NIP5;1 encodes a boron channel; this work shows that the 5′ untranslated region of NIP5;1 is required for mRNA accumulation in response to boron deficiency and mRNA degradation in response to high-boron conditions.
Abstract
Boron (B) is an essential plant micronutrient that is toxic at higher levels. NIP5;1 is a boric acid channel required for B uptake and growth under B deficiency. Accumulation of the NIP5;1 transcript is upregulated under B deficiency in Arabidopsis thaliana roots. To elucidate the mechanism of regulation, the 5′ untranslated region (UTR) of NIP5;1 was tested for its ability to confer B-dependent regulation using β-glucuronidase and green fluorescent protein as reporters. This analysis showed that the 5′ UTR was involved in NIP5;1 transcript accumulation in response to B conditions. We also found that high-B conditions trigger NIP5;1 mRNA degradation and that the sequence from +182 to +200 bp in the 5′ UTR is required for this mRNA destabilization. In the nip5;1-1 mutant background, a NIP5;1 complementation construct without the 5′ UTR produced high levels of mRNA accumulation, increased B concentrations in tissues, and reduced growth under high-B conditions. These data suggest that the 5′ UTR controls B-dependent NIP5;1 mRNA degradation and that NIP5;1 mRNA degradation is important for plant acclimation to high-B conditions.
INTRODUCTION
Boron (B) was established as an essential micronutrient for plants in 1923 (Warington, 1923). Since then, B was shown to be essential for diatoms (Smyth and Dugger, 1981) and cyanobacteria (Bonilla et al., 1990) as well as for animals, including zebra fish (Danio rerio) (Rowe and Eckhert, 1999), trout (Oncorhynchus mykiss) (Eckhert, 1998; Rowe et al., 1998), frogs (Xenopus laevis) (Fort et al., 1998), and mice (Mus musculus) (Lanoue et al., 2000). Although the precise physiological role of B has not been established, it is beneficial for human health (Nielsen, 2000).
B in neutral solution is predominantly present as boric acid (H3BO3), an uncharged molecule. Boric acid is a very weak Lewis acid with a pKa of 9.24 (Woods, 1996), and plants take up B from the soil mainly in the form of boric acid. B availability in soils is limited in many high rainfall areas because boric acid is easily leached out from the soil due to its high solubility (Shorrocks, 1997). B, however, is toxic to plants when present in excess, and arid or semiarid areas often have B toxicity problems. Water reaches the topsoil by capillary action and then evaporates, causing B to accumulate to a high concentration (Yau et al., 1995). Because B is relatively phloem immobile in most crop plants, B deficiency symptoms often occur in the growth of apical meristems (both shoots and roots), and toxicity symptoms often appear as necrosis along the margins of old leaves (Marschner, 1995; Dell and Huang, 1997; Shorrocks, 1997). Since both B deficiency and B toxicity reduce crop yield, managing B availability in soils is important to maintain high crop productivity.
The main known function of B in plants is to maintain cell wall structure, and B is a component of rhamnogalacturonan-II, a complex pectic polysaccharide. Cross-linking of rhamnogalacturonan-II by borate is essential for normal expansion of leaves (O’Neill et al., 2001, 2004).
After B is taken up from root surface, it must be transported across the plasma membranes of various cells. Boric acid, the major B form in the plant, is a small, uncharged molecule that can diffuse relatively easily across membranes. Under optimum B conditions, plants can acquire enough B for normal growth mainly by passive diffusion (Dordas et al., 2000; Dordas and Brown, 2001; Stangoulis et al., 2001). By contrast, under low B conditions, two types of B transporters, BORs and the nodulin 26-like intrinsic proteins (NIPs), are necessary for effective B movement from roots to shoots and for effective B uptake from soils to roots.
BOR1, identified as the first B efflux transporter in Arabidopsis thaliana, is localized to the plasma membrane and is involved in xylem loading of B in roots under B limitation (Takano et al., 2002). BOR1 is polarly localized on the inner side of the cells (Takano et al., 2010), and BOR1 accumulation is regulated by posttranscriptional mechanisms in response to B deficiency (Takano et al., 2005). Higher accumulation of BOR1 fused with green fluorescent protein (GFP) was observed in transgenic plants grown under low-B conditions compared with those under high-B conditions. BOR1-GFP was internalized through endocytosis and degraded in vacuoles in response to high-B supply (Takano et al., 2005, 2010). A BOR1 paralog, BOR4, also encodes a functional efflux B transporter in Arabidopsis. BOR4 is stable under high-B conditions and excludes B from the roots under such conditions. Overexpression of BOR4 was found to significantly improve excessive B stress tolerance in Arabidopsis plants (Miwa et al., 2007).
Another type of membrane protein, NIP5;1, was discovered as a boric acid channel (Takano et al., 2006). NIP5;1 is a member of the major intrinsic protein family; major intrinsic protein is known to facilitate the passive flow of water and small uncharged molecules and is widely present in various organisms, such as mammals, amphibians, yeast, bacteria, and plants (Johanson et al., 2001; Zardoya, 2005; reviewed in Tyerman et al., 2002; Maurel, 2007; Tanaka and Fujiwara, 2008).
Under B limitation, NIP5;1 is required for B uptake for normal growth in Arabidopsis. NIP5;1 is localized to the plasma membrane on the outer side of epidermal cells in the roots (Takano et al., 2006, 2010). Accumulation of NIP5;1 mRNA is upregulated by 10-fold in response to B deprivation. NIP6;1, the Arabidopsis protein most similar to NIP5;1, is also a boric acid channel and functions in preferential B distribution to shoot sink tissues under B deficiency (Tanaka et al., 2008). NIP6;1 is localized to the plasma membrane in the vascular bundle, especially in phloem parenchyma cells and companion cells in the nodal region in shoots. NIP6;1 mRNA accumulates significantly even under high B supply and is upregulated by approximately twofold in response to B deprivation.
To adjust to the environmental changes required for growth and development, gene expression can be regulated at multiple levels. Posttranscriptional control of mRNA stability can be beneficial, providing a rapid response to changes in the intracellular and extracellular environments (reviewed in Abler and Green, 1996). In many cases, mRNA stability is regulated by sequences in the 3′ untranslated region (UTR). One A-rich element located in the 3′ UTR is well known as an mRNA stability element for translation-dependent destabilization in eukaryotic cells (Ohme-Takagi et al., 1993; Chen et al., 2001; Sarkar et al., 2003). The downstream element, containing an mRNA instability sequence located in the 3′ UTR, was also characterized in plants (Gil and Green, 1996; Pérez-Amador et al., 2001). In some cases, mRNA stability is controlled by sequences in the 5′ UTR. For example, dark-dependent FERREDOXIN-1 mRNA destabilization in transgenic tobacco (Nicotiana tabacum) is regulated by multiple CAUU sequences located in the 5′ UTR (Bhat et al., 2004). Lhcb1*4 encodes light-harvesting complex II proteins in peas (Pisum sativum), and the Lhcb1*4 transcript is destabilized by blue light through a 5′ UTR sequence (Anderson et al., 1999).
In this study, to obtain insight into the mechanisms regulating NIP5;1 in response to B conditions, we performed deletion analyses of the NIP5;1 promoter and the 5′ UTR and investigated the physiological role of NIP5;1 regulation. We demonstrated that B-responsive NIP5;1 expression is regulated through sequences in the 5′ UTR and that a conserved element located in the 5′ UTR controls mRNA stability in a B-dependent manner in Arabidopsis roots. This regulatory mechanism plays an important role in maintaining B concentration in plants within the range that allows normal growth, even during rapid changes in B availability.
