This study shows that the role of Polycomb Group (PcG) proteins in the regulatory network determining tissue-specific expression is not identical for all PcG-target genes. The data suggest that a positive regulatory factor produced in differentiated phloem companion cells sets a prerequisite for FT expression.
Abstract
The Polycomb Group (PcG) pathway represses transcription through a mechanism conserved among plants and animals. PcG-mediated repression can determine spatial territories of gene expression, but it remains unclear whether PcG-mediated repression is a regulatory requirement for all targets. Here, we show the role of PcG proteins in the spatial regulation of FLOWERING LOCUS T (FT), a main activator of flowering in Arabidopsis thaliana exclusively expressed in the vasculature. Strikingly, the loss of PcG repression causes down-regulation of FT. In addition, our results show how the effect of PcG-mediated regulation differs for target genes and that, for FT expression, it relies primarily on tissue differentiation.
INTRODUCTION
Two main complexes are involved in the repression mediated by the Polycomb Group (PcG) pathway. Polycomb Repressive Complex2 (PRC2) tri-methylates lysine 27 of histone 3 (H3K27me3) and recruits PRC1, which further contributes to the repression of target genes (Hennig and Derkacheva, 2009; Simon and Kingston, 2009). The expression of homeotic (Hox) genes is the paradigm of PcG regulation in animals. Hox genes are expressed only in specific regions of the animal body; however, in PcG mutants their expression extends to other regions, causing homeotic changes in body patterning (Simon and Kingston, 2009). In plants, the best known example of a Hox gene spatially regulated by PcG proteins is the Arabidopsis thaliana MADS box gene AGAMOUS (AG). In the wild-type plants, AG is expressed only in the flowers, whereas in PcG mutants, it is ectopically expressed in leaves and other parts of the plant, correlating with a decrease in H3K27me3 (Goodrich et al., 1997; Chanvivattana et al., 2004; Schubert et al., 2006; Calonje et al., 2008). H3K27me3 target genes in Arabidopsis often show marked tissue-specific expression patterns (Turck et al., 2007; Zhang et al., 2007). Two recent tissue-specific, genome-wide distribution studies of H3K27me3 have revealed that the H3K27me3 is mostly absent from targets in the tissues where these are expressed (Weinhofer et al., 2010; Lafos et al., 2011). On the other hand, changes in H3K27me3 levels are not always correlated with changes in transcription (Schubert et al., 2006; Schwartz and Pirrotta, 2007; Kwon et al., 2009; Adrian et al., 2010).
H3K27me3 is widely dispersed across the FLOWERING LOCUS T (FT) locus (Turck et al., 2007; Zhang et al., 2007), and the histone mark is proposed to delimit the accessibility of transcription factors to certain cis-elements at the FT promoter (Adrian et al., 2010). Chromatin-mediated repression is required to confer photoperiod-dependent control of FT expression (Goodrich et al., 1997; Takada and Goto, 2003). FT encodes a plant florigen and is induced by long-day conditions through the activator CONSTANS (CO) (Turck et al., 2008; Imaizumi, 2010). CO has been shown to bind DNA sequences specifically in vitro, and CO consensus binding sites within the proximal FT promoter are functional cis-elements required for FT expression (Adrian et al., 2010; Tiwari et al., 2010). However, these elements and the proximal promoter alone are not sufficient to drive expression of reporter genes in transgenic plants, but require the presence of enhancer elements that are located more than 4 kb from the transcription start (Adrian et al., 2010). One striking aspect of FT transcriptional regulation is its spatial pattern: prior to flowering, FT is expressed only in the phloem companion cells of the cotyledons and the distal blade of rosette leaves (Takada and Goto, 2003; Yamaguchi et al., 2005; Adrian et al., 2010); after flowering, the gene is also expressed in other organs (cauline leaves, sepals, and fruits) but is still strongly limited to the vascular tissue (Adrian et al., 2010). FT expression is limited to the vasculature even if CO is provided ectopically (Yamaguchi et al., 2005; Adrian et al., 2010). In plants carrying mutations in LIKE HETEROCHROMATIN PROTEIN1 (LHP1), a FT expression pattern similar to that of CO-overexpressing plants is observed (Takada and Goto, 2003). LHP1 directly represses FT as part of a plant PRC1, but in lhp1 mutants the PcG pathway is only partially affected, and the level of H3K27me3 at FT does not change (Turck et al., 2007; Farrona et al., 2008); therefore, the analysis of this mutant is not sufficient to understand the role of PcG proteins in FT regulation. As neither CO nor LHP1 are responsible for the characteristic tissue-specific expression pattern of FT, other regulatory components must be involved.
Here we investigate whether the H3K27me3 mark is required for the tissue-specific expression pattern of FT as part of a PcG-mediated repression mechanism that is independent of LHP1. We show that the loss of PcG repression differentially affects targets, and that the domain of FT expression depends on tissue differentiation, even in the absence of a PcG-mediated chromatin configuration.
