Plant genomes contain a large number of short interspersed nuclear elements, which so far have been detected only randomly. A software tool was created integrating weakly conserved structural motifs for the analysis of genome sequences. Thousands of sequences from many plant taxa were identified, giving insights into their variability, localization along chromosomes, and evolution.
Abstract
Short interspersed nuclear elements (SINEs) are non-long terminal repeat retrotransposons that are highly abundant, heterogeneous, and mostly not annotated in eukaryotic genomes. We developed a tool designated SINE-Finder for the targeted discovery of tRNA-derived SINEs. We analyzed sequence data of 16 plant genomes, including 13 angiosperms and three gymnosperms and identified 17,829 full-length and truncated SINEs falling into 31 families showing the widespread occurrence of SINEs in higher plants. The investigation focused on potato (Solanum tuberosum), resulting in the detection of seven different SolS SINE families consisting of 1489 full-length and 870 5′ truncated copies. Consensus sequences of full-length members range in size from 106 to 244 bp depending on the SINE family. SolS SINEs populated related species and evolved separately, which led to some distinct subfamilies. Solanaceae SINEs are dispersed along chromosomes and distributed without clustering but with preferred integration into short A-rich motifs. They emerged more than 23 million years ago and were species specifically amplified during the radiation of potato, tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum). We show that tobacco TS retrotransposons are composite SINEs consisting of the 3′ end of a long interspersed nuclear element integrated downstream of a nonhomologous SINE family followed by successfully colonization of the genome. We propose an evolutionary scenario for the formation of TS as a spontaneous event, which could be typical for the emergence of SINE families.
INTRODUCTION
Retrotransposons have been identified in all plants investigated and, together with degenerated derivatives, often account for more than the half of the nuclear genome (SanMiguel et al., 1996; Bennetzen et al., 2005; Baucom et al., 2009).
LTR-retrotransposons flanked by long terminal repeats (LTRs) are represented by the main types Copia and Gypsy elements (Wicker et al., 2007). Non-LTR retrotransposons include long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). SINEs are short nonautonomous retroelements that range in size up to 500 nucleotides and have a composite structure, and their retrotransposition relies on proteins encoded by a partner LINE (Singer, 1982; Weiner et al., 1986; Boeke, 1997; Kajikawa and Okada, 2002; Dewannieux et al., 2003). Similarly to LINEs, they are usually terminated by a poly(A) stretch, poly(T) stretch, or simple sequence motifs at the 3′ end and flanked by target site duplications (TSDs). Most SINEs are ancestrally derived from tRNAs and transcribed by RNA polymerase III from degenerated internal promoters (Galli et al., 1981; Weiner et al., 1986; Okada, 1991a, 1991b; Sun et al., 2007). By contrast, some mammalian SINEs, such as B1 and Alu, originate from 7SL RNAs, while the zebra fish SINE3 is derived from 5S rRNA genes (Ullu and Tschudi, 1984; Kapitonov and Jurka, 2003).
Degenerated primers for coding regions, such as the reverse transcriptase open reading frames, enabled the identification of LTR retrotransposons and LINEs from numerous plant genomes by PCR (Hirochika et al., 1992; Wright et al., 1996; Kubis et al., 1998; Suoniemi et al., 1998). However, SINEs are noncoding and extremely heterogeneous. Therefore, the identification of complete SINEs is difficult, and application of PCR to target the RNA polymerase III promoter regions is not straightforward (Borodulina and Kramerov, 1999). Nevertheless, SINEs have been well studied in animal genomes where they form prominent sequence families (reviewed in Kramerov and Vassetzky, 2005).
Only a small number of SINE families from a limited group of plant taxa, such as Poaceae, Cruciferae, and Solanaceae, have been investigated to date. The first plant SINEs identified were p-SINE1 of rice (Oryza sativa) and TS in tobacco (Nicotiana tabacum; Umeda et al., 1991; Yoshioka et al., 1993). Subsequently, several SINE families from two grasses and a few Cruciferae species, in particular from Brassica napus and Arabidopsis thaliana, have been reported (Deragon et al., 1994; Goubely et al., 1999; Tikhonov et al., 2001; Yasui et al., 2001; Zhang and Wessler, 2005; Deragon and Zhang, 2006; Baucom et al., 2009). Most plant SINEs have been randomly identified, for example, by insertional polymorphisms, and a systematic and targeted search for SINEs is not possible, even in the era where complete genome sequences are available in an ever increasing rate.
Here, we report the targeted identification and characterization of tRNA-derived SINEs from numerous plant genomes. Focusing on potato and related Solanaceae species, we investigated SINE families with respect to their family structure, diversity, and frequent truncation. By comparative blot hybridization, we show that SINE families were differently amplified during the phylogeny of Solanaceae species. Fluorescent in situ hybridization (FISH) was applied to visualize the SINE distribution along chromosomes, while bioinformatic analysis was used to unravel the genomic dispersal pattern.
RESULTS
Mining of SINEs in Plant Genome Sequences
We analyzed the molecular structure of known plant SINEs to identify common features. The comparison of their RNA polymerase III promoter boxes A and B (Galli et al., 1981) revealed high sequence divergence and a size ranging from 95 to 352 bp (Zhang and Wessler, 2005; Deragon and Zhang, 2006). However, two degenerated consensus motifs (box A: RVTGG and box B: GTTCRA) were identified (Umeda et al., 1991; Yoshioka et al., 1993; Lenoir et al., 1997; Yasui et al., 2001; Xu et al., 2005; Tsuchimoto et al., 2008). The distance between both boxes A and B is limited to 24 to 43 nucleotides (Myouga et al., 2001; Baucom et al., 2009), and SINEs are usually terminated downstream by a poly(A) or poly(T) stretch or simple sequence repeat followed by the 3′ TSD of variable lengths (reviewed in Kramerov and Vassetzky, 2005; Wicker et al., 2007). The 5′ TSD is located 5 to 15 nucleotides upstream of box A (Mochizuki et al., 1992; Deragon et al., 1994). These weakly conserved features were compiled in a sequence-based algorithm and implemented in a tool designated SINE-Finder (see Supplemental Data Set 1 online), which was used to analyze Solanaceae genome sequence entries. Potato (Solanum tuberosum) was used as reference species, where two SINE families, designated TS and AU, have been reported (Yoshioka et al., 1993; Pozueta-Romero et al., 1998; Yasui et al., 2001).
We investigated public genome sequences of potato consisting of 386,309 accessions corresponding to ~350 Mbp and representing 40% of the ~850 Mbp potato genome (www.potatogenome.net/index.php/Main_Page; Arumuganathan and Earle, 1991).