RESULTS
Deletion Analysis of the NIP5;1 Promoter to Identify Cis-Acting Elements for B-Regulated Expression
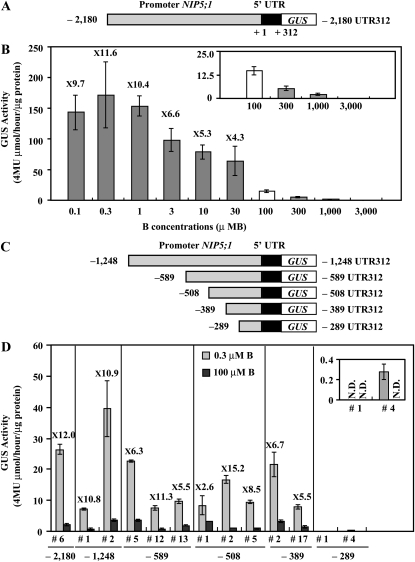
In a previous report, Takano et al. (2006) demonstrated that β-glucuronidase (GUS) activity driven by the NIP5;1 promoter (2180 bp) and 5′ UTR (312 bp) was higher in Arabidopsis roots grown under low-B (0.3 μM B) conditions than those under high-B (100 μM B) conditions. To further investigate the induction pattern, the same transgenic plants containing the full-length promoter with the 5′ UTR fused to GUS, referred to as P-2,180 UTR312-GUS in this article (see Supplemental Table 1 online for the nomenclature of the constructs), were grown for 14 d on solid medium containing a wide range of B concentrations (0.1 to 3000 μM B), and the GUS activity in the roots was determined (Figures 1A and 1B). The –2180 refers to the nucleotide number from the transcriptional start site (+1) of NIP5;1 (based on The Arabidopsis Information Resource [TAIR] database), and 312 is the length of the 5′ UTR in NIP5;1 (Figure 1A). GUS activity strongly increased under B-deficient conditions at 11.6- and 10.4-fold higher at the 0.1 and 0.3 μM B, respectively, than at the 100 μM B condition and gradually declined as the B concentration in the medium increased (Figure 1B). At 3 mM B, GUS activity was below the detection limit.
Figure 1.
GUS Activity and Deletion Analysis of the NIP5;1 Promoter in Response to B in Roots.
(A) Schematic representation of the P-2,180 UTR312-GUS construct. The nucleotides are numbered from the transcription start site (+1).
(B) Transgenic plants (#6) were grown for 14 d on plates with various B concentrations, and the GUS activity in roots was measured. The small window represents the scale-up of the data for 100, 300, 1000, and 3000 μM B. The numerals in the figure indicate ratios of the GUS activity in roots grown under 0.1, 0.3, 1, 3, 10, and 30 μM B to the GUS activity in roots grown under the 100 μM B condition (induction ratio). Means of three biological replicates ± sd (n = 3) are shown.
(C) Schematic representation of transformation constructs. P-1,248 UTR312-GUS, P-589 UTR312-GUS, P-508 UTR312-GUS, P-389 UTR312-GUS, and P-289 UTR312-GUS constructs are shown as –1248, –589, –508, –389, and –289 UTR 312, respectively. The nucleotides are numbered from the transcription start site (+1).
(D) Transgenic plants were grown for 7 d on plates with 0.3 or 100 μM B and the GUS activity in roots was measured. The small window represents the scale-up of the data for P-289 UTR312-GUS constructs. The numerals in the figure indicate induction ratios of the GUS activity in roots grown under the 0.3 μM B condition to the GUS activity in roots grown under the 100 μM B condition. Means of three biological replicates ± sd (n = 3) are shown.
To clarify the regulatory mechanism of expression by the NIP5;1 promoter fragment, a 5′-deletion series (–1248, –589, –508, –389, and –289 bp) from the transcriptional start site (+1) containing the 5′ UTR was constructed and fused to the GUS reporter gene (referred to as P-1,248 UTR312-GUS, P-589 UTR312-GUS, P-508 UTR312-GUS, P-389 UTR312-GUS, and P-289 UTR312-GUS, respectively) and introduced into Columbia-0 (Col-0) plants (Figure 1C). Independent T3 transgenic lines homozygous for the T-DNA insertion were grown for 7 d on solid medium containing 0.3 or 100 μM B. For each construct, GUS activity was measured in the roots of two to three independent lines (Figure 1D). The measured GUS activity was lower in Figure 1D than in Figure 1B presumably because plants were grown for 14 and 7 d in Figures 1B and 1D, respectively. GUS activity at 0.3 μM B was around 12-fold higher than at 100 μM B in transgenic plants containing –2180 promoter in Figure 1D, and the ratio (12.0) is almost similar to that in Figure 1B (11.6), suggesting that plants grown for 7 d are sufficient to detect induction of GUS activity under the low-B condition compared with that under the high-B condition. GUS activity in the transgenic lines carrying P-1,248 UTR312-GUS, P-589 UTR312-GUS, P-508 UTR312-GUS, and P-389 UTR312-GUS was elevated under B deficiency similarly to the transgenic lines carrying the full-length P-2,180 UTR312-GUS. The GUS activity was ~2.6- to 15.2-fold higher under the 0.3 μM B condition compared with the 100 μM B condition in these lines. By contrast, GUS activity in the transgenic line carrying P-289 UTR312-GUS was low and close to the detection limit. These results suggest that a cis-acting element that acts in the response to B status is likely to exist downstream of –389.
Role of the 5′ UTR in the Regulation of Gene Expression in Response to B Concentration in Arabidopsis Roots
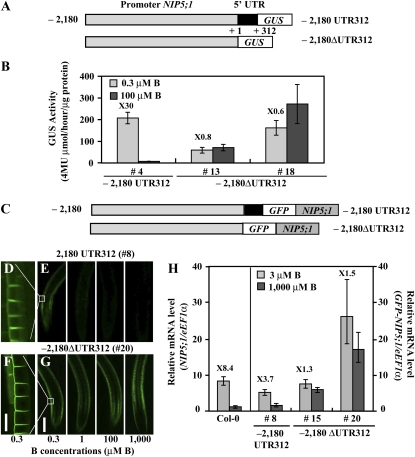
The above results suggest that regions of NIP5;1 required for regulation of B-dependent expression may be present in the promoter region between −389 and +1 or in the 5′ UTR. It is known that the 5′ UTR can affect regulation of gene expression, including regulation of translation efficiency and mRNA stability. To investigate the contribution of the 5′ UTR to NIP5;1 expression in response to B status, we tested the NIP5;1 promoter fragment without the 5′ UTR by making a construct that fused the promoter to the GUS reporter gene (P-2,180 ΔUTR312-GUS; Figure 2A) and then introducing that construct into Col-0 plants. Independent T3 transgenic lines homozygous for the T-DNA insertion were grown for 11 d on 0.3 or 100 μM B. The GUS activity controlled by P-2,180 UTR312 construct containing the 5′ UTR was higher under low-B conditions (0.3 μM B) than under high-B conditions (100 μM B), but the GUS activity controlled by the P-2,180 ΔUTR312 construct without the 5′ UTR was similar under both conditions (Figure 2B). This result suggests that the 5′ UTR is required for B-dependent expression of NIP5;1.
Figure 2.
The 5′ UTR Is Involved in B-Dependent Gene Expression in Roots.
(A) Schematic representation of transformation constructs. P-2,180 UTR312-GUS and P-2,180 ΔUTR312-GUS constructs are shown as 2180 UTR312 and –2180 ΔUTR312, respectively. The nucleotides are numbered from the transcription start site (+1).
(B) Transgenic plants were grown for 11 d on plates with 0.3 or 100 μM B and GUS activity in roots was measured. The numerals in the figure indicate induction ratios of the GUS activity in roots grown under the 0.3 μM B conditions to the GUS activity in roots grown under the 100 μM B conditions. Means of three biological replicates ± sd (n = 3) are shown.
(C) Schematic representation of transformation constructs. P-2,180 UTR312-GFP-NIP5;1 and P-2,180ΔUTR312-GFP-NIP5;1 are shown as –2180 UTR312 and –2180 ΔUTR312, respectively. The nucleotides are numbered from the transcription start site (+1).
(D) to (G) Transgenic plants were grown for 14 d on plates containing 0.3, 1, 100, or 1000 μM B and visualized by confocal microscopy ([E] and [G]). The images show GFP-derived fluorescence in the root differentiation zone. Bar = 200 μm. (D) and (F) represent the scale-up of the images of 0.3 μM B in (E) and (G), respectively. Bar = 25 μm.