RESULTS
FT Expression Is Strongly Reduced in the swn-7 clf-28 PcG Mutant
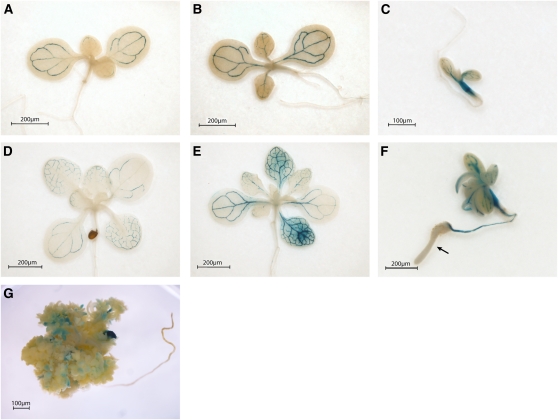
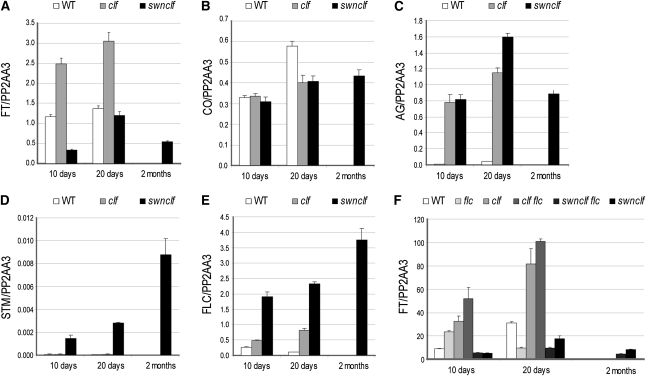
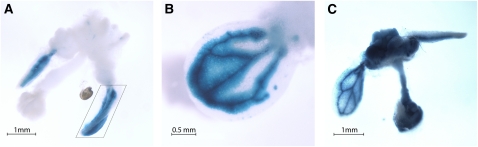
To analyze PcG-mediated FT spatial expression, a transgene carrying an 8.1-kb FT promoter fragment fused to the β-Glucuronidase gene (FTprom:GUS) (Adrian et al., 2010) was introduced into the single clf-28 (clf) and double swn-7 clf-28 (swn clf) PcG mutants (Figure 1). CURLY LEAF (CLF) and SWINGER (SWN) are part of the histone methyltransferase SET family responsible for H3K27me3 deposition. CLF and SWN show partially overlapping functions and are involved in FT repression (Jiang et al., 2008; Liu et al., 2010). In the wild-type plants, the FTprom:GUS signal was obtained only at the vascular tissue of cotyledons and leaves, as expected (Figures 1A and 1D) (Takada and Goto, 2003; Adrian et al., 2010). In clf mutant plants, the signal was stronger (Figures 1B and 1E), which supported the quantitative RT-PCR (qRT-PCR) data (Figure 2A) (Barrero et al., 2007; Jiang et al., 2008), but it was still limited to the vascular tissue. Mutant seeds of swn clf germinate, giving rise to a small seedling that degenerates to a callus-/embryo-like structure (Chanvivattana et al., 2004). In swn clf seedlings, the FT signal was observed at the veins of cotyledons and in leaf-like structures (Figures 1C and 1F). In addition, the roots are also stained, and a strong, diffused signal is observed at the hypocotyls. Strikingly, as the tip of the root develops a swollen and opaque embryo-like tissue, known as a pickle (pkl)-like root phenotype (Ogas et al., 1997), FT promoter-driven signal completely disappears (Figure 1F). In 2-month-old swn clf callus, the GUS signal strongly decreased and was obtained only in a punctate pattern (Figure 1G). Expression analysis confirmed a reduction in FT in swn clf (Figure 2A) despite a maintained expression of CO (Figure 2B). By contrast, AG is strongly expressed in clf and swn clf mutants, indicating that its regulation depends mostly on CLF; whereas STM is up-regulated only in the swn clf background and, therefore, is more dependent on SWN (Figures 2C and 2D) (Schubert et al., 2006).
Figure 1.
FT Spatial Expression Changes in the Wild Type, clf, and swn clf.
(A) to (G) Histochemical localization of GUS activity in 10- and 20-day-old wild-type, clf, and swn clf seedlings and 2-month-old swn clf callus carrying an FTprom:GUS construct. Ten-day-old wild type (A), clf (B), and swn clf (C); 20-d-old wild type (D), clf (E), and swn clf (F); 2-month-old swn clf (G). Arrow in (F) indicates the loss of GUS signal in the pkl-like root.
Figure 2.
Expression Analysis in the Wild Type, clf, and swn clf.
(A) to (E) qRT-PCRs in 10- and 20-d-old wild type (WT), clf, and swn clf and 2-month-old swn clf callus. qRT-PCRs of FT (A), CO (B), AG (C), STM (D), and FLC (E).