Alignments of SINE-Finder output sequences of potato, tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum) revealed many SINEs that grouped into seven distinctly different families designated SolS-I to SolS-VII (Solanaceae SINE-familiy I to VII). SINEs were assigned to a single family when they shared at least 60% similarity; they were used as queries for BLAST searches to retrieve the majority of all family members, resulting in 1489 full-length and 870 5′ truncated copies in potato. Analysis of the complete Solanaceae sequence data set including many novel members of the AU and TS family, revealed a similar number of full-length (8153) and 5′ truncated (7431) SINEs. A summary of these results is provided in Table 1, Supplemental Data Set 2 online, and Supplemental Data Set 3 online.
Table 1.
SINE Families in Solanaceae
| Family | Species | No. (Full-Length) | 5′ Truncated Copies | Similarity (%)a | Consensus (bp)b | poly(A) (bp)c |
| SolS-Ia | Potato | 274 | 139 | 67 | 179 | 10 |
| Tomato | 231 | 76 | 64 | |||
| Tobacco | 352 | 244 | 64 | |||
| SolS-Ib | Potato | 217 | 57 | 67 | 184 | 9 |
| Tomato | 135 | 61 | 62 | |||
| Tobacco | 280 | 379 | 69 | |||
| SolS-II | Potato | 309 | 253 | 69 | 213 | 8 |
| Tomato | 548 | 361 | 60 | |||
| Tobacco | 977 | 1004 | 67 | |||
| SolS-IIIa | Potato | 213 | 186 | 87 | 231 | 11 |
| Tomato | 316 | 291 | 79 | |||
| Tobacco | n.d. | – | – | |||
| SolS-IIIb | Potato | 19 | 26 | 80 | 244 | 10 |
| Tomato | 3 | 41 | 78 | |||
| Tobacco | 72 | 72 | 61 | |||
| SolS-IV | Potato | 216 | 128 | 77 | 193 | 11 |
| Tomato | 223 | 99 | 63 | |||
| Tobacco | 555 | 310 | 66 | |||
| SolS-V | Potato | 221 | 44 | 76 | 106 | 10 |
| Tomato | 193 | 52 | 68 | |||
| Tobacco | 108 | 38 | 71 | |||
| SolS-VI | Potato | 17 | 34 | 99 | 225 | 17 |
| Tomato | n.d. | – | – | |||
| Tobacco | 98 | 158 | 93 | |||
| SolS-VII | Potato | 3 | 3 | 96 | 210 | 15 |
| Tomato | 12 | 12 | 76 | |||
| Tobacco | 10 | 19 | 89 | |||
| AU | Potato | 71 | 169 | 66 | ||
| Tomato | 107 | 134 | 65 | |||
| Tobacco | 1578 | 2335 | 74 | |||
| TS | Potato | n.d. | – | – | ||
| Tomato | n.d. | – | – | |||
| Tobacco | 795 | 706 | 87 |
Averaged identity of full-length SINEs.
Consensus sequence without poly(A).
Averaged length.
n.d., not detected.
Furthermore, we conducted a search in 10 angiosperm genome sequences of different taxonomic clades to investigate the occurrence of SINEs in diverse plant genomes (Table 2; see Supplemental Data Set 2 online). We identified 22 SINE families comprising of 2197 copies in species of the Brassicaceae (BraS-I), Cucurbitaceae (CucuS-I and CucuS-II), Euphorbiaceae (EuphS-I), Fabaceae (FabaS-I to -IX), Salicaceae (SaliS-I to -V), Scrophulariaceae (ScroS-I), Vitaceae (VitaS-I), and Poaceae (PoaS-I and PoaS-II). SINEs were also found in the basal angiosperm Nuphar advena (NymS-I) and in three gymnosperm species (PinS-I in Picea glauca, Picea sitchensis, and Pinus taeda). Identification of the CypS-I family in the zebra fish (Cyprinidae) demonstrated the successful application of the SINE-Finder for an animal genome sequence. Direct SINE-Finder output sequences and resulting consensus sequences of SINE families are shown in Supplemental Data Set 2 online and Supplemental Data Set 4 online, respectively.
Table 2.
SINE Families in Genomes of Diverse Plant Species and Danio rerio
| Family | Species | No. | Similarity (%)a | Consensus (bp)b | poly(A) (bp)c |
| BraS-I | A. lyrata | 37 | 90 | 134 | 15 |
| CucuS-I | C. sativus | 41 | 80 | 206 | 9 |
| CucuS-II | C. sativus | 16 | 87 | 98 | 11 |
| EuphS-I | M. esculenta | 247 | 86 | 83 | 12 |
| FabaS-I | M. truncatula | 198 | 89 | 131 | 12 |
| FabaS-II | M. truncatula | 133 | 81 | 116 | 11 |
| FabaS-III | M. truncatula | 99 | 85 | 166 | 12 |
| FabaS-IV | M. truncatula | 80 | 90 | 205 | 13 |
| FabaS-V | M. truncatula | 17 | 91 | 185 | 9 |
| FabaS-VI | M. truncatula | 16 | 87 | 186 | 8 |
| FabaS-VII | M. truncatula | 11 | 84 | 124 | 10 |
| FabaS-VIII | M. truncatula | 11 | 75 | 115 | 9 |
| FabaS-IX | G. max | 66 | 77 | 107 | 9 |
| SaliS-I | P. trichocarpa | 666 | 87 | 186 | 11 |
| SaliS-II | P. trichocarpa | 127 | 75 | 169 | 7 |
| SaliS-III | P. trichocarpa | 75 | 84 | 168 | 10 |
| SaliS-IV | P. trichocarpa | 44 | 82 | 185 | 12 |
| SaliS-V | P. trichocarpa | 30 | 94 | 268 | 16 |
| ScroS-I | M. guttatus | 95 | 92 | 210 | 20 |
| VitaS-I | V. vinifera | 143 | 86 | 93 | 13 |
| PoaS-I | B. distachyon | 25 | 86 | 156 | 8 |
| PoaS-II | B. distachyon | 20 | 81 | 115 | 8 |
| NymS-I | N. advena | 30 | 79 | 131 | 12 |
| PinS-I | P. glauca | 10 | 72 | 227 | 7 |
| P. sitchensis | 5 | 68 | 7 | ||
| P. taeda | 3 | 75 | 9 | ||
| CypS-I | D. rerio | 270 | 79 | 186 | 9 |
Averaged identity.
Consensus sequence without poly(A).
Averaged length.
Comparative Analysis of Solanaceae SINEs
We analyzed nine SINE families, including SolS-I to -VII, and TS and AU elements of potato, tomato, and tobacco using potato SINEs as a reference group. The Solanaceae SINE families are highly different in abundance, and the number of full-length members per family across the Solanaceae genomes investigated ranged from three (SolS-VII in potato) to 1578 copies (AU in tobacco). Reverse transcription of non-LTR retrotransposons starts at the 3′ end and may be aborted before the 5′ end of the RNA intermediate is reached. Therefore, we searched for 5′ truncated copies of Solanaceae SINEs that can be clearly identified by the poly(A) tail. We identified 1039 potato, 1127 tomato, and 5265 tobacco truncated SINEs. This is most likely an underestimation that does not include 3′ end truncated copies resulting from genomic rearrangements. We investigated the break points of truncated copies and did not observe a preferred region or site of truncation but a random dispersion of length variants by comparison of SINE families within and across potato, tomato, and tobacco (see Supplemental Figure 1 online).