(H) B-dependent NIP5;1 or GFP-NIP5;1 mRNA accumulation in roots was quantified by qRT-PCR analysis. Transgenic plants were grown for 11 d on plates containing 3 or 1000 μM B. The numerals in the figure indicate induction ratios of the NIP5;1 or GFP-NIP5;1 mRNA accumulation in roots grown under the 3 μM B conditions to the NIP5;1 or GFP-NIP5;1 mRNA accumulation in roots grown under the 1000 μM B conditions, respectively. Means of three biological replicates ± sd (n = 3) are shown.
To further investigate the requirement of 5′UTR for B-dependent expression of NIP5;1 protein, NIP5;1 cDNA was fused to GFP (GFP-NIP5;1) and placed downstream of the NIP5;1 promoter fragment with or without the 5′ UTR (referred to as P-2,180 UTR312-GFP-NIP5;1 and P-2,180 ΔUTR312-GFP-NIP5;1, respectively, in Figure 2C). The constructs were introduced into the NIP5;1 T-DNA insertion line (nip5;1-1). Independent T3 transgenic lines homozygous for the T-DNA were grown for 14 d on solid medium containing 0.3, 1, 100, or 1000 μM B. GFP fluorescence in the root differentiation zone was detected by confocal laser scanning microscopy. Three independent transgenic lines were observed, and representative images are shown in Figures 2D to 2G. The GFP-NIP5;1 fluorescence in the transgenic plants carrying P-2,180 UTR312 is localized in the outer (distal) plasma membrane domain of the epidermal cells under low-B conditions as previously reported (Figure 2D; Takano et al., 2010), and the signal intensity gradually declined as B increased (Figure 2E). The GFP-NIP5;1 fluorescence in the transgenic plants carrying P-2,180 ΔUTR312 was also localized to the outer (distal) plasma membrane domain of epidermal cells (Figure 2F). In contrast with the transgenic plants carrying P-2,180 UTR312, the signal intensity of transgenic plants carrying P-2,180 ΔUTR312 was maintained even under excess B conditions (1000 μM B) (Figure 2G). These results suggested that the 5′ UTR controls B-dependent expression of NIP5;1.
We also determined the accumulation of the GFP-NIP5;1 transcript in the roots of these transgenic plants by quantitative RT-PCR (qRT-PCR) (Figure 2H). Plants were grown for 11 d on solid medium containing 3 or 1000 μM B. The primers used in this study detect both the NIP5;1 and GFP-NIP5;1 transcripts, but in nip5;1-1, the NIP5;1 mRNA is reduced to around 1% of that in Col-0 plants (Takano et al., 2006); thus, the endogenous NIP5;1 mRNA in these transgenic plants is negligible. GFP-NIP5;1 mRNA accumulation in transgenic plants carrying P-2,180 UTR312 decreased under the excess B conditions (1000 μM B) compared with the low-B conditions (3 μM B) as with NIP5;1 mRNA accumulation in Col-0 plants, while GFP-NIP5;1 mRNA accumulation in transgenic plants carrying P-2,180 ΔUTR312 remained high under the excess B conditions compared with the low-B conditions. These findings suggest that the NIP5;1 5′ UTR regulates mRNA accumulation in response to B and that posttranscriptional regulation may be important for this regulation.
Identification of a Region in the 5′ UTR Required for B Response
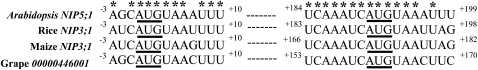
To search for regulatory element(s) in response to B status in the 5′ UTR, the 5′ UTR of Arabidopsis NIP5;1 was compared with the corresponding sequences in Os NIP3;1, the rice (Oryza sativa) NIP5;1 ortholog, which is also increased by B deficiency (Hanaoka and Fujiwara, 2007). Note that the 5′ UTR regions of these two genes are highly conserved (see Supplemental Figure 1 online). Both sequences contain three upstream open reading frame (uORF). The distances of the uORFs from the putative transcriptional start site (+1) are +1, +80, and +191 bp, and the uORFs consist of one, six, and one codon(s), respectively, in NIP5;1. In Os NIP3;1, the corresponding ORFs are at +1, +85, and +190 bp and consist of one, three, and one codon(s), respectively. The sequence (CAUGUAA) including the first uORF and the sequence (UCAAAUCAUGUAA) including the third uORF were identical between the Arabidopsis and rice genes (Figure 3).
Figure 3.
Sequence Comparison of the 5′ UTR of Arabidopsis NIP5;1, Rice NIP3;1, Maize NIP3;1, and Grape 00000446001.
5′ UTR sequences of At NIP5;1, Os NIP3;1, Zm NIP3;1, and Vv 00000446001 were compared. Two conserved sequences are shown. The nucleotide number corresponds to that from the putative transcription start site (+1). For Os NIP3;1, Zm NIP3;1, and Vv 00000446001, the transcription start site was deduced based on the alignment and sequence identity around the transcriptional start site of the At NIP5;1. Based on the TAIR database, the transcription start site of At NIP5;1 is the first A in AUGUAA; this is also the transcription start site of the rice, maize, and grape genes, and the nucleotides are numbered accordingly. Asterisks indicate conserved sequence among At NIP5;1, Os NIP3;1, Zm NIP3;1, and Vv 00000446001. Short uORFs are underlined.
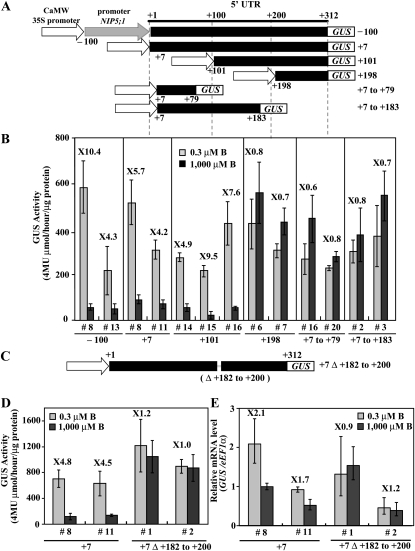
To confirm whether the 5′UTR is sufficient to control B-dependent expression, the NIP5;1 5′ UTR was fused to the cauliflower mosaic virus 35S RNA promoter (P35S) (referred to as P35S-100UTR+312-GUS and P35SUTR+7-GUS in Figure 4A) and introduced into Col-0 plants. In this experiment, we used P35S to eliminate other possible transcriptional regulation in response to B. For each construct, two to three independent T3 transgenic lines homozygous for the T-DNA insertion were established and used for the analysis of B-regulated expression. Plants were grown for 11 d under 0.3 or 1000 μM B conditions, and the GUS activity in roots was determined (Figure 4B). GUS activity in transgenic plants carrying P35S-100UTR312 and P35SUTR+7 was higher under the 0.3 μM B condition compared with the 1000 μM B condition. Ratios of GUS activity in B-deficient roots to that in B-sufficient roots were ~4.2 to 10.4, indicating that NIP5;1 5′ UTR is sufficient to regulate B-dependent expression. These findings suggested that B-dependent expression is regulated by the 5′ UTR through posttranscriptional mechanisms.
Figure 4.
Identification of the 5′ UTR Region Required for B Response in Roots.
(A) Schematic representation of transformation constructs. P35S-100UTR312-GUS, P35SUTR+7-GUS, P35SUTR+101-GUS, P35SUTR+198-GUS, P35SUTR+7 to 79-GUS, and P35SUTR+7 to +183-GUS constructs are shown as –100, +7, +101, +198, +7 to +79, and +7 to +183, respectively. The nucleotides are numbered from the transcription start site (+1).
(B) Transgenic plants were grown for 11 d on plates with 0.3 or 1000 μM B, and the GUS activity in roots was measured. The numerals in the figure indicate induction ratios of the GUS activity in roots grown under the 0.3 μM B conditions to the GUS activity in roots grown under the 1000 μM B conditions. Means of three biological replicates ± sd (n = 3) are shown.