(F) qRT-PCRs of FT in 10- and 20-d-old wild type, flc, clf, clf flc, swn clf flc, and swn clf and 2-month-old swn clf flc and swn clf callus.
Error bars represent se of the mean based on three technical replicates. Similar results were obtained in at least two independent experiments. A biological replicate of the analysis is included as Supplemental Figure 6 online.
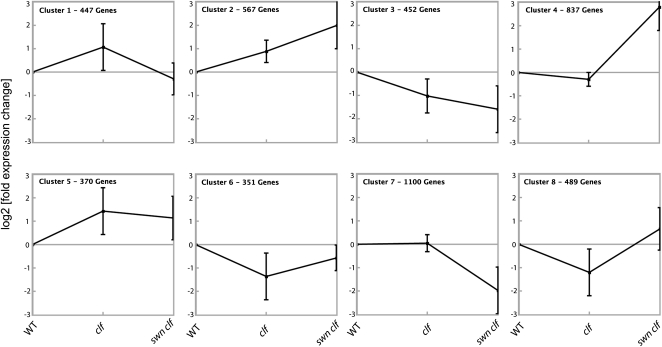
To further characterize the expression pattern of other PcG targets and to investigate whether other genes mimic FT expression in PcG mutants, the expression of H3K27me3-enriched genes in the wild type, clf, and swn clf was analyzed by expression microarray analysis and clustered according to the different patterns of expression. Although an important percentage of PcG targets was strongly up-regulated in swn clf plants, the clustering showed that FT, which is placed in cluster 1, is not an exception among PcG target genes, because 9.7% of all H3K27me3-enriched genes follow a similar pattern of expression. In addition, other clusters showed only slight expression changes between the wild-type and mutant plants (Figure 3; see Supplemental Figure 1 and Supplemental Data Set 1 online).
Figure 3.
K-Means Clustering of Expression Data of H3K27me3-Enriched Genes in PcG Mutant Seedlings.
The expression data of the H3K27me3-enriched set of genes (n = 4599) were analyzed by K-means clustering (k = 8) using the software Genesis. Expression values for each gene were calculated as fold change compared with the wild type (WT). Clustering was performed on log2-transformed fold-change values. The Pearson correlation coefficient was used as a distance measurement to cluster according to patterns rather than absolute values. Each graph represents one cluster. Squares represent the average expression, and bars indicate the normalized sd. FT is located in Cluster 1.
In summary, despite the general role of PcG in establishing a repressive chromatin configuration and thereby participating in tissue-specific gene regulation, loss of PcG repression is not synonymous with transcriptional activation, and no general rule predicts how expression might change in PcG mutants.
FLC Overexpression Is Not the Cause of FT Repression in the swn clf Mutant
The MADS box transcription factors FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) directly repress FT (Li et al., 2008), and the expression of FLC itself is regulated by CLF and SWN as part of different PRC2 complexes (Hennig and Derkacheva, 2009; Liu et al., 2010). Like FLC, the SVP locus is covered by the H3K27me3 mark, indicating repression by PcG proteins, although such regulation has not been described (Zhang et al., 2007). FLC was up-regulated in clf plants compared with the wild-type plants (Figure 2E) (Jiang et al., 2008) and was further increased in swn clf plants, indicating that FLC repression depends on both PcG proteins (Figure 2E). By contrast, SVP is not up-regulated in clf or swn clf plants (see Supplemental Figure 2A online). AP2-like proteins TEMPRANILLO1 (TEM1), TEM2, and SCHLAFMÜTZE (SMZ) repress FT expression (Imaizumi, 2010) but are not marked by H3K27me3 and are down-regulated in swn clf (see Supplemental Figures 2B to 2D online). Therefore, FLC, but neither SVP nor TEM1 and 2 or SMZ, might cause FT down-regulation in swn clf. To investigate the role of FLC, FT expression was analyzed in the triple mutant swn-7 clf-28 flc-3 (swn clf flc). Although loss of FLC caused an increase in FT expression in the clf mutant, no up-regulation was observed in the swn clf background (Figure 2F), demonstrating that FLC is not involved in FT down-regulation in this genetic background.
Chromatin Changes at FT Are Not Sufficient to Explain Its Down-Regulation
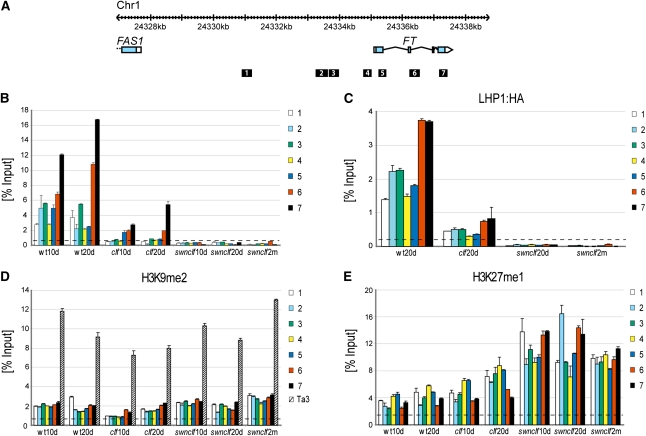
FT expression data obtained in clf and swn clf mutants were correlated to changes in FT chromatin (Figure 4A). It has been reported that H3K27me3 is absent at FT in clf (Jiang et al., 2008); however, our ChIP data show that this loss is not homogeneous for the whole FT locus, because significant enrichment of H3K27me3 can still be detected at the 3′ region. In swn clf plants, the H3K27me3 signal completely disappeared (Figure 4B). LHP1 binding to FT followed almost exactly the same pattern as H3K27me3 in the single and double mutants (Figure 4C). Similar results were obtained for the AG locus (see Supplemental Figure 3A online).
