DELLA proteins, which have a JAZ-like function in GA signaling, reduce binding of their bHLH partners, PIF3 (At1g09530) and PIF4 (At2g43010), to their target promoters (Davière et al., 2008). Accordingly, at least for EIN3, in vivo association with a target promoter is enhanced after JA treatment, presumably due to JAZ degradation (Zhu et al., 2011).
JAZ Targets
Since their discovery, an astonishing number of TFs and other proteins have been identified as binding targets of the JAZ proteins, establishing them as important interfaces, not only for JA signaling but also for other (hormonal) signal transduction pathways. Similarly, a single JAZ protein can possess multiple distinct protein binding domains (i.e., at least three in JAZ1; Figure 1), each responsible for the recruitment of an ample set of interactors.
Most of the protein–protein interactions discussed in this review have been detected with the Y2H system and sometimes complemented with coimmunoprecipitation, pull-down, or bimolecular fluorescence complementation assays. Y2H is a convenient approach to map the domains, motifs, and amino acid residues within the JAZ proteins that are necessary and sufficient to bind a particular TF or other protein. Certainly, more JAZ interactors will be identified in the future through Y2H, but other screening techniques, such as the purification of whole protein complexes from Arabidopsis cell extracts, combined with mass spectrometry analysis, will add value for the identification of bona fide protein interactions in planta (Pauwels et al., 2010). Technical advances, such as increases in the sensitivity of mass spectrometry, permit downscaling of the required sample material and, in the future, the use of whole-plant organs, instead of plant cell cultures. Hence, in contrast with Y2H, such techniques will allow characterizing organ- or cell type–specific JAZ complexes and studying alterations in JAZ complex formation upon internal or external cues. Chemical genetics approaches to identify small compounds that disrupt specific molecular interactions may offer additional, unprecedented approaches to the specific dissection of hormonal signaling pathways. Such compounds may represent alternatives to chemicals that are currently used to study signaling pathways but often have pleiotropic effects, such as MG132 and staurosporine for inhibition of proteasome and kinase activity, respectively.
JAZ Regulation
The determination of the crystal structure of the JAZ degron in complex with COI1 (Sheard et al., 2010) has provided invaluable information to dissect the JA signal transduction cascade. So far, phosphorylation has not been implicated in JAZ–COI1 interaction (Katsir et al., 2008b; Sheard et al., 2010). Similarly, binding of the Aux/IAA proteins to the TIR1 F-box protein does not seem to require Aux/IAA phosphorylation (Dharmasiri et al., 2003; Tan et al., 2007). Nonetheless, a tobacco homolog of JAZ3, NbPPS3, has been identified as a kinase target (Katou et al., 2005) and, at least for JAZ12 (At5g20900), phosphopeptides have been detected (PhosPhAt 3.0) (Durek et al., 2010), suggesting that JAZ function is regulated by phosphorylation. No physical interactions between JAZ and any of the phosphatases or kinases involved in JA signaling (Gfeller et al., 2010) have been reported; hence, the responsible mediators of JAZ (de)phosphorylation still need to be identified.
Surprisingly, little is known about (poly)ubiquitination in plants. Although Aux/IAA proteins have been known for many years to be regulated by SCFTIR1, their polyubiquitination has been verified experimentally only recently (dos Santos Maraschin et al., 2009). As more and more targets of E3 ubiquitin ligases are being identified, techniques to detect and study poly- and deubiquitination of plant proteins should be optimized to characterize these highly regulated processes. Indeed, as a large number of JAZ interactors has been established, the next step, and undoubtedly one of the most challenging ones, will be to determine how phosphorylation, ubiquitination, and possibly other types of posttranslational modifications affect JAZ function and the complexes in which they are involved.
JAZ Role in Planta
The JAZ protein–protein interaction data thus far support the idea that most of the 12 JAZ proteins are functionally redundant for the processes now understood to be controlled by this protein family. This is consistent with the observation that T-DNA insertion lines corresponding to single knockouts in the JAZ1, JAZ2, JAZ5, JAZ7, and JAZ9 loci failed to produce any obvious JA-related phenotype (Thines et al., 2007; Demianski et al., 2011). Likewise, identification and characterization of insertion mutants in 12 of the 29 Aux/IAA family members revealed little on their function. The single Aux/IAA mutants showed no visible developmental defects compared with the wild type and even double or triple mutants exhibited wild-type phenotypes (Overvoorde et al., 2005). Information obtained from gene expression studies may prove useful to revisit phenotypic analysis of jaz mutants and eventually establish subtle JAZ-specific phenotypes. For instance, of all JAZ genes, only JAZ1 expression is early responsive to auxin, and the JAZ1 promoter was found active during lateral root initiation. Phenotypic analysis of transgenic lines with reduced JAZ1 expression showed that root growth was more inhibited by JAs, and the number of lateral roots increased in the presence of either JAs or auxins (Grunewald et al., 2009). Knockout and knockdown lines of JAZ10 also exhibited increased sensitivity of JA on root growth (Yan et al., 2007; Demianski et al., 2011), besides being more susceptible to P. syringae DC3000 infection (Demianski et al., 2011).