(C) Schematic representation of transformation constructs. P35SUTR+7Δ+182-+200-GUS constructs is shown as +7Δ+182 to +200.
(D) Transgenic plants were grown for 11 d on plates with 0.3 or 1000 μM B and the GUS activity in roots was measured. The numerals in the figure indicate induction ratios of the GUS activity in roots grown under the 0.3 μM B conditions to the GUS activity in roots grown under the 1000 μM B conditions. Means of three biological replicates ± sd (n = 3) are shown.
(E) Transgenic plants were grown for 11 d on plates with 0.3 or 1000 μM B and GUS mRNA was quantified by qRT-PCR. The numerals in the figure indicate induction ratios of the GUS mRNA accumulation in roots grown under the 0.3 μM B conditions to the GUS mRNA accumulation in roots grown under the 1000 μM B conditions. Means of three biological replicates ± sd (n = 3) are shown.
To identify the element responsible for B-dependent regulation in the 5′ UTR, a series of truncated 5′ UTRs in NIP5;1 was generated and fused to the GUS reporter gene under the control of P35S (referred to as P35SUTR+101-GUS, P35SUTR+198-GUS, P35SUTR+7 to +79-GUS, and P35SUTR+7 to +183-GUS in Figure 4A) and introduced into Col-0 plants. For each construct, two to three independent T3 transgenic lines homozygous for the T-DNA insertion were established and used for the analysis of B-regulated expression. Plants were grown for 11 d under 0.3 or 1000 μM B conditions, and the GUS activity in roots was determined (Figure 4B). GUS activity in transgenic plants carrying P35SUTR+101 as well as P35S-100UTR312 and P35SUTR+7 was higher under the 0.3 μM B condition compared with the 1000 μM B condition. Ratios of GUS activity in B-deficient roots to that in B-sufficient roots were ~4.9 to 9.5. By contrast, the GUS activity in transgenic plants carrying P35SUTR+198, P35SUTR+7 to +79, and P35SUTR+7 to +183 was not downregulated by external B concentration (0.3 or 1000 μM B). Ratios of the GUS activity in B-deficient roots to that in B-sufficient roots were ~0.6 to 0.8. These results suggest that the 13-bp region from +184 to +197 bp in the 5′ UTR is important in the regulation of NIP5;1 expression in response to low B.
To confirm the above results, transgenic plants carrying P35SUTR+7 harboring an internal deletion between +182 and +200 bp were established (referred to as P35SUTR+7Δ+182 to 200-GUS in Figure 4C) and introduced into Col-0 plants. Two independent T3 transgenic lines homozygous for the T-DNA insertion were grown for 11 d under 0.3 and 1000 μM B conditions, and GUS activity in the roots was determined (Figure 4D). GUS activity in the roots of transgenic plants carrying P35SUTR+7 increased 4.5- to 4.8-fold in 0.3 μM B compared with 1000 μM B, whereas GUS activity in the roots of transgenic plants carrying P35SUTR+7Δ+182 to +200 grown under 0.3 μM B was similar to that under the 1000 μM B condition. This result demonstrated that the sequence from +182 to +200 bp in the 5′ UTR is required for the regulation of NIP5;1 expression in response to B.
To investigate whether NIP5;1 5′UTR can affect the mRNA accumulation driven by a heterogeneous promoter, P35S, GUS mRNA accumulation in roots was measured by qRT-PCR (Figure 4E). GUS mRNA accumulation in transgenic plants carrying P35SUTR+7 increased 1.7- to 2.1-fold under the 0.3 μM B condition compared with the 1000 μM B condition, suggesting that the 5′ UTR affects the mRNA accumulation driven by the P35S. By contrast, GUS mRNA accumulation in transgenic plants carrying P35SUTR+7Δ+182 to +200 grown in 0.3 μM B was similar to that in the 1000 μM B condition, indicating that the sequence from +182 to +200 bp in the 5′ UTR influences the regulation of mRNA accumulation in response to B in Arabidopsis roots.
The 5′ UTR Affects NIP5;1 mRNA Stability in a B-Dependent Manner
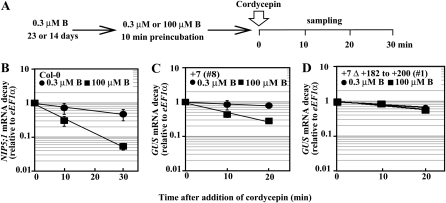
A possible mechanism of NIP5;1 transcript regulation in response to B by the 5′ UTR is mRNA decay. To test this possibility, the half-life of NIP5;1 mRNA in intact Col-0 roots was estimated by determining a time course for mRNA decay after exposure to 3′-deoxyadenosine (cordycepin) (Seeley et al., 1992), an RNA synthesis inhibitor, under high- and low-B conditions. Col-0 plants were grown with 0.3 μM B for 23 d and then preincubated for 10 min in hydroponic culture medium containing 0.3 or 100 μM B. After the preincubation, cordycepin was added to a final concentration of 0.6 mM. Roots were harvested at 0, 10, and 30 min after cordycepin treatment, and the NIP5;1 mRNA level was measured by qRT-PCR (Figures 5A and 5B).
Figure 5.
Time Course of B-Dependent NIP5;1 mRNA Degradation in Col-0 Roots and Effect of +182 to +200 bp Substitution in the 5′ UTR.
(A) A schematic representation of growth conditions. Col-0 plants were grown with 0.3 μM B for 21 d on solid medium and then transferred to hydroponic culture containing 0.3 μM B for 2 d. Transgenic plants were grown with 0.3 μM B for 14 d on solid medium. Plants were transferred to hydroponic medium containing 0.3 or 100 μM B for 10 min and then cordycepin was applied in the hydroponic culture. Root samples for Col-0 and whole-plant samples for transgenic plants were harvested at 0, 10, and 30 min, and 20 min, respectively, after cordycepin treatment, and mRNA levels were measured by qRT-PCR.
(B) Col-0.
(C) and (D) Transgenic plants carrying P35SUTR+7-GUS shown as +7 (C) and P35SUTR+7Δ+182 to +200-GUS shown as +7Δ+182 to +200 (D). The RNA levels at each time point were normalized to the amount at the zero time point and plotted on a log scale to show mRNA decay. Means of three biological replicates ± sd (n = 3) are shown.
After cordycepin treatment, the NIP5;1 mRNA level in Col-0 plants exposed to high-B conditions decreased rapidly compared with that in Col-0 plants exposed to low-B conditions (Figure 5B). The estimated half-lives of NIP5;1 mRNA were 5 and 17 min in Col-0 plants exposed to high-B and low-B conditions, respectively, suggesting that the rate of NIP5;1 transcript degradation is more than 3 times faster under high-B conditions than under low-B conditions.
Requirement of the +182- to +200-bp Region in the 5′ UTR for the B-Dependent Regulation of mRNA Stability
To determine if the sequence from +182 to +200 bp in the 5′ UTR is involved in mRNA degradation in response to B, the half-lives of GUS mRNA in transgenic plants carrying P35SUTR+7 and P35SUTR+7Δ+182 to +200 were measured under high- and low-B conditions (Figures 5C and 5D). Transgenic plants were grown in 0.3 μM B for 14 d, then cordycepin was added and whole plants were harvested at 0, 10, and 20 min after cordycepin treatment. The GUS mRNA level in transgenic plants carrying P35SUTR+7 exposed to high-B condition decreased faster than in transgenic plants carrying P35SUTR+7 exposed to low-B condition (Figure 5C). The estimated half-lives of GUS mRNA were 10 and 55 min in the transgenic plants exposed to high-B and low-B conditions, respectively, suggesting that the rate of GUS transcript degradation is 5 times faster under high-B condition than under low-B condition. In P35SUTR+7Δ+182 to +200-GUS transgenic plants, the rate of GUS mRNA accumulation did not significantly differ under high- and low-B conditions (Figure 5D). The estimated half-lives of the GUS mRNA were 24 and 38 min in transgenic plants exposed to high-B and low-B conditions, respectively. These results indicate that the sequence from +182 to +200 is involved in B-dependent mRNA degradation.