Figure 4.
Chromatin Changes at FT in the Wild Type, clf, and swn clf.
(A) Genome browser view of the FT locus at chromosome 1. Exons of FT and upstream gene FAS1 are represented as blue boxes, and untranslated regions are represented as white boxes. Positions of amplicons used in the ChIP analysis are presented as black boxes and are numbered.
(B) H3K27me3 at FT locus. ChIP experiments were performed with chromatin from 10-d-old (10d) and 20-d-old (20d) wild type (wt), clf, and swn clf and 2-month-old (2m) swn clf plants. Signals detected along the FT locus were normalized against the input.
(C) Binding of LHP1 at FT locus. ChIP experiments were performed with chromatin from 20-d-old (20d) wild type, clf, and swn clf and 2-month-old (2m) swn clf plants carrying a 35Sprom:LHP1:HA transgene. Normalization as in (B).
(D) H3K9me2 at FT locus and the Ta3 retrotransposon. Data are based on the same chromatin extract and analysis as used in (B).
(E) H3K27me1 at FT locus. Data are based on the same chromatin extract and analysis as used in (B).
Error bars represent se of the mean based on three technical replicates. Dashed lines represent background calculated as average of qPCR signals obtained from each sample with Ta3 primers ([B] and [C]) or RBCS1A primers ([D] and [E]), as described in Methods. A biological replicate of the analysis is included as Supplemental Figure 7 online.
Loss of H3K27me3 and LHP1 promotes a more permissive chromatin structure, which cannot explain the down-regulation of FT observed in swn clf. Therefore, we analyzed whether, in the absence of H3K27me3, the repressive histone marks H3K9me2 and H3K27me1 were recruited to FT. H3K9me2 ChIP data at FT showed some increase of this mark over the FT locus only in swn clf plants. However, the H3K9me2 increase at FT seems nonsignificant when compared with levels at the Ta3 retrotransposon, although no studies have shown threshold levels for effective H3K9me2-mediated repression (Figure 4D). For H3K27me1, the ChIP data showed an inverse correlation between changes in this mark and H3K27me3 changes at FT (Figures 4B and 4E). An increase in H3K27me1 was also observed at AG, despite the strong up-regulation of this gene in swn clf (see Supplemental Figure 3B online).
H3K4me3 has been shown to act as a bivalent mark together with H3K27me3 at the FT and FLC loci (Jiang et al., 2008; Carles and Fletcher, 2009). Increased H3K4me3 levels were reported in the 5′ region of FT and AG in the clf mutant (Carles and Fletcher, 2009). Our ChIP experiments did not show any significant changes for H3K4me3 in the clf background either at FT or AG (see Supplemental Figures 3C and 3D online). An increase for this mark was observed in swn clf plants, despite the repression of FT (see Supplemental Figure 3D online).
In conclusion, despite their divergent expression, FT and AG are subjected to similar changes in chromatin conformation in the PcG mutants. Therefore, chromatin changes are not sufficient to explain FT down-regulation in swn clf.
FT Expression Is Strongly Reduced upon Loss of Vascular Identity in Induced Callus
In swn clf plants, development of the vascular tissue is particularly affected. The characteristic reticulated pattern of the veins is lost in the leaf-like structures, and the linear pattern is interrupted in the roots (Figure 1G; see Supplemental Figure 4 online). To analyze the importance of vascular development in the down-regulation of FT in swn clf calli, the FTprom:GUS line in the wild-type background was germinated on callus-inducing medium. In hormone-induced callus, FT was expressed only in the cotyledons (Figure 5A), which are formed during embryogenesis and were therefore less affected by the hormone treatment. In these organs, FT was strongly expressed in the veins from where the signal diffused to the surrounding cells (Figure 5B). In the hormone-induced opaque embryo-like tissue that lacked differentiation of veins, the GUS signal disappeared, similar to the results obtained in pkl-like roots and callus of the swn clf mutant (Figures 1F, 1G and 5A). The loss of GUS signal in the hormone-induced callus occurred despite a high and ectopic up-regulation of CO (Figure 5C) and the presence of CO protein (see Supplemental Figure 5 online). Strong expression of CO in the cotyledons might cause high FTprom:GUS signal in these organs in the callus-inducing medium.
Figure 5.