Possibly due to the posttranslational regulation of JAZ proteins, transgenic Arabidopsis lines in which JAZ proteins are overproduced exhibit no recognizable phenotypes (Thines et al., 2007). Only when stabilized JAZ proteins are produced, JA-insensitive phenotypes are observed (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Expressing such mutant JAZ proteins under the control of their own promoter may turn out to be a powerful method to uncover JAZ-specific functions and reveal why Arabidopsis has evolved to harbor 12 JAZ proteins.
Acknowledgments
We thank Jelle Van Leene for helpful discussions and Martine De Cock for help in preparing the manuscript. L.P. is a postdoctoral fellow of the Research Foundation-Flanders.
References
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Meng Y., Huang D., Qi Y., Chen M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 [DOI] [PubMed] [Google Scholar]
- Balbi V., Devoto A. (2008). Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Balkunde R., Pesch M., Hülskamp M. (2010). Trichome patterning in Arabidopsis thaliana: From genetic to molecular models. Plant Development, Current Topics in Developmental Biology, Vol. 91, Timmermans M.C.P., (Amsterdam: Elsevier; ), pp. 299–321 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J.D.G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Berger S., Bell E., Mullet J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111: 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009a). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009b). The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chico J.M., Chini A., Fonseca S., Solano R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Chini A., Boter M., Solano R. (2009a). Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 276: 4682–4692 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009b). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Niu Y., Browse J., Howe G.A. (2009). Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., de Lucas M., Prat S. (2008). Transcriptional factor interaction: A central step in DELLA function. Curr. Opin. Genet. Dev. 18: 295–303 [DOI] [PubMed] [Google Scholar]
- Demianski A.J., Chung K.M., Kunkel B.N. (June 15, 2011). Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. http://dx.doi.org/10.1111/j.1364-3703.2011.00727.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Jones A.M., Estelle M. (2003). Auxin action in a cell-free system. Curr. Biol. 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X. (2010). PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38 (Database issue): D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Ma L., Wang X., Xie D., Dinesh-Kumar S.P., Wei N., Deng X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009a). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Geerinck J., Pauwels L., De Jaeger G., Goossens A. (2010). Dissection of the one-MegaDalton JAZ1 protein complex. Plant Signal. Behav. 5: 1039–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A., Liechti R., Farmer E.E. (2010). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3: cm4. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Grant M.R., Jones J.D.G. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inzé D., Gheysen G., Goossens A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Hentrich M., Pollmann S. (2011). Auxin-oxylipin crosstalk: Relationship of antagonists. J. Integr. Plant Biol. 53: 429–445 [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou S., Yoshioka H., Kawakita K., Rowland O., Jones J.D.G., Mori H., Doke N. (2005). Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol. 139: 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J.K., Howe G.A. (2008a). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K.T., Walcher C.L., Nemhauser J.L. (2009). Cross-regulatory mechanisms in hormone signaling. Plant Mol. Biol. 69: 375–381 [DOI] [PubMed] [Google Scholar]
- Lacatus G., Sunter G. (2009). The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 392: 196–202 [DOI] [PubMed] [Google Scholar]
- Lee B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., Chook Y.M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae R., Long J. (2011). Interactions between TOPLESS and histone-modifying enzymes. 22nd International Conference on Arabidopsis Research. (Madison, WI: The North American Arabidopsis Steering Committee; ). [Google Scholar]
- Maes L., Inzé D., Goossens A. (2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148: 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Maraschin Fdos, S., Memelink J., Offringa R. (2009). Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J. (2009). Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Mosblech A., Thurow C., Gatz C., Feussner I., Heilmann I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65: 949–957 [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D.G. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P.J., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2008). Fine-tuning of early events in the jasmonate response. Plant Signal. Behav. 3: 846–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Inzé D., Goossens A. (2009). Jasmonate-inducible gene: What does it mean? Trends Plant Sci. 14: 87–91 [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C., Springer A., Samol I., Reinbothe S. (2009). Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276: 4666–4681 [DOI] [PubMed] [Google Scholar]
- Ren C., Pan J., Peng W., Genschik P., Hobbie L., Hellmann H., Estelle M., Gao B., Peng J., Sun C., Xie D. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42: 514–524 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Willige B.C. (2009). Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11: 274–280 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M., Matsuda Y., Ando K., Nishii A., Takemura M., Yokota A., Kohchi T. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55: 631–639 [DOI] [PubMed] [Google Scholar]
- Shikata M., Takemura M., Yokota A., Kohchi T. (2003). Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci. Biotechnol. Biochem. 67: 2495–2497 [DOI] [PubMed] [Google Scholar]
- Smolen G.A., Pawlowski L., Wilensky S.E., Bender J. (2002). Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13: 66–71 [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R.J., Chiou S.-T., Frye R.A., York J.D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 102: 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I.A., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Traw M.B., Bergelson J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T.S., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Van der Ent S., Van Wees S.C.M., Pieterse C.M.J. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Vanholme B., Grunewald W., Bateman A., Kohchi T., Gheysen G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.W.R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang H. (2008). Jasmonate-Responsive Transcriptional Regulation in Catharanthus roseus. PhD dissertation (Leiden, The Netherlands: University of Leiden; ). [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu F., Zhang Y., Cheng Y.T., Wiermer M., Li X., Zhang Y. (2010). Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34 [DOI] [PubMed] [Google Scholar]