Physiological Roles of the Regulation of NIP5;1 Expression by the 5′ UTR in Roots under Low- and Excess-B Conditions
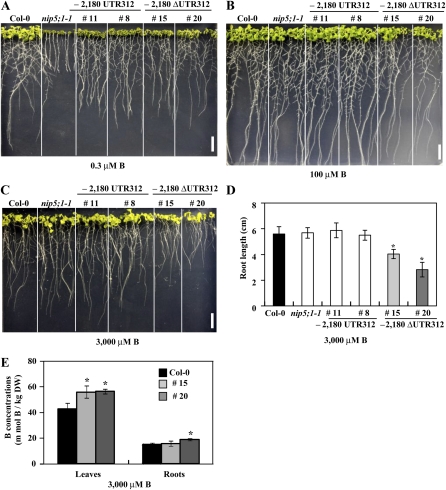
To investigate the role of the 5′ UTR on growth characteristics, three independent T3 lines carrying P-2,180 ΔUTR312-GFP-NIP5;1 and P-2,180 UTR312-GFP-NIP5;1 were grown on plates containing 0.3, 100, and 3000 μM B. The growth patterns of the three independent transgenic lines were very similar, and images of two of the transgenic lines are shown in Figures 6A to 6C. All transgenic plants showed growth recovery compared with nip5;1-1 mutant plants under low-B conditions (0.3 μM B; Figure 6A), suggesting that GFP-NIP5;1 functions as a B transporter and complements the nip5;1-1 mutation. Under high-B conditions (100 μM B), growth was similar among all plants (Figure 6B). Under excess-B conditions (3000 μM B), growth of the transgenic plants carrying the P-2,180 ΔUTR312 construct was reduced in both the roots and rosette leaves compared with Col-0 and nip5;1-1, whereas growth of the transgenic plants carrying P-2,180 UTR312 was similar to that of Col-0 and nip5;1-1 (Figure 6C). Moreover, leaves of the transgenic plants carrying the P-2,180 ΔUTR312 construct were yellower and smaller compared with other plant lines (Figure 6C). These results established that the construct lacking the 5′ UTR inhibited growth under excess-B condition, whereas the construct with the 5′ UTR did not.
Figure 6.
The 5′ UTR Affects Plant Growth and B Accumulation.
(A) to (C) Transgenic plants carrying P-2,180 UTR312-GFP-NIP5;1 or P-2,180ΔUTR312-GFP-NIP5;1, Col-0, and nip5;1-1 were grown on plates containing 0.3 μM B for 11 d, 100 μM B for 12 d, and 3000 μM B for 14 d. Bars = 10 mm.
(D) Root length of Col-0, nip5;1-1, and transgenic plants grown under 3000 μM B for 14 d was measured (n = 4 to 5). Means of four to five biological replicates ± sd are shown. Asterisks indicate a significant difference from Col-0 plants (P < 0.05, Student’s t test).
(E) B concentrations in roots and rosette leaves of plants grown under 3000 μM B for 14 d were determined using inductively coupled plasma–mass spectrometry (n = 3). Means of three biological replicates ± sd are shown. Asterisks indicate a significant difference from Col-0 plants (P < 0.05, Student’s t test).
To further characterize the phenotype of transgenic plants carrying P-2,180 ΔUTR312 under excess-B conditions, the root length and B concentration were determined in transgenic plants grown under 3000 μM B for 14 d (Figures 6D and 6E). Root length in transgenic plants carrying P-2,180 ΔUTR312 was reduced compared with Col-0, nip5;1-1, and transgenic plants carrying P-2,180 UTR312 (Figure 6D). B concentrations in shoots (two lines) and roots (one line) were significantly higher in transgenic plants carrying P-2,180 ΔUTR312 than in Col-0 plants (Figure 6E). Because boric acid is an uncharged small molecule that can relatively easily diffuse across membranes, under high-B conditions, B can be absorbed from roots without specific B transporters. It is considered that the additional B uptake by NIP5;1 resulted in higher B accumulation in leaves and caused B toxicity in the transgenic plants. Therefore, these data further confirmed the importance of the 5′ UTR in avoiding the negative effects of NIP5;1 expression under excess B conditions.
DISCUSSION
In this study, we demonstrated that changes in mRNA stability are involved in B-dependent accumulation of the NIP5;1 transcript and that destabilization in response to high-B conditions is regulated by the sequence in the 5′ UTR from +182 to +200 bp. The B-dependent regulatory mechanism controlled by the 5′UTR plays an important role in growth of Arabidopsis, taking up B from soils under low-B conditions and avoiding excess-B uptake under high-B conditions.
The NIP5;1 Transcript Is Destabilized under High-B Conditions through the 5′ UTR
We demonstrated that the NIP5;1 5′ UTR acts to repress transcript accumulation under high-B conditions but not to induce transcript accumulation under low-B conditions. In addition to the involvement of the 5′ UTR in response to B, measurement of the mRNA half-life revealed that the sequence from +182 to +200 bp in the 5′ UTR is required for B-dependent NIP5;1 mRNA destabilization (Figures 2 and 5).
The region from +182 to +200 bp in the 5′ UTR contained a highly conserved sequence (UCAAAUCAUGUAA) found in both Arabidopsis and rice (Figure 3). From the database, the maize (Zea mays) and grape (Vitis vinifera) orthologs of NIP5;1 also contained the conserved sequence, suggesting that it may also function as a boric acid channel and that its expression is repressed under high-B conditions in the roots (Figure 3). We also investigated whether a highly conserved sequence exists in the 5′ UTR of Arabidopsis NIP6;1, a boric acid channel responsible for B distribution in shoots (Tanaka et al., 2008). No conserved sequence was detected in the 5′ UTR of NIP6;1 compared with that of NIP5;1 (see Supplemental Figure 2 online), suggesting that the identified regulatory mechanism for B via mRNA decay is specific to NIP5;1. Consistently, B-dependent regulation of NIP6;1 mRNA levels is not as drastic as that of NIP5;1 (Tanaka et al., 2008). Orthologs of NIP5;1, which are expressed in the roots, might have retained these elements during plant evolution.
So far, mRNA stability controlled by the 5′ UTR has been characterized for a few genes, including light-regulated genes (Gil and Green, 1996; Pérez-Amador et al., 2001; Bhat et al., 2004) and salt and heat stress–regulated genes (Hua et al., 2001) in plants. Here, we identified an example of mRNA stability control by nutrient conditions through the 5′ UTR in Arabidopsis. Multiple transcriptional and posttranscriptional regulatory mechanisms of gene expression have been reported for acclimation to nutrient conditions. Our study demonstrated that mRNA stability control through the 5′ UTR is another important mechanism for plant acclimation to nutritional conditions.
Possible Mechanisms of mRNA Stability Regulation via the 5′ UTR in NIP5;1
The mechanism by which the NIP5;1 5′ UTR modulates mRNA stability in a B-dependent manner remains to be studied. Several different mechanisms of regulating mRNA stability through the 5′ UTR are reported, including the influence of the uORF, the 5′ UTR–specific RNA binding protein, the riboswitch and ribozyme, and microRNA (miRNA). A brief description of the current understanding of each mechanism is as follows: The uORF-dependent degradation can be classified into nonsense-mediated mRNA decay (NMD)–dependent or –independent degradation in yeast (Linz et al., 1997; Vilela et al., 1999). NMD is a well-studied mRNA surveillance system that degrades aberrant mRNA with premature termination codons. The NMD-independent pathway was shown in yeast, but the details are not well understood. In yeast, RNA binding proteins interfere with ribosome–mRNA interaction, which results in translational inhibition. This inhibition then leads to mRNA destabilization (Linz et al., 1997). Binding of metabolites and/or metals (Mg+) to riboswitches induces conformational changes in mRNA. The altered structures could affect transcriptional control, translational control, alternative splicing, and mRNA processing or stability in bacteria (Sudarsan et al., 2003; Cromie et al., 2006; Henkin, 2008). Ribozymes efficiently cleave specific sites in mRNA, resulting in the inhibition of gene expression in Gram-positive bacteria (Winkler et al., 2004). miRNAs can interact with a target region, mainly in the 3′ UTR, that influences mRNA degradation in melanoma cells (Goswami et al., 2010).