FT Spatial Expression in Hormone-induced Callus.
(A) and (B) Histochemical localization of GUS activity in FTprom:GUS plants in the wild-type background grown for 10 d on GM medium supplemented with 4.5 μM 2,4-D and 0.45 μM kinetin.
(B) Detail of the adaxial surface of the cotyledon marked on (A) with a square.
(C) Histochemical localization of GUS activity in COprom:GUS plants in the wild-type background grown as in (A).
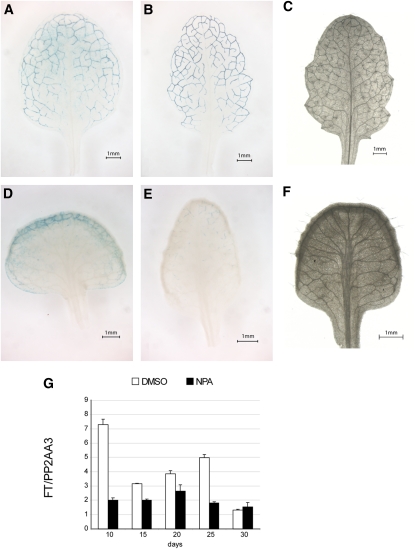
We next assessed whether an increase in the number of leaf veins could lead to higher expression of FT. 1-N-Naphtylphtalamic acid (NPA) affects vascular patterning by inhibiting auxin efflux (Wenzel et al., 2008). NPA treatment increases the number and width of vascular bundles but impairs vein differentiation, because the cells within the vascular strand are improperly aligned and the petiole vessels are not connected to those of the stem (Wenzel et al., 2008). FT expression decreased in NPA-treated plants, despite the apparent increase in vasculature (Figure 6).
Figure 6.
Expression Analysis of FT in the Presence of an Auxin Transporter Inhibitor.
(A) and (B) Histochemical localization of GUS activity in leaves of 25-d-old 8.1kbFTprom:GUS plants grown on GM plates supplemented with DMSO.
(C) Vasculature organization of a 25-d-old leaf grown on GM plates supplemented with DMSO.
(D) and (E) Histochemical localization of GUS activity in leaves of 25-d-old 8.1kbFTprom:GUS plants grown on GM plates supplemented with NPA.
(F) Vasculature organization of a 25-d-old leaf grown on GM plates supplemented with NPA.
(G) qRT-PCRs in plants grown on GM plates supplemented with DMSO or NPA. Data were normalized to PP2AA3. Error bars represent se of the mean based on three technical replicates.
Taken together, the results confirm the necessity of proper vascular development and differentiation to establish the domain of FT expression.
DISCUSSION
FT Expression Domain Is Reduced in Strong PcG Mutants
Previous studies have shown the requirement for PcG repression to define territories of expression of different target genes in animals and plants. Consequently, in PcG mutants the expression of such target genes extends to other domains (Goodrich et al., 1997; Chanvivattana et al., 2004; Schubert et al., 2006; Calonje et al., 2008; Simon and Kingston, 2009; Bratzel et al., 2010). We have focused on FT, a PcG target with a well-known and defined spatial pattern, to characterize this regulation further. Although in clf mutants, FT is up-regulated (Figure 2A) (Barrero et al., 2007; Jiang et al., 2008), our results showed that FT expression in clf is confined to the vascular tissue, its normal domain of expression. However, in calli of the strong PcG double mutant swn clf, FT domain of expression is not ectopically extended, but rather is strongly reduced to a small group of cells. Clustering analysis of PcG targets divided them in eight different clusters according to their pattern of expression in the wild-type, clf, and swn clf seedlings. Cluster 2, Cluster 4, Cluster 5, and Cluster 8, with almost 50% of the targets, include genes with a very strong to moderate up-regulation in swn clf plants, as expected after the loss of a general repressive pathway. Some targets differentially depend on CLF and SWN (Figure 2) (Schubert et al., 2006). Indeed, Cluster 5 includes genes that depend mainly on CLF, and genes in Cluster 2 depend on both proteins, whereas for genes in Cluster 4, the absence of SWN has a strong effect. Strikingly, ~10% of PcG targets show similar regulation as FT, with high expression in clf and low expression in swn clf (Cluster 1). The dependency of FT on signals lost in the de-differentiated swn clf callus may therefore be a model for the regulation of many PcG targets.
FT down-regulation in swn clf could be due to the up-regulation of a repressor. FT plays a central role in flowering and, therefore, is tightly regulated by different flowering pathways (Turck et al., 2008; Imaizumi, 2010). TEM1 and TEM2 are partially overlapping AP2-like transcription factors that repress FT directly by binding to the region encoding the 5′ untranslated region (Castillejo and Pelaz, 2008). SMZ, which belongs to a different AP2 clade (Aukerman and Sakai, 2003), also represses FT expression but interacts with a region several kilobases downstream of the coding region (Mathieu et al., 2009). The MADS box factors FLC and SVP form a complex that represses FT transcription, interacting with CArG cis-elements located in the first intron and FT proximal promoter region (Searle et al., 2006; Li et al., 2008). TEM1, TEM2, and SMZ are not PcG targets, and the microarray analysis showed that these genes are not up-regulated, but rather are down-regulated in swn clf plants. Therefore, we discarded them as the potential cause for FT down-regulation in swn clf plants. SVP is enriched in H3K27me3 but belongs to Cluster 3, which includes genes that are down-regulated in clf and swn clf plants. The results were confirmed by qRT-PCR, indicating that SVP is probably not involved in FT down-regulation in swn clf mutants.