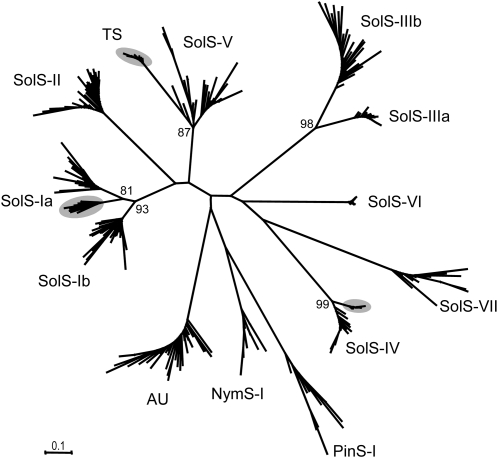
To visualize the divergence and grouping into SINE families, an unrooted dendrogram was constructed from representative copies (Table 1; see Supplemental Data Set 5 online) of each family, which showed the highest similarity to the consensus sequence. Each SolS family forms a separate branch consisting of SINEs from potato, tomato, and tobacco, indicating conservation of families across species (Figure 1). The families SolS-I and SolS-III were assigned to two subfamilies and grouped on separate branches supported by high bootstrap values. Consensus sequences of SolS-Ia/b and SolS-IIIa/b show identity values of 83 and 77%, respectively. In addition, tobacco-specific occurrence was observed for some SINE families (gray-shaded branches in Figure 1). SINEs from N. advena and Pinaceae (Table 2) were involved as distant outgroup sequences.
Figure 1.
Dendrogram of Representative Copies Showing the Lowest Divergence to the Consensus of the SINE Families in Solanaceae, Nymphaeaceae, and Pinaceae.
Ten SolS and AU from potato, tomato, and tobacco, NymS (N. advena), PinS (P. glauca, P. sitchensis, and P. taeda), as well as TS (tobacco) were chosen. For low-copy families, <10 elements had to be used (Tables 1 and 2; see Supplemental Data Set 5 online). Gray shaded branches indicate tobacco-specific SINE subsets. Bootstrap values >80 are shown.
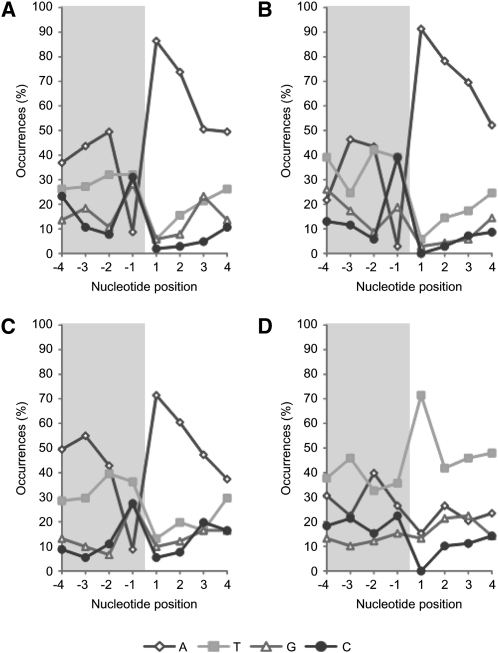
The length of the poly(A) region is very variable, and inspection of the 3′ flanking region of 1023 potato SINEs of all SolS families revealed average poly(A) tails from eight to 17 within families (Table 1). Extreme values ranged from 0 to 45 adenosine residues without family specificity. Next, we characterized preferential integration sites of Solanaceae SINEs exemplarily analyzed for 263 potato SINEs of the families SolS-IIIa, SolS-IV, and SolS-V (Figure 2; see Supplemental Data Set 6 online). Four nucleotides of the flanking region upstream of the 5′ TSD and the first four nucleotides of the 5′ TSD were examined. Depending on the family, 71 to 91% of the potato SINEs integrated upstream of an adenine or short adenine motif (Figures 2A to 2C). By contrast, analysis of 98 TS copies that possess a repeated GTT motif at their 3′ end showed preferential integration (71%) upstream of thymine or short thymine stretches (Figure 2D).
Figure 2.
Insertion Site Preference of Solanaceae SINEs.
Nucleotide frequencies at the 5′ nicking sites of potato SolS-IIIa (A), SolS-IV (B), SolS-V (C) and tobacco TS (D) were analyzed. Positions −4 to −1 (gray shaded) are nucleotides of the flanking DNA directly upstream of the 5′ TSD. Positions 1 to 4 mark the first four nucleotides of the 5′ TSD.
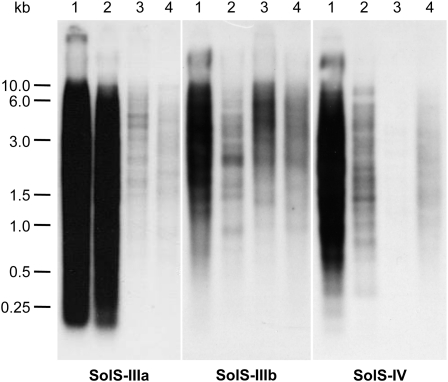
Because bioinformatic identification of SINEs is based on and limited by available public sequences, we performed comparative DNA gel blot hybridization using potato SINEs as probes to investigate their genomic abundance. Pepper (Capsicum annuum; Capsiceae) was included because of its intermediate taxonomic position between the Solaneae (potato and tomato) and Nicotianoideae (tobacco). Blot hybridization revealed that the SolS families are present in potato, tomato, pepper, and tobacco with extreme differences in abundance between species. Seven SINE families and subfamilies (SolS-Ia, Ib, II, IIIa, IIIb, IV, and V) were highly amplified in potato. Examples are shown in Figure 3.
Figure 3.
Distribution and Genomic Organization of SINE Families in Solanaceae.
DNA gel blot hybridization to BsmAI-digested genomic DNA of potato (1), tomato (2), pepper (3), and tobacco (4) using probes derived from consensus elements of SolS-IIIa, SolS-IIIb, and SolS-IV.