Each of these mechanisms could play a role in NIP5;1 regulation. Regarding the possibility of the involvement of a uORF in regulation, the NIP5;1 5′ UTR contains three short uORFs located at positions +1 (one codon), +80 (six codons), and +191 bp (one codon). The identified sequence (182 to 200 bp in the 5′ UTR) includes the third uORF position at +191 bp (Figure 3). According to Nyikó et al. (2009), longer uORFs (>50 amino acids) can trigger NMD, but shorter uORFs cannot effectively activate NMD. In NIP5;1, all uORFs are short and, per this mechanism, mRNA decay is to be controlled by the third uORF encoding 1 amino acid. However, we think that this does not necessarily exclude the possibility of involvement of the NMD-mediated pathway because if the stop codon of the third uORF was read through, a peptide with 51 amino acids overlapping the main ORF could be produced. Considering that boric acid can bind with cis-diols (Springsteen and Wang, 2002), it may be possible that boric acid binding causes a stop codon read-through. It could then inhibit translation from the main ORF and induce NIP5;1 mRNA destabilization by the NMD-dependent pathway. Alternatively, NMD-independent mechanisms may be involved in the regulation of mRNA stability through the NIP5;1 5′ UTR. The excess B may induce and/or activate an RNA binding protein(s) that recognizes the identified sequence in the NIP5;1 5′ UTR, which in turn leads to destabilization. A third possibility is that high-B conditions may trigger induction of miRNAs that interact with the identified sequence in the NIP5;1 5′ UTR, causing mRNA degradation. However, no miRNA that targets the NIP5;1 5′ UTR was found by massively parallel signature sequencing (Brenner et al., 2000) and none are in the plant miRNA database (Zhang et al., 2010).
Possible Involvement of Translational Regulation
In addition to controlled mRNA degradation, modulation of translation efficiency may also be involved in the regulation of B-dependent expression through NIP5;1 5′ UTR. In Figures 4D and 4E, ratios of GUS activity in two independent transgenic plants carrying the NIP5;1 5′ UTR were 4.8- and 4.5- fold higher under low-B than high-B conditions; by contrast, ratios of GUS mRNA accumulation in these transgenic plants were only 2.1- and 1.7-fold higher under low-B than high-B conditions.
To further support this hypothesis, immunoblot analysis of microsomal protein from transgenic plants carrying P-2,180 UTR312-GFP-NIP5;1 was also performed, and the NIP5;1-GFP protein accumulation in roots was at least 16-fold higher under low-B than high-B conditions, but ratios of GFP-NIP5;1 mRNA accumulation in roots were 6.7-fold higher under low-B than high-B conditions (see Supplemental Figure 3 online). These findings imply that translational regulation reduces protein accumulation under high-B conditions. Such a difference was not observed in the constructs carrying the deletion between +182 to +200 bp in the 5′ UTR (Figures 4C and 4D), suggesting that the deleted sequence may be required for the regulation of translation efficiency as well as mRNA stability in response to B in Arabidopsis roots. Further experiments are needed to confirm this hypothesis.
The Control of mRNA Degradation Plays an Important Role in Plant Growth in Acclimating to B Conditions
Growth defects in nip5;1-1 plants under low-B conditions are complemented by introduction of not only P-2,180 UTR312 GFP-NIP5;1 but also P-2,180 ΔUTR312-GFP-NIP5;1, suggesting that NIP5;1 produced from the construct without the 5′ UTR functions similarly to that from the construct with the 5′ UTR (Figure 6A). The nip5;1-1 plants carrying P-2,180 ΔUTR312 GFP-NIP5;1 showed higher sensitivity to toxic B conditions than the nip5;1-1 plants carrying P-2,180 UTR312 GFP-NIP5;1 (Figures 6C to 6E). This suggests that the 5′ UTR is essential in circumventing B toxicity.
In general, nutrients, including B, are nonuniformly distributed in soils. Rainfall often reduces the B content in soil, while B accumulates on the soil surface under dry conditions. Furthermore, the optimum B range for plant growth is reportedly very narrow (Marschner, 1995). To maintain optimum B concentrations for normal growth, roots must recognize the B concentration in the soil and control B uptake through NIP5;1 expression. mRNA degradation may be advantageous for the rapid change in gene expression to acclimate to changing B conditions.
In conclusion, our study demonstrated the importance of NIP5;1 transcript stability, controlled by a conserved sequence in the 5′ UTR, in a B-dependent manner. Our study provides further understanding of B transport wherein NIP5;1 is downregulated through controlled mRNA decay mediated by 5′ UTR sequences under high-B conditions to avoid excessive B transport.
METHODS
Plant Materials
The Col-0 ecotype of Arabidopsis thaliana was derived from our laboratory stocks. Information on the T-DNA insertion lines (nip5;1-1) was obtained from the Salk Institute Genomic Analysis Laboratory (SIGnAL) database (Alonso et al., 2003), and the seeds were obtained from the ABRC. The genotype of plants was determined by PCR using left border T-DNA–specific primers and gene-specific primers (Takano et al., 2006).
Plant Growth Conditions
For phenotypic analysis, GUS activity, confocal imaging experiments, mRNA analysis, immunoblot analysis, and B concentration measurement, plants were grown on solid medium (Fujiwara et al., 1992) containing 1% (w/v) Suc and 1.5% (w/v) Gellan Gum (Wako Pure Chemical) with different concentrations of boric acid (Wako Pure Chemicals) and with identical pH. Surface-sterilized seeds were sown on the plates and incubated for 1 to 2 d at 4°C, and the plates were placed vertically at 22°C in a growth chamber under long-day conditions (16-h/8-h light/dark cycle). For the mRNA half-life experiment in Col-0, plants were grown on solid medium containing 1% (w/v) Suc and 0.15% (w/v) Gellan Gum containing 0.3 μM B and then transferred to hydroponic culture containing 0.3 μM B for 2 d at 22°C in a growth chamber under short-day conditions (10-h/14-h light/dark cycle). For the mRNA half-life experiment in transgenic plants, plants were grown on solid medium containing 1% (w/v) Suc and 1.5% (w/v) Gellan Gum containing 0.3 μM B at 22°C in a growth chamber under long-day conditions.
Quantification of Transcript Accumulation by qRT-PCR
Total RNA was extracted using an RNeasy plant mini kit (Qiagen) as recommended by the manufacturer. Total RNA (500 ng) was reverse transcribed into cDNA in a 20-μL reaction using an ExScript RT reagent kit (Takara Bio) with oligo-d(T)16 primers. The cDNA was amplified by PCR in a Thermal Cycler Dice Real-Time System TP800 (Takara Bio) with a SYBR Premix Ex Taq kit (Takara Bio). The eukaryotic elongation factor 1-α (eEF1-α) gene was used as a control for quantification. The primers used in qRT-PCR are shown in Supplemental Table 2 online. Specific amplification of target genes was confirmed by electrophoresis of PCR reactions and by melting curve analysis of PCR products using the Thermal Cycler Dice instrument.
Plasmid Construction and Plant Transformation
The NIP5;1 promoter was serially deleted and fused to the GUS reporter gene in the binary vector pTkan+ (kindly provided by K. Schumacher, University of Heidelberg). The promoter and 5′ UTR (from –2180 to +312 bp) of the NIP5;1 gene was previously isolated (Takano et al., 2006). Briefly, the region from –2180 to + 312 bp of the NIP5;1 gene fused to the GUS reporter gene was subcloned into the pUC18 vector (named pMW1). The nucleotide number corresponding to that from the transcription start site and the 5′ end of the full-length cDNA (TAIR database) was used as the predicted transcript start site (+1). Using the pMW1 construct as a template, the 5′-upstream regions of the NIP5;1 gene, starting from the –1248, –589,–508, –389, and –289 bp to +312 bp and –2180 to +1 bp, were amplified by PCR. The primers used in plasmid construction are shown in Supplemental Table 2 online. The amplified fragments were digested using a BamHI site at the 5′ end of the primers (underlined) and a NotI site at the 3′ end of the GUS gene (underlined). The BamHI-NotI fragment of the resulting plasmids contains deleted promoter fragments fused to the GUS gene. The fragments were then subcloned into the BamHI-Bsp120-I vector fragment from the binary vector pTkan+.