FLC is the best characterized PcG target in Arabidopsis. Different PcG complexes have been shown to regulate FLC. The VRN-PRC2 complex participates in the vernalization pathway that stably represses FLC after a prolonged exposure to cold temperatures, whereas the EMF-PRC2 complex seems to regulate FLC independently of vernalization (Hennig and Derkacheva, 2009). CLF and SWN are components of both complexes (Hennig and Derkacheva, 2009; Liu et al., 2010). The expression analysis indicated an additive effect of both proteins in FLC regulation, and, therefore, FLC was a candidate for FT down-regulation in swn clf. However, the genetic data concluded that FLC is not the factor responsible for the decrease in FT expression in the strong PcG mutants. Our results do not exclude that other putative repressors could down-regulate FT in swn clf plants, but the known direct repressors do not have a role in FT down-regulation in the absence of PcG regulation.
Chromatin Changes Are Not Key Factors in Determining FT Spatial Expression
The PcG pathway modifies chromatin conformation, which is believed to repress transcription by interfering with the binding of transcription factors to cis-elements, affecting the recruitment of the RNA polymerase and blocking transcriptional initiation and/or elongation (Simon and Kingston, 2009). However, transcription also feeds back on chromatin conformation, and it is not yet fully resolved under which conditions the chromatin plays a determining regulatory role in the regulation of individual target genes (Buzas et al., 2011). Therefore, we investigated whether changes in FT and AG chromatin correlated with the transcriptional changes observed in swn clf plants.
We observed a reduction of H3K27me3 in clf at both loci and a complete loss in swn clf, pointing out that a SWN-PRC2 complex is also involved in FT methylation. LHP1, as part of a PRC1, recognizes and binds to its targets by direct interaction with the H3K27me3 mark (Exner et al., 2009). It has been suggested that H3K27me3 contributes to, but is not fully responsible for, PRC1 targeting (Simon and Kingston, 2009). Our data showed that LHP1 binding mirrors H3K27me3 occurrence, and, therefore, that at FT and AG, H3K27me3 acts as obligatory docking site for LHP1.
In the absence of the PcG signatures H3K27me3 and LHP1, other repressive marks could be recruited and spread over the FT locus. In Arabidopsis, H3K9me2 is a repressive mark characteristic of heterochromatic regions that is excluded from H3K27me3-enriched regions (Turck et al., 2007). Although the minimum level of H3K9me2 required to promote down-regulation is unknown, the slight increase in this mark observed at FT in swn clf mutants seems nonsignificant when compared with the H3K9me2 levels at the Ta3 retrotransposon. Consequently, we do not favor a role for H3K9me2 in FT down-regulation. H3K27me1 is also considered as a repressive histone mark and is believed to be the precursor for H3K27me3 (Campos and Reinberg, 2009). In Arabidopsis, a decrease in H3K27me3 does not affect nuclear H3K27me1 levels (Lafos et al., 2011). We did observe local increase in H3K27me1, but, considering that similar changes in H3K27me1 were observed at FT and AG (Figure 4; see Supplemental Figure 3 online), this mark neither predicts nor follows the transcriptional state in swn clf plants. Because H3K27me1 reflects the reduced activity of PRCs, it might also be a PRC2 substrate in Arabidopsis.
H3K4me3 is part of the active Trithorax Group pathway, which antagonizes PcG regulation (Köhler and Hennig, 2010). In animal stem cells, active and repressive chromatin marks are present at the same developmental loci and poise their targets for activation or repression during differentiation (Fisher and Fisher, 2011). In Arabidopsis, bivalent chromatin was demonstrated for FT and FLC (Jiang et al., 2008), and a role of bivalent chromatin in fine-tuning FLC and FT expression was suggested (Jiang et al., 2008; Carles and Fletcher, 2009; Jeong et al., 2009). However, genome-wide data in Arabidopsis did not show a general association between H3K4me3 and H3K27me3 (Ha et al., 2011). Despite previous results (Jiang et al., 2008; Carles and Fletcher, 2009; Jeong et al., 2009), our ChIP experiments did not confirm any significant change for H3K4me3 in the clf background either at FT or AG. An increase for this mark was observed in swn clf plants, despite the repression of FT. Lafos et al. have recently shown that H3K4me3 levels increase globally in swn clf calli, which may explain why higher levels were detected at the FT locus (Lafos et al., 2011). In comparison to H3K27 methylation marks, the levels of H3K4me3 at FT and AG were very low (see Supplemental Figure 3 online), which could explain why the number of genes that are detected as positive for the H3K4me3 mark genome-wide does not change in mutant seedlings that have lost PRC2 function (Bouyer et al., 2011).