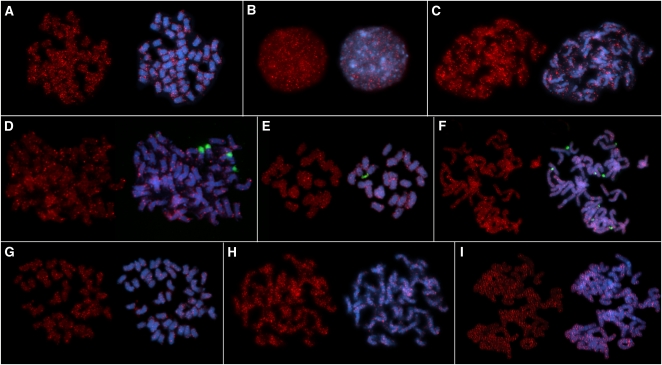
The chromosomal distribution of selected Solanaceae SINEs was investigated by FISH on potato, tomato, and tobacco chromosomes. For both potato and tomato, dispersed signals of the abundant SolS-IIIa are detectable on all 48 and 24 mitotic metaphase chromosomes (Figures 4A and 4C), respectively. SolS-IIIa is predominantly localized in distal euchromatic chromosome regions and often detectable on both chromatides. Chromosomes studied at interphase showed that SolS-IIIa SINEs are mostly excluded from heterochromatic 4′,6–diamidino–2-phenylindole–positive blocks (Figure 4B). SolS-II was chosen as example to compare the chromosomal distribution between species (Figures 4D to 4F). In potato, tomato, and tobacco, SolS-II is dispersed localized on all chromosomes; however, the 18S-5.8S-25S rDNA regions are depleted from SolS-II in all three species. The dispersed distribution along chromosomes has also been observed for SolS-V on potato chromosomes (Figure 4G) and for the remaining SolS families (see Supplemental Figure 2 online).
Figure 4.
Physical Mapping of SINE Families and SolRTE-I LINEs by FISH.
SolS-IIIa was localized on potato metaphase (A), interphase (B), and tomato early metaphase (C) chromosomes. Dispersed distribution of SolS-II is shown on metaphase chromosomes from potato (D), tomato (E), and tobacco (F). SolS-V was mapped on potato metaphase chromosomes (G) and SolRTE-I on early metaphase chromosomes from potato (H) and tobacco (I). Red signals are sites of retrotransposon hybridization, green signals label the 18S-5.8S-25S rRNA gene arrays, and blue fluorescence shows 4′,6–diamidino-2-phenylindole–stained DNA.
A detailed investigation of the genomic distribution of SolS SINEs at high resolution was performed using 50 BACs of both potato and tomato. Examples of BACs carrying most SINEs are shown for potato (see Supplemental Table 1 online) and tomato (see Supplemental Table 2 online). Up to nine or seven copies of different SINE families were found of a single potato or tomato BAC, respectively. However, no family-specific preference or clustering was observed. The number of SINEs per sequence accession for the whole Solanaceae sequence data set is shown in Supplemental Table 3 online.
Origin and Evolution of Solanaceae SINEs
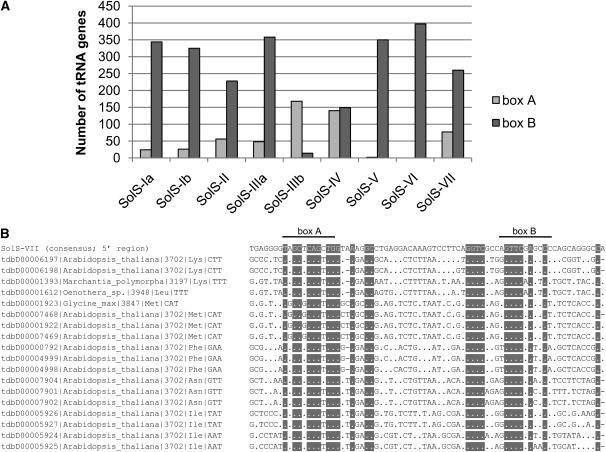
The 5′ regions of all SolS families were compared with 702 Viridiplantae tRNA genes (Jühling et al., 2009). No significant similarity to specific tRNA genes was found. However, positions of diverged boxes A and B and spaces between them of SolS SINEs resemble tRNA polymerase III promoters. We conducted an assignment of individual SINE boxes A and B with 70 to 100% similarity to corresponding tRNA motifs (Figure 5A). Generally, SINE B boxes are significantly more highly conserved to the corresponding motif of tRNA genes than SINE A boxes. The SolS-VII family showed the highest similarity to various whole tRNA genes of different plants, particularly in the region of boxes A and B (Figure 5B). Taken together, the similarity observed suggests a tRNA-derived origin of SolS SINEs.
Figure 5.
Similarity of Potato SolS SINEs to Viridiplantae tRNA Genes.
(A) RNA polymerase III promoter boxes A and B of 702 Viridiplantae tRNA genes were assigned to corresponding boxes A and B of SolS-I to VII consensus sequences, respectively, based on similarity (minimal eight identical nucleotides out of eleven).
(B) Full-length tRNA genes were aligned with the 5′ region of the potato SolS-VII consensus sequence showing 55 to 62% identity. Nucleotides fully conserved between tRNA promoter regions including boxes A and B and corresponding boxes of SolS-VII are gray-boxed. Dots show nucleotides identical between tRNA genes and SolS-VII, and dashes indicate gaps.
Short branches in the dendrogram (Figure 1) indicate ancestry and close phylogenetic relationships of some SINE subfamilies, such as SolS-Ia/b and SolS-IIIa/b. For example, the SolS-IIIa subfamily was not detectable in tobacco. By contrast, SolS-IIIb SINEs were identified in all three Solanaceae species (Table 1) and hence are likely to be more ancestral and evolutionarily older. Species-specific amplification and exclusive colonization of the tobacco genome has been detected for TS and subsets of the SolS-Ia and SolS-IV clades (gray-shaded branches in Figure 1). Both tobacco-specific SolS SINE groups contain 16 and 15 diagnostic nucleotide positions, respectively.
We also suggest an evolutionary scenario for the formation of the tobacco TS SINEs. The TS family is highly abundant in tobacco, and we isolated 1501 full-length and truncated copies from sequence entries in public databases (Table 1). However, we could not identify TS in potato and tomato. Analysis of the 5′ end of TS (99 bp) revealed an 84% similarity to the SolS-V family. Surprisingly, using the remaining 102 bp of the 3′ end of TS as query for similarity search in the potato genome revealed many sequences that are identical to the 3′ ends of a previously unknown LINE family, which we designated SolRTE-I. Therefore, TS SINEs are composed at the 5′ end of SolS-V and at the 3′ end of the 3′ untranslated region (UTR) of SolRTE-I. Moreover, TS SINEs share the poly(GTT) tail with SolRTE-I (Figure 6; see Supplemental Data Set 7 online). A typical member of the SolRTE-I family is SolRTE-I_St1, which is 4174 bp long and belongs to the RTE clade (Malik and Eickbush, 1998) of LINEs (see Supplemental Figures 3 and 4 and Supplemental Data Set 7 online). FISH on metaphase chromosomes of potato and tobacco using the reverse transcriptase gene of SolRTE-I_St1 as probe revealed that Solanaceae SolRTE-I is widely dispersed along chromosomes, in particular in distal regions (Figures 4H and 4I).