To construct P-2,180 ΔUTR312-GFP-NIP5;1, the NIP5;1 promoter region from –2180 to –1 bp was amplified by PCR from pMW1. The amplified fragments were digested using a BamHI site at the 5′ end of the primers and an NcoI site at the 3′ end of the primers. Using BamHI and NcoI, the amplified fragment was subcloned into the BamHI-NcoI vector fragment from the binary vector P-2,180 UTR312-GFP-NIP5;1 (Takano et al., 2010).
A plasmid carrying a truncated 5′ UTR fused to the GUS reporter gene under the control of P35S was constructed as described below. The truncated 5′ UTR, the 5′-upstream regions of the 5′ UTR, starting from –100, +7, +101, and +198 to +312 and +7 to +79, +7 to +183, and +7 Δ+182 to +200 bp was amplified from pMW1 by PCR using primers (see Supplemental Table 2 online). Accuracy of the cloned PCR product was confirmed by DNA sequencing. The amplified fragment was subcloned into the pENTR/D-TOPO vector via the TOPO cloning reaction (Gateway technology; Invitrogen). The cloned promoter fragment was subsequently subcloned into pMDC140 (Curtis and Grossniklaus, 2003), which contains the dual P35S, gusA, and terminator of the gene for nopaline synthase (Nos T) using the Gateway system according to the manufacturer’s instructions (Invitrogen).
Arabidopsis Col-0 and nip5;1-1 plants were used for plant transformation using the floral dip method (Clough and Bent, 1998). Homozygous T3 plants were established for each transgenic line and used for analysis.
Fluorometric Assays of GUS Activity
T3 (homozygous) transgenic lines carrying the P-2,180 UTR312-GUS construct were grown with solid medium containing 0.1, 0.3, 1, 3, 10, 30, 100, 300, 1000, and 3000 μM B for 14 d to observe the NIP5;1 expression pattern by GUS assay. T3 (homozygous) transgenic lines carrying the promoter deletion series constructs were grown with solid medium containing 0.3 or 100 μM B for 7 or 11 d to observe the NIP5;1 expression pattern by GUS assay. T3 (homozygous) transgenic lines carrying the 5′ UTR deletion series constructs were grown on solid medium containing 0.3 or 1000 μM B for 11 d to observe the NIP5;1 expression pattern by GUS assay. Quantitative GUS assays were conducted as described by Jefferson et al. (1987). Five to ten transgenic seedlings were collected as one sample and used for the GUS assay. The seedlings were homogenized in 300 μL of extraction buffer (50 mM sodium-phosphate, pH 7.0, 10 mM Na2EDTA, 0.1% Na-sarcosyl, and 0.1% Triton X-100). The samples were centrifuged for 10 min at 4°C, and the supernatant was used for subsequent analysis. The enzyme activity was measured fluorometrically using 4-methylumbelliferyl-β-d-glucuronide as the substrate, and the reaction product 4-methylumbelliferone was fluorometrically detected. The protein concentration of extracts was measured using a Bio-Rad protein assay kit (Bio-Rad Laboratories). GUS activity was shown in units of micromole 4-methylumbelliferone produced per hour per microgram of protein.
Measurement of B Concentration
Plants were grown on solid medium containing 3000 μM B for 14 d. Roots and rosette leaves were harvested, and B concentrations of these samples were determined using inductively coupled plasma–mass spectrometry (SPQ-9000; Seiko Instruments) as previously described (Takano et al., 2006).
Half-Life Measurement of mRNA
mRNA half-lives were determined according to Seeley et al. (1992) with a few modifications. The growth conditions for hydroponic culture in Col-0 plants were described previously (Takano et al., 2005). Briefly, seeds were sown on nylon mesh (10 × 10 cm2 and 300-μm pore size) on solid medium (Fujiwara et al., 1992) containing 1% (w/v) Suc and 0.15% (w/v) Gellan Gum for 21 d under short-day conditions. Plants were then transferred to liquid medium supplied with 0.3 μM B and grown for 2 d. For transgenic plants, seeds were sown on solid medium containing 1% (w/v) Suc and 1.5% (w/v) Gellan Gum for 14 d under long-day conditions. Col-0 and transgenic plants were preincubated with hydroponic culture solution (Fujiwara et al., 1992) containing 0.3 or 100 μM B for 10 min. After preincubation, cordycepin (Funakoshi) was added to a final concentration of 0.6 mM (0 min) into the hydroponic medium, and vacuum infiltration was immediately applied for 45 s. Root samples in Col-0 and whole-plant samples in transgenic plants were harvested at 0, 10, or 30 and 20 min, respectively, and frozen in liquid nitrogen. In this study, cordycepin was directly added to the hydroponic medium supplemented with 0.3 or 100 μM B instead of using incubation buffer (Seeley et al., 1992), and total RNA was isolated and analyzed by qRT-PCR. The RNA remaining at each time point was determined relative to the amount at the zero time point and plotted on a log scale. To test whether cordycepin conferred a strong block to transcription in plants grown under hydroponic culture, NIA2 (At1g37130) encoding nitrate reductase identified as an unstable transcript was used as a control for cordycepin activity and measured mRNA accumulation (Gutierrez et al., 2002) (see Supplemental Figure 4 online). Similar results were observed and the mRNA levels rapidly dropped in hydroponic culture supplied with 0.3 μM B. We thus used this condition in further experiments.
Expression of GFP-Tagged NIP5;1 in Arabidopsis Roots
The transgenic plants carrying P-2,180 UTR312-GFP-NIP5;1 and P-2,180 ΔUTR312-GFP-NIP5;1 were grown on vertically placed solid medium containing 0.3, 1, 100, or 1000 μM B for 14 d. Confocal laser scanning microscopy was performed using an LSM 510 (Carl Zeiss) with an excitation wavelength of 488 nm and a detection wavelength of 515 to 545 nm for GFP imaging in the root differentiation zone.
The relative expression of NIP5;1 in transgenic plants carrying P-2,180 UTR312-GFP-NIP5;1 was assayed by immunoblot analyses of microsomal proteins with an anti-GFP antibody as described previously (Takano et al., 2005). The membrane was then stripped and reprobed with plasma membrane marker and plasma membrane aquaporin antibody (Ohshima et al., 2001) as loading control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GeneBank/EMBL data libraries under the following accession numbers: At NIP5;1, At4g10380; At NIP6;1, At1g80760; NIA2, At1g37130; Os NIP3;1, Os10g36924; Zm NIP3;1, AF326486; and Vv 00000446001, CAO62847.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Comparison of the 5′ UTR of Arabidopsis NIP5;1 and Rice NIP3;1.
Supplemental Figure 2. Sequence of the NIP6;1 5′ UTR.
Supplemental Figure 3. NIP5;1-GFP Protein and NIP5;1-GFP mRNA Accumulation under Low- and High-B Conditions.
Supplemental Figure 4. Time-Course Analysis of mRNA Degradation of NIA2 in Col-0 Plants.
Supplemental Table 1. List of Construct Names.
Supplemental Table 2. List of Primers.
Acknowledgments
The T-DNA insertion lines were kindly provided by the ABRC at Ohio State University. Plasma membrane aquaporin antibody was kindly provided by M. Maeshima (Nagoya University). We thank Y. Kawara and K. Aizawa for excellent technical assistance and K. Miwa (Hokkaido University) for inspiring discussion. This work is supported in part by a Grant-in-Aid for Scientific Research S (to T.F.) and a Grant-in-Aid for Scientific Research on Innovative Areas (to T.F.) from the Ministry of Education, Culture, Sports, Science and Technology and from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation Grant IPG-0005 (to T.F.).