Tissue Differentiation Bypasses Lack of PcG Repression
Considering that chromatin changes are not sufficient to explain FT down-regulation in strong PcG mutants and that vascular tissue is the normal domain of FT expression, its alteration could be the basis for FT down-regulation in swn clf callus. Auxin is key to the development of the vascular tissue, and important genes involved in the biosynthesis, transportation, perception, and signaling of this hormone are among PcG targets (Notaguchi et al., 2008; Lafos et al., 2011; Rizzardi et al., 2011). Several genes that play an important role in the auxin pathway are affected in swn clf (Figure 3; see Supplemental Data Set 1 online) (Lafos et al., 2011). One possible result of misregulating the auxin pathway is the alteration of vasculature development. This hypothesis is supported by the results in hormone-induced calli and in plants grown in NPA-containing medium. In both situations, the development of the vascular tissue is affected; in hormone-enriched medium by reducing the differentiation of the veins, and in the presence of NPA by increasing the amount and distribution of vascular bundles while impairing their integrity (Figures 5 and 6) (Wenzel et al., 2008). Based on microrarray studies performed by the AtGenExpress consortium, neither auxin nor NPA affect FT transcription directly (Goda et al., 2008). Therefore, the results confirm the sensitivity of FT expression to the appropriate development and differentiation of the vascular tissue. The absence of proper vasculature development prevents the activation of FT by CO, which is expressed at normal levels in swn clf and is strongly up-regulated in hormone-induced calli.
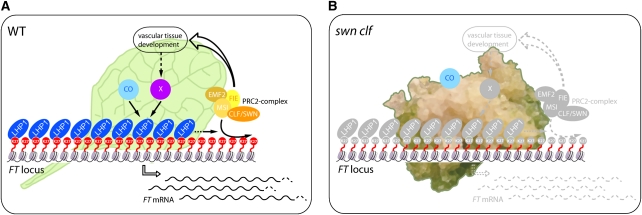
In summary, our data show that the role of PcG proteins in the regulatory network determining tissue-specific expression is not identical for all PcG-target genes. Although almost one quarter of PcG targets are strongly up-regulated in the swn clf mutant (Figure 3) and some have been shown to be ectopically expressed in different PcG mutants (Goodrich et al., 1997; Chanvivattana et al., 2004; Schubert et al., 2006; Calonje et al., 2008; Bratzel et al., 2010), the lack of the repressive H3K27me3 mark is not always synonymous with an increase in the expression domain or increased expression levels. This indicates that the lack of chromatin-mediated repression leads to elevated expression only if additional positive regulators are present. Specifically, the spatial expression of FT depends on the proper development and differentiation of the vascular system, and the removal of the PcG-mediated repressive chromatin structure, even in the presence of the activator CO, is not sufficient to overcome this requirement (Figure 7).
Figure 7.
The Impaired Differentiation of Vascular Tissue Bypasses the Lack of PcG Repression.
(A) In a wild-type (WT) leaf, the FT expression pattern is the result of the interplay among repressors, such as LHP1 and the PRC2 complex, and activators, such as CO. The PcG pathway is not only involved in the direct repression of FT but is also necessary to promote the proper development of the vascular tissue that is essential for FT activation through an unknown “X” factor.
(B) In a swn clf callus, where the differentiation of the vascular tissue is strongly impaired, CO is not able to activate FT expression due to the absence of the vascular “X” factor, despite the loss of PcG repressive marks in the FT chromatin.
METHODS
Plant Materials and Growth Conditions
All the plants used in this work were the Columbia (Col-0) ecotype. clf-28 (SALK_139371) (Doyle and Amasino, 2009), swn-7 (SALK_109121), and swn-7 clf-28 mutants were kindly provided by Daniel Schubert. The flc-3 mutant has been previously described (Michaels and Amasino, 1999). Due to the sterility of the double mutant, the 8.1kbFTprom:GUS and 35S:LHP1:HA transgenic lines (Adrian et al., 2010) were crossed to swn-7−/−clf-28+/− and were segregated until plants were obtained carrying each transgene in homozygosis in a swn-7−/−clf-28+/− background. The progenies from these lines were analyzed to find the corresponding phenotypes for the wild-type, clf, and swn clf mutants.
Plants were grown on GM medium under long-day conditions (16 h light/8 h dark) at 20°C. The callus-inducing GM medium was supplemented with 4.5 μM 2,4D and 0.45 μM kinetin (Hu et al., 2000). Overgrowth of the vascular tissue was induced with GM medium supplemented with 10 μM 1-N-naphtylphtalamic acid (Wenzel et al., 2008).