Figure 6.
Composite Structure and Origin of TS.
Sequence similarities revealed by comparative alignment of tobacco TS and the 3′ UTR of SolRTE-I of potato, tomato, and tobacco (above) and TS to SolS-V (below). Nucleotides are color coded: adenine (green), thymine (red), guanine (yellow), and cytosine (blue). TS_Ntcons is the TS consensus (Yoshioka et al., 1993). SolRTE-I_Nt is reconstructed from 1640 assembled tobacco sequences (see Supplemental Data Set 8 online). Dots indicate identical nucleotides. Dashes show gaps introduced to optimize the alignment. Gray shaded rectangles indicate the RNA polymerase III promoters. St, S. tuberosum; Sl, S. lycopersicum; Nt, N. tabacum. Accessions for compared elements are as follows: SolRTE-I_Sl, EF647616; SolRTE-I_St1, AC233388; SolRTE-I_St3, AC232878; SolRTE-I_St4, AC219014; TS_Nt1, FH227950; TS_Nt2, ET774464; SolS-V_St1, AC234546; SolS-V_Sl1, CU459061; and SolS-V_Nt1, ET727010.
We conclude that TS evolved from two independent retrotransposon families, namely, SolS-V and SolRTE-I, and hence has a composite structure. Although the SolRTE-I family has also been identified in tobacco, potato, and tomato, TS evolved only in the tobacco genome.
DISCUSSION
Plant SINEs have been characterized only in a very limited number of taxa, namely, Cruciferae (Arabidopsis, B. napus, and Brassica oleracea), Poales (rice, Aegilops umbellulata, and maize), and one species in the Solanaceae, tobacco. Here, we identified SINE families of many plants and focused on the comparative study of Solanaceae SINEs.
Using the SINE-Finder, we identified a wealth of SINEs of 16 plant species showing their widespread occurrence in higher plants (Tables 1 and 2). As a proof of principle, we analyzed the Arabidopsis genome sequence (1C = 157 Mbp; Bennett et al., 2003) and found all abundant SINE families described as AtSB2, AtSB3, AtSB4, and AtSB6 (Deragon and Zhang, 2006). Members of the AtSB5 family are only detectable applying modified SINE-Finder parameters (reduction of the TSD score cutoff to eight), while AtSB7, a low-copy family with only three members, is not detectable because it harbors no or only diverged crucial sequence motifs such as poly(A) or TSDs. However, we were also able to identify the four SINE families from the maize (Zea mays) genome reported so far (Baucom et al., 2009). Recently, four SINE families from Medicago truncatula were identified by screening of public genome sequences (Gadzalski and Sakowicz, 2011). In addition to these SINEs (designated here FabaS-I to -IV), we were able to identify four novel SINE families (FabaS-V to -VIII) in the M. truncatula genome demonstrating the potential of the method. SINE-Finder is based on relatively weakly conserved sequence motifs. To test the specificity and to exclude the identification of sequences with random similarity to SINEs, we applied SINE-Finder to a contiguous 350-Mbp sequence consisting of the randomly shuffled potato sequence data analyzed in this study. We did not find any SINE or SINE-like family.
Due to the heterogeneity in sequence and length, the criteria for family assignment suggested by Wicker et al. (2007) could not be applied in this study. Nevertheless, a clear family structure was observed and the identified plant SINEs share similar structural features.
A number of relatively highly conserved SINE families has been described for rice and Brassica species (Deragon et al., 1994; Myouga et al., 2001; Xu et al., 2005; Zhang and Wessler, 2005), such as BoS elements from B. oleracea and S1 SINEs from rapeseed. By contrast, most SolS families described here have sequence similarities below 80% among members. In particular, abundant SolS families with widespread appearance in potato, tomato, and tobacco exist as heterogeneous families (Table 1).
SINEs may attain high copy numbers in small genomes such as rice (1C = 420 Mbp) where 10,178 SINEs were identified (Paterson et al., 2009). Based on the genome sequence information, we calculated that ~6500 SINEs exist in the potato genome. These SINEs represent ~0.15% of the genome, which is in a similar range as in B. oleracea (1C = 600 Mbp) with 4290 Bos elements (Zhang and Wessler, 2005; Deragon and Zhang, 2006). The maize genome contains 1991 SINE copies (Baucom et al., 2009). Nevertheless, the maize genome is 2.7 times larger than the potato genome; hence, the average SINE density is higher in potato and in B. oleracea.
These data suggest that there is no stringent correlation between plant genome size and number of SINEs as has also been observed for LTR retrotransposons (Vitte and Bennetzen, 2006). Only a small number of active SINEs are required as master elements for constant amplification, while most copies are transpositional inactive (Deininger and Batzer, 2002). However, the strong differences in copy numbers also indicate transpositional bursts in some SINE families (e.g., SolS-IIIa in potato and tomato) (Figure 3). In potato, 315 SINE copies (~20% of 1560 full-length elements) from all SolS families were identified in ESTs, including two out of three SolS-VII elements. SINEs adjacent to genes may be cotranscribed during transcription of these genes by RNA polymerase II. Alternatively, the relatively high number of transcribed SolS SINEs copies found in potato ESTs does not exclude the possible activity, which is in line with the low divergence found in SolS-IIIa, SolS-VI, and SolS-VII.
Extreme variation in abundance between closely related species has also been observed for other retrotransposons, such as Ty1-copia retrotransposons and LINEs (Pearce et al., 1996; Vitte and Bennetzen, 2006). However, LTR retrotransposons and LINEs are autonomous, while SINEs are noncoding and depend on the reverse transcriptional machinery of LINEs. SINE retrotransposition requires active LINEs and the sequence-dependent recognition of the SINE 3′ end by the LINE reverse transcriptase. We found an approximately equal number of full-length and truncated SINEs (8153 versus 7431) in three Solanaceae species. The relatively high number of full-length SINEs might be explained by the fact that SINEs, although using the same enzymes, are much shorter than LINEs and the probability of truncation during reverse transcription increases with the length of the RNA intermediate.
A partnership between SINEs and LINE-like elements has been found in invertebrates, fish, reptiles, mammals, and recently in maize (Ohshima and Okada, 2005; Baucom et al., 2009; reviewed in Kramerov and Vassetzky, 2005). Members of many LINE clades are considered as parental elements of SINEs; however, the relation of RTE LINEs to SINEs has not been demonstrated in plants so far. The retrotransposons TS and SolRTE-I share a common poly(GTT) and have considerable similarity of their target sites. The RTE type of LINEs, only poorly investigated in plants (Zupunski et al., 2001), has the potential to provide the enzymes for SINE transposition. SolRTE-I probably belongs to the stringent group LINEs, which can initiate reverse transcription only for elements exhibiting homologous 3′-terminal sequences (Okada et al., 1997).