AUTHOR CONTRIBUTIONS
M.T., J.T., Y.C., H.O., S.N., and T.F. designed the research. M.T. and Y.O. performed the research. M.T., J.T., Y.C., F.L., H.O., S.N., and T.F. analyzed data. M.T., J.T., S.N., and T.F. wrote the article.
References
- Abler M.L., Green P.J. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32: 63–78 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson M.B., Folta K., Warpeha K.M., Gibbons J., Gao J., Kaufman L.S. (1999). Blue light-directed destabilization of the pea Lhcb1*4 transcript depends on sequences within the 5′ untranslated region. Plant Cell 11: 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S., Tang L., Krueger A.D., Smith C.L., Ford S.R., Dickey L.F., Petracek M.E. (2004). The Fed-1 (CAUU)4 element is a 5′ UTR dark-responsive mRNA instability element that functions independently of dark-induced polyribosome dissociation. Plant Mol. Biol. 56: 761–773 [DOI] [PubMed] [Google Scholar]
- Bonilla I., Garcia-González M., Mateo P. (1990). Boron requirement in cyanobacteria : its possible role in the early evolution of photosynthetic organisms. Plant Physiol. 94: 1554–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. (2001). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cromie M.J., Shi Y., Latifi T., Groisman E.A. (2006). An RNA sensor for intracellular Mg(2+). Cell 125: 71–84 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell B., Huang L. (1997). Physiological response of plants to low boron. Plant Soil 193: 103–120 [Google Scholar]
- Dordas C., Brown P.H. (2001). Evidence for channel mediated transport of boric acid in squash (Cucurbita pepo). Plant Soil 235: 95–103 [Google Scholar]
- Dordas C., Chrispeels M.J., Brown P.H. (2000). Permeability and channel-mediated transport of boric acid across membrane vesicles isolated from squash roots. Plant Physiol. 124: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhert C.D. (1998). Boron stimulates embryonic trout growth. J. Nutr. 128: 2488–2493 [DOI] [PubMed] [Google Scholar]
- Fort D.J., Propst T.L., Stover E.L., Strong P.L., Murray F.J. (1998). Adverse reproductive and developmental effects in Xenopus from insufficient boron. Biol. Trace Elem. Res. 66: 237–259 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P., Green P.J. (1996). Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: The 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 15: 1678–1686 [PMC free article] [PubMed] [Google Scholar]
- Goswami S., Tarapore R.S., Teslaa J.J., Grinblat Y., Setaluri V., Spiegelman V.S. (2010). MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J. Biol. Chem. 285: 20532–20540 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gutierrez R.A., Ewing R.M., Cherry J.M., Green P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Fujiwara T. (2007). Channel-mediated boron transport in rice. Plant Cell Physiol. 48: S227 [Google Scholar]
- Henkin T.M. (2008). Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes Dev. 22: 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X.J., Van de Cotte B., Van Montagu M., Verbruggen N. (2001). The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J. 26: 157–169 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., Karlsson M., Johansson I., Gustavsson S., Sjövall S., Fraysse L., Weig A.R., Kjellbom P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue L., Trollinger D.R., Strong P.L., Keen C.L. (2000). Functional impairments in preimplantation mouse embryos following boron deficiency. FASEB J. 14A: 539 [Google Scholar]
- Linz B., Koloteva N., Vasilescu S., McCarthy J.E. (1997). Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272: 9131–9140 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants, 2nd ed. (San Diego, CA: Academic Press). [Google Scholar]
- Maurel C. (2007). Plant aquaporins: Novel functions and regulation properties. FEBS Lett. 581: 2227–2236 [DOI] [PubMed] [Google Scholar]
- Miwa K., Takano J., Omori H., Seki M., Shinozaki K., Fujiwara T. (2007). Plants tolerant of high boron levels. Science 318: 1417. [DOI] [PubMed] [Google Scholar]
- Nielsen F.H. (2000). The emergence of boron as nutritionally important throughout the life cycle. Nutrition 16: 512–514 [DOI] [PubMed] [Google Scholar]
- Nyikó T., Sonkoly B., Mérai Z., Benkovics A.H., Silhavy D. (2009). Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71: 367–378 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Taylor C.B., Newman T.C., Green P.J. (1993). The effect of sequences with high AU content on mRNA stability in tobacco. Proc. Natl. Acad. Sci. USA 90: 11811–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Iwasaki I., Suga S., Murakami M., Inoue K., Maeshima M. (2001). Low aquaporin content and low osmotic water permeability of the plasma and vacuolar membranes of a CAM plant Graptopetalum paraguayense: Comparison with radish. Plant Cell Physiol. 42: 1119–1129 [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Eberhard S., Albersheim P., Darvill A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. (2004). Rhamnogalacturonan II: Structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55: 109–139 [DOI] [PubMed] [Google Scholar]
- Pérez-Amador M.A., Lidder P., Johnson M.A., Landgraf J., Wisman E., Green P.J. (2001). New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13: 2703–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe R.I., Bouzan C., Nabili S., Eckhert C.D. (1998). The response of trout and zebrafish embryos to low and high boron concentrations is U-shaped. Biol. Trace Elem. Res. 66: 261–270 [DOI] [PubMed] [Google Scholar]
- Rowe R.I., Eckhert C.D. (1999). Boron is required for zebrafish embryogenesis. J. Exp. Biol. 202: 1649–1654 [DOI] [PubMed] [Google Scholar]
- Sarkar B., Xi Q., He C., Schneider R.J. (2003). Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23: 6685–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley K.A., Byrne D.H., Colbert J.T. (1992). Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell 4: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrocks V.M. (1997). The occurrence and correction of boron deficiency. Plant Soil 193: 121–148 [Google Scholar]
- Smyth D.A., Dugger W.M. (1981). Cellular changes during boron deficient culture of the diatom Cylindrotheca fusiformis. Plant Physiol. 51: 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springsteen G., Wang B. (2002). A detailed examination of boronic acid-diol complexation. Tetrahedron 58: 5291–5300 [Google Scholar]
- Stangoulis J.C.R., Reid R.J., Brown P.H., Graham R.D. (2001). Kinetic analysis of boron transport in Chara. Planta 213: 142–146 [DOI] [PubMed] [Google Scholar]
- Sudarsan N., Barrick J.E., Breaker R.R. (2003). Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Noguchi K., Yasumori M., Kobayashi M., Gajdos Z., Miwa K., Hayashi H., Yoneyama T., Fujiwara T. (2002). Arabidopsis boron transporter for xylem loading. Nature 420: 337–340 [DOI] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., von Wirén N., Fujiwara T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18: 1498–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Fujiwara T. (2008). Physiological roles and transport mechanisms of boron: Perspectives from plants. Pflugers Arch. 456: 671–677 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Wallace I.S., Takano J., Roberts D.M., Fujiwara T. (2008). NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20: 2860–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman S.D., Niemietz C.M., Bramley H. (2002). Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25: 173–194 [DOI] [PubMed] [Google Scholar]
- Vilela C., Ramirez C.V., Linz B., Rodrigues-Pousada C., McCarthy J.E. (1999). Post-termination ribosome interactions with the 5’UTR modulate yeast mRNA stability. EMBO J. 18: 3139–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warington K. (1923). The effect of boric acid and borax on the broad bean and certain other. Ann. Bot. (Lond.) 27: 629–672 [Google Scholar]
- Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. (2004). Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286 [DOI] [PubMed] [Google Scholar]
- Woods W.G. (1996). Review of possible boron speciation relating to its essentiality. J. Trace Elem. Exp. Med. 9: 153–163 [Google Scholar]
- Yau S.K., Nachit M.M., Hamblin J., Ryan J. (1995). Phenotypic variation in boron-toxicity tolerance at seedling stage in durum wheat (Triticum durum). Euphytica 83: 185–191 [Google Scholar]
- Zardoya R. (2005). Phylogeny and evolution of the major intrinsic protein family. Biol. Cell 97: 397–414 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., Wang T., Ling Y., Su Z. (2010). PMRD: Plant microRNA database. Nucleic Acids Res. 38(Database issue): D806–D813 [DOI] [PMC free article] [PubMed] [Google Scholar]