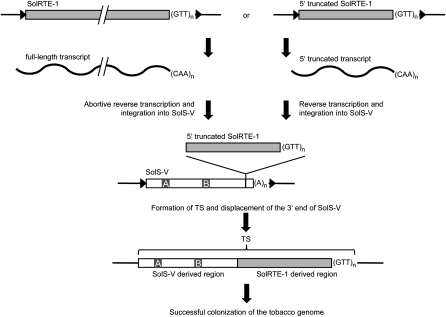
Based on the similarity to the tobacco LINE SolRTE-I, we propose the following evolutionary scenario for the origin of TS (Figure 7). Abortive target-primed reverse transcription (Luan et al., 1993; Ostertag and Kazazian, 2001) of a full-length SolRTE-I transcript may have resulted in the formation of a heavily truncated copy consisting of 102 bp of the partial 3′ UTR and the poly(GTT) tail. Alternatively, the partial 3′ UTR and the poly(GTT) tail may originate from transcription of a rearranged 5′ truncated SolRTE-I. The SolRTE-I–derived fragment was integrated into the 3′ end of a SolS-V SINE, thereby displacing the SolS-V poly(A) region, which was then dispensable for transposition and diverged. The resulting chimeric element was a founder TS and contained the RNA polymerase III promoters needed for transcription and the 3′ end/poly(GTT) necessary for binding of the RTE reverse transcriptase. Subsequent retrotransposition mediated by SolRTE-I resulted in the massive proliferation of TS SINEs in tobacco after splitting from the last common ancestor. The spontaneous emergence of TS in tobacco could be a typical mode of evolution of SINE families in genomes.
Figure 7.
Model of the Evolutionary Origin of the TS SINE Family.
The model shows the possible evolution of the tobacco TS based on the strong similarity of the SINE to the 3′ region of SolRTE-I, which is a LINE of the RTE clade. Polymerase III promoter boxes A and B are designated. Black triangles indicate TSD.
The comprehensive analyses described here enabled insights into the evolution and age of Solanaceae SINEs. Solanaceae SINEs are most likely derived from diverse tRNA genes, suggesting a polyphyletic origin. Subsequent amplification and diversification resulted in the heterogeneity and family structure observed. All SolS families identified in potato are also detectable in tobacco, which is the most distantly related Solanaceae species investigated in this study. Therefore, we assume that the SolS families emerged in the genome of the most recent common ancestor of potato, tomato, pepper, and tobacco at least 23.7 million years ago (MYA) (Wu and Tanksley, 2010). The subfamily SolS-IIIa is highly repetitive in potato and tomato, and the weak hybridization in pepper and tobacco indicates a massive amplification after separation of these two species 19.6 MYA (Figure 3, left panel). Furthermore, SolS-IV is not detectable in pepper and might have decayed in copy number or sequence after species separation. After splitting of potato and tomato 7.3 MYA, the SINE families SolS-IV (Figure 3, right panel) and SolS-IIIb (Figure 3, middle panel) were highly amplified in potato.
Because SINEs have considerable impact on genome organization, diversification, and transcriptional activity, knowledge of SINEs populating a genome is crucial. Next-generation sequencing is a rapidly evolving technique and generates vast amounts of genome sequences. However, the annotation of raw data still lags behind and is a cumbersome process. Among the many sequenced plant genomes, SINEs have only been annotated in Arabidopsis, rice, and maize in contrast to other transposable elements, such as LTR retrotransposons, LINEs, and DNA transposons (reviewed in Velasco et al., 2010). The approach described here may support the annotation of plant genomes by the identification of sequence families that consist of short, heterogeneous, and highly abundant transposable elements.
METHODS
SINE-Finder Tool
The SINE-Finder is a computational tool that we developed for the identification of SINEs in genome sequences of any species.
Based on structural features of SINEs, we designed a search algorithm as follows: at the beginning, a 5′ TSD region of 40 nucleotides, followed by the box A motif RVTGG, a spacer of 25 to 50 nucleotides, the box B motif GTTCRA, a spacer of 20 to 500 nucleotides, a poly(A) stretch of six adenines, and a 3′ TSD region of 40 nucleotides. The search algorithm designated as pattern is as follows: pattern = (('TSD_region_1', “{,40}”), ('a_box', “[GA][CGA]TGG”), ('spacer_1', “{25,50}”), ('b_box', “GTTC[AG]A”), ('spacer_2', “{20,500}”?), ('polyA', “A{6,}|T{6,}”), ('TSD_region_2', “{,40}”)). Lengths of TSD regions, spacers, and poly(A)/poly(T) as well as sequence motifs can be modified by replacements of numbers and nucleotides in the script of the tool.
The SINE-Finder (see Supplemental Data Set 1 online) is a Python script (http://www.python.org/). The only requirement for application is installation of the Python software (at least version 2.5). The file sine_finder.py and sequence files to be analyzed must be deposited in the same folder. The required nucleotide sequence format is FASTA (a sequence starts with a single-line description having the greater than [>] symbol as the first followed by sequence lines). Sequence files with extensions (fas, FASTA, and mfa) are accepted as input. The tool may be started using the command line mode by calling with or without arguments (e.g., calling for help: python sine_finder.py -h), the interactive mode (Python's integrated development environment) by calling without arguments or as a module by including it in other scripts. Before the program starts to analyze the sequences, several parameters will be prompted. The application flow is self-explanatory, and recommended settings (default) are given. Depending on the capacity of the computer, large sequences or sequences of whole chromosomes can be split into smaller segments (chunk-wise function) by the program. Output sequences will be continuously saved in a separate file (FASTA format). The SINE-Finder tool also can be obtained from http://tu-dresden.de/die_tu_dresden/fakultaeten/fakultaet_mathematik_und_naturwissenschaften/fachrichtung_biologie/botanik/zellmolbiopflanzen.
Data Mining and Resources
Sequence data were obtained from public entries (GenBank and EMBL) as follows: 386,309 potato (Solanum tuberosum) accessions (350 Mbp), 641,862 tomato (Solanum lycopersicum) accessions (722 Mbp), 1,758,009 tobacco (Nicotiana tabacum) accessions (1420 Mbp), 22,402 Nymphaeales accessions (27 Mbp), and 1,062,987 Pinaceae accessions (843 Mbp). Genome sequences were downloaded via the phytozome homepage (http://www.phytozome.net/; Joint Genome Institute [JGI]) for Arabidopsis thaliana (ftp://ftp.Arabidopsis.org/sequences/whole_chromosomes/ Swarbreck et al., 2008), Arabidopsis lyrata (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Alyrata/; JGI), Brachypodium dystachion (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Bdistachyon/assembly/sequences/; JGI), Cucumis sativus (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Csativus/assembly/; JGI), Glycine max (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Gmax/assembly/sequences/ Schmutz et al., 2010), Manihot esculenta (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Mesculenta/; JGI), Mimulus guttatus (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Mguttatus/assembly/; JGI), Medicago truncatula (http://www.medicago.org/genome/downloads/Mt3/; Medicago Genome Sequence Consortium), Populus trichocarpa (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Ptrichocarpa/assembly/; Tuskan et al., 2006), and Vitis vinifera (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v5.0/Vvinifera/assembly/). The Zea mays genome sequence was obtained from the PlantGDB (http://zoneannfu.gdcb.iastate.edu/XGDB/phplib/download.php?GDB=Zm) and the Danio rerio genome from the Danio rerio Sequencing Project (http://hgdownload.cse.ucsc.edu/goldenPath/danRer6/bigZips/; Wellcome Trust Sanger Institute). A list of the analyzed species and corresponding amount of sequence data is provided in Supplemental Table 4 online.
Alignments and BLAST searches were performed using stand-alone versions of MUSCLE (Edgar, 2004) and FASTA (ftp://ftp.ebi.ac.uk/pub/software/unix/fasta/fasta36/), respectively. Dendrograms were constructed by MEGA 5 software (Tamura et al., 2007), applying the neighbor-joining distance method and the maximum composite likelihood nucleotide model. Bootstrap values were calculated using 1000 replicates. Randomizing of the complete set of Solanaceae genomic sequences was performed using the “random.shuffle” function of Biopython 1.57 (http://www.biopython.org/wiki/Biopython).
Genomic Organization and Chromosomal Localization
Genomic DNA were extracted using the CTAB protocol (Saghai-Maroof et al., 1984) from young leaves of S. tuberosum cv Gala, S. lycopersicum cv Tamina, Capsicum annuum cv De cayenne, and N. tabacum cv Virginia. DNA gel blot hybridization was performed according to Sambrook and Russell (2001) using radioactively labeled probes. Preparation of chromosomes, labeling of probes with biotin-11-dUTPs, and FISH were performed as described by Wenke et al. (2009) according to Schwarzacher and Heslop-Harrison (2000). Probes were cloned from potato DNA using PCR and primers listed in Supplemental Table 5 online.
Accession Numbers
Sequences of representative SINEs of each SINE family and species from this article can be found in the GenBank/EMBL data libraries under accession numbers HE583424 to HE583588.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Frequency and Length of 5′ Truncated SINEs (SolS, AU, and TS) Detected in the Data Sets of Potato, Tomato, and Tobacco.
Supplemental Figure 2. Physical Mapping of SolS SINE Families by FISH.
Supplemental Figure 3. Structure and Amino Acid Alignments of SolRTE-I Sequences.
Supplemental Figure 4. Assignment of the SolRTE-I Family to the RTE Clade of LINEs.
Supplemental Table 1. Number of SINEs in Large Sequence Accessions of Potato.
Supplemental Table 2. Number of SINEs in Large Sequence Accessions of Tomato.
Supplemental Table 3. Number of SINEs (SolS, AU, and TS) per Sequence Accession Detected in the Sequence Data Sets of Potato, Tomato, and Tobacco.
Supplemental Table 4. Genome Data Sets Analyzed in This Study.
Supplemental Table 5. Primers Used for the Generation of SolS SINE Probes for DNA Gel Blot and Fluorescent in Situ Hybridization.
Supplemental Data Set 1. Python Script of the SINE-Finder.
Supplemental Data Set 2. Sequence File (FASTA Format) Containing Identified SINEs.
Supplemental Data Set 3. Sequence File (FASTA Format) Containing the 5′ Truncated SolS, AU, and TS SINEs of Solanaceae.
Supplemental Data Set 4. Sequence File (FASTA Format) Containing the Consensus Sequences of Identified SINE Families.
Supplemental Data Set 5. Sequence File (FASTA Format) Containing the Aligned SINEs without TSDs, Poly(A) Tails, Poly(T) Tails, or Poly(GTT) Tails from Which the Dendrogram (Figure 1) Was Constructed.
Supplemental Data Set 6. Sequence File (FASTA Format) Containing SINEs of the Potato SolS-IIIa, IV, V, and the Tobacco TS Families Used for the Analysis of Insertion Sites (Figure 2) Inclusive of Poly(A) Tails, Poly(GTT) Tails, and TSDs.
Supplemental Data Set 7. Sequence File (FASTA Format) Showing Tobacco TS SINEs and SolS-V from Potato, Tomato, and Tobacco Aligned with SolRTE-I LINEs of Potato, Tomato, and Tobacco.
Supplemental Data Set 8. Sequence File (FASTA Format) Showing an Assembly of 1640 Sequences of the Tobacco SolRTE-I Family Used to Derive the Consensus Sequence.
Acknowledgments
We thank F. Ludwig for assistance in programming and our colleagues for discussion and critical reading of the manuscript. This work was supported by the projects Kleine und Mittlere Unternehmen Innovativ 0315425B and the Coordinated Research Program D23028.
AUTHOR CONTRIBUTIONS
T.W., T.D. and T.R.S. performed the research and analyzed the data. H.J. selected, cultivated, and contributed the plant material. T.W., B.W., and T.S. wrote the article.
References
- Arumuganathan K., Earle E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218 [Google Scholar]
- Baucom R.S., Estill J.C., Chaparro C., Upshaw N., Jogi A., Deragon J.M., Westerman R.P., Sanmiguel P.J., Bennetzen J.L. (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.D., Leitch I.J., Price H.J., Johnston J.S. (2003). Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb. Ann. Bot. (Lond.) 91: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L., Ma J., Devos K.M. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. (Lond.) 95: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D. (1997). LINEs and Alus—The polyA connection. Nat. Genet. 16: 6–7 [DOI] [PubMed] [Google Scholar]
- Borodulina O.R., Kramerov D.A. (1999). Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 457: 409–413 [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A. (2002). Mammalian retroelements. Genome Res. 12: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Deragon J.M., Landry B.S., Pélissier T., Tutois S., Tourmente S., Picard G. (1994). An analysis of retroposition in plants based on a family of SINEs from Brassica napus. J. Mol. Evol. 39: 378–386 [DOI] [PubMed] [Google Scholar]
- Deragon J.M., Zhang X.Y. (2006). Short interspersed elements (SINEs) in plants: Origin, classification, and use as phylogenetic markers. Syst. Biol. 55: 949–956 [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. (2003). LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadzalski M., Sakowicz T. (2011). Novel SINEs families in Medicago truncatula and Lotus japonicus: Bioinformatic analysis. Gene 480: 21–27 [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M.L. (1981). Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294: 626–631 [DOI] [PubMed] [Google Scholar]
- Goubely C., Arnaud P., Tatout C., Heslop-Harrison J.S., Deragon J.M. (1999). S1 SINE retroposons are methylated at symmetrical and non-symmetrical positions in Brassica napus: Identification of a preferred target site for asymmetrical methylation. Plant Mol. Biol. 39: 243–255 [DOI] [PubMed] [Google Scholar]
- Hirochika H., Fukuchi A., Kikuchi F. (1992). Retrotransposon families in rice. Mol. Gen. Genet. 233: 209–216 [DOI] [PubMed] [Google Scholar]
- Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. (2009). tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37(Database issue): D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M., Okada N. (2002). LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111: 433–444 [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2003). A novel class of SINE elements derived from 5S rRNA. Mol. Biol. Evol. 20: 694–702 [DOI] [PubMed] [Google Scholar]
- Kramerov D.A., Vassetzky N.S. (2005). Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 247: 165–221 [DOI] [PubMed] [Google Scholar]
- Kubis S.E., Heslop-Harrison J.S., Desel C., Schmidt T. (1998). The genomic organization of non-LTR retrotransposons (LINEs) from three Beta species and five other angiosperms. Plant Mol. Biol. 36: 821–831 [DOI] [PubMed] [Google Scholar]
- Lenoir A., Cournoyer B., Warwick S., Picard G., Deragon J.M. (1997). Evolution of SINE S1 retroposons in Cruciferae plant species. Mol. Biol. Evol. 14: 934–941 [DOI] [PubMed] [Google Scholar]
- Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. (1993). Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H. (1998). The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 15: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Umeda M., Ohtsubo H., Ohtsubo E. (1992). Characterization of a plant SINE, p-SINE1, in rice genomes. Jpn. J. Genet. 67: 155–166 [DOI] [PubMed] [Google Scholar]
- Myouga F., Tsuchimoto S., Noma K., Ohtsubo H., Ohtsubo E. (2001). Identification and structural analysis of SINE elements in the Arabidopsis thaliana genome. Genes Genet. Syst. 76: 169–179 [DOI] [PubMed] [Google Scholar]
- Ohshima K., Okada N. (2005). SINEs and LINEs: Symbionts of eukaryotic genomes with a common tail. Cytogenet. Genome Res. 110: 475–490 [DOI] [PubMed] [Google Scholar]
- Okada N. (1991a). SINEs. Curr. Opin. Genet. Dev. 1: 498–504 [DOI] [PubMed] [Google Scholar]
- Okada N. (1991b). SINEs: Short interspersed repeated elements of the eukaryotic genome. Trends Ecol. Evol. (Amst.) 6: 358–361 [DOI] [PubMed] [Google Scholar]
- Okada N., Hamada M., Ogiwara I., Ohshima K. (1997). SINEs and LINEs share common 3′ sequences: A review. Gene 205: 229–243 [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H., Jr (2001). Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pearce S.R., Harrison G., Li D., Heslop-Harrison J., Kumar A., Flavell A.J. (1996). The Ty1-copia group retrotransposons in Vicia species: Copy number, sequence heterogeneity and chromosomal localisation. Mol. Gen. Genet. 250: 305–315 [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J., Houlné G., Schantz R. (1998). Identification of a short interspersed repetitive element in partially spliced transcripts of the bell pepper (Capsicum annuum) PAP gene: New evolutionary and regulatory aspects on plant tRNA-related SINEs. Gene 214: 51–58 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M.A., Soliman K.M., Jorgensen R.A., Allard R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- SanMiguel P., Tikhonov A., Jin Y.K., Motchoulskaia N., Zakharov D., Melake-Berhan A., Springer P.S., Edwards K.J., Lee M., Avramova Z., Bennetzen J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison P. (2000). Practical in Situ Hybridization. (Oxford, UK: BIOS Scientific Publishers: ). [Google Scholar]
- Singer M.F. (1982). Highly repeated sequences in mammalian genomes. Int. Rev. Cytol. 76: 67–112 [DOI] [PubMed] [Google Scholar]
- Sun F.J., Fleurdépine S., Bousquet-Antonelli C., Caetano-Anollés G., Deragon J.M. (2007). Common evolutionary trends for SINE RNA structures. Trends Genet. 23: 26–33 [DOI] [PubMed] [Google Scholar]
- Suoniemi A., Tanskanen J., Schulman A.H. (1998). Gypsy-like retrotransposons are widespread in the plant kingdom. Plant J. 13: 699–705 [DOI] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36(Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tikhonov A.P., Lavie L., Tatout C., Bennetzen J.L., Avramova Z., Deragon J.M. (2001). Target sites for SINE integration in Brassica genomes display nuclear matrix binding activity. Chromosome Res. 9: 325–337 [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S., Hirao Y., Ohtsubo E., Ohtsubo H. (2008). New SINE families from rice, OsSN, with poly(A) at the 3′ ends. Genes Genet. Syst. 83: 227–236 [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. (1984). Alu sequences are processed 7SL RNA genes. Nature 312: 171–172 [DOI] [PubMed] [Google Scholar]
- Umeda M., Ohtsubo H., Ohtsubo E. (1991). Diversification of the rice Waxy gene by insertion of mobile DNA elements into introns. Jpn. J. Genet. 66: 569–586 [DOI] [PubMed] [Google Scholar]
- Velasco R., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vitte C., Bennetzen J.L. (2006). Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103: 17638–17643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A.M., Deininger P.L., Efstratiadis A. (1986). Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 55: 631–661 [DOI] [PubMed] [Google Scholar]
- Wenke T., Holtgräwe D., Horn A.V., Weisshaar B., Schmidt T. (2009). An abundant and heavily truncated non-LTR retrotransposon (LINE) family in Beta vulgaris. Plant Mol. Biol. 71: 585–597 [DOI] [PubMed] [Google Scholar]
- Wicker T., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Wright D.A., Ke N., Smalle J., Hauge B.M., Goodman H.M., Voytas D.F. (1996). Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142: 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Tanksley S.D. (2010). Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.H., Osawa I., Tsuchimoto S., Ohtsubo E., Ohtsubo H. (2005). Two new SINE elements, p-SINE2 and p-SINE3, from rice. Genes Genet. Syst. 80: 161–171 [DOI] [PubMed] [Google Scholar]
- Yasui Y., Nasuda S., Matsuoka Y., Kawahara T. (2001). The Au family, a novel short interspersed element (SINE) from Aegilops umbellulata. Theor. Appl. Genet. 102: 463–470 [Google Scholar]
- Yoshioka Y., Matsumoto S., Kojima S., Ohshima K., Okada N., Machida Y. (1993). Molecular characterization of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific tRNAs. Proc. Natl. Acad. Sci. USA 90: 6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wessler S.R. (2005). BoS: A large and diverse family of short interspersed elements (SINEs) in Brassica oleracea. J. Mol. Evol. 60: 677–687 [DOI] [PubMed] [Google Scholar]
- Zupunski V., Gubensek F., Kordis D. (2001). Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 18: 1849–1863 [DOI] [PubMed] [Google Scholar]