Gene Expression Analysis
Total RNA was extracted with the RNeasy mini kit (Quiagen). Five micrograms of RNA were DNase treated using the DNA-free kit (Ambion) to cDNA synthesis. Real time-qPCR was performed using a BioRad iQ5 apparatus and SYBR Green II detection. Expression of PP2AA3 (At1g13320) (Hong et al., 2010) was used for normalization. Primer sets can be found in Supplemental Table 1 online. GUS staining was performed as previously described (Adrian et al., 2010).
Microarray Analysis and Clustering of Expression Data
For genome-wide distribution of H3K27me3 in the ecotype Col-0, the data set described elsewhere (Göbel et al., 2010; Reimer and Turck, 2010) was used. H3K27me3-positive regions (chers) were determined with the implementation of RINGO (Toedling et al., 2007) in the R package ChIPR (half-window-size: 100, dist_CUT_off:200) and were subsequently mapped to genes according to the TAIR8 genome annotation (minimal number of probes per gene: 3). Genes were considered as H3K27me3 targets if at least 20% of the gene or 500 bp were covered by a positive cher. For microarray expression analysis, total RNA was extracted from 10-d-old Col and clf-28 and 12-d-old swn-7 clf-28 seedlings grown on 0.5× MS medium. Affymetrix ATH1 arrays were hybridized with two biological replicates per genotype and processed using the MAS 5.0 procedure (NASCARRAYS-425). For each gene, mean fold-change values compared with Col-0 were calculated. Clustering analysis was performed with all H3K27me3 target genes that were present in the expression data sets (4614 genes).
Clustering was performed with the software Genesis (Sturn et al., 2002). As a distance measurement, the Pearson correlation coefficient was used after computing log2 fold changes using the wild type as reference. For an overview on the major groups of patterns in the data set, hierarchical clustering was performed on a randomly selected subset of 500 H3K27me3 target genes and revealed eight major groups of expression patterns (see Supplemental Figure 1 online). Therefore, k = 8 was used for the K-means clustering analysis shown in Figure 3.
Chromatin Immunoprecipitation
ChIP experiments were performed as previously described (Searle et al., 2006) with anti-H3K27me3 (07-449; Millipore), anti-H3K9me2 (pAb-060-050; Diagenode), anti-H3K27me (pAB-045-050; Diagenode), anti-H3K4me3 (ab1012; Abcam), and anti-HA (H6908; Sigma-Aldrich). A small aliquot of untreated sonicated chromatin was used as the total input DNA. qPCR data were normalized against the input and represented as means of three technical replicates. For background, qPCRs with Ta3 primers were used for H3K27me3, H3K4me3, and LHP1:HA ChIPs, and the average of the signal obtained in each sample was plotted (dashed line in graphs). A similar method was used for H3K9me2 and H3K27me1, but with RBSC1A primers (a highly expressed gene) as background measure. Primer sets can be found in Supplemental Table 1 online.
Vascular Staining
Staining of the vasculature was performed by incubating ethanol-clarified plant tissues for 30 min in Basic Fuchsin (0.05%) after 10 min of incubation in 0.2 M NaOH. Excess stain was removed by rinsing in lactic acid.
Immunoblotting
Eleven-day-old 35Sprom:CO (Jang et al., 2008) seedlings were ground in lysis buffer (50 mM MES, pH 8.5, 25 mM KCl, 5 mM MgCl2, 5% Suc, 30% Glycerol, 10 mM β-mercaptoethanol, 1 mM DTT, 0.3% Triton X-100, 1:1000 Proteinase Inhibitor Cocktail [PIC, Sigma-Aldrich], and 1 mM PMSF). Nuclei were filtered through Miracloth, washed with wash buffer (6 mM MgCl2, 33 mM KCl, 17 mM HEPES, pH 7.5, 13% Suc, 13% glycerol, 13 mM β-mercaptoethanol, 1 mM DTT, 1.5 mM PMSF, 0.3% Triton X-100, 1:500 PIC), and resuspended in 500 μL Talon buffer (300 mM NaCl, 50 mM NaPO4, 6 M guanidine HCl, 100 μM ZnSO4, 100 mM DTT). Proteins were precipitated by addition of ethanol and guanidine to final concentrations of 95% and 0.3 M guanidine, respectively, and were resuspended in 25 μL of resuspension buffer (50 mM Tris HCl, pH 7.5, 150 mM KCl, 10 μM ZnSO4) and 25 μL of 2× Laemmli buffer. Immunoblots were developed with primary antibodies anti-CO (Valverde et al., 2004) and anti-Histone 3 (ab1791, Abcam) as loading control, and secondary anti-Rabbit IgG-HRPO conjugated (711-035-152, Dianova). The membrane was incubated with a mix of SuperSignal West Dura Chemiluminescent Substrate and SuperSignal West Femto Chemiluminescent Substrate (Pierce Chemical Co.), and the positive signals were detected by a cooled–charge-coupled device camera detection system.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data libraries under the following accession numbers: AG (At4g18960), CO (At5g15840), FLC (At5g10140), FT (At1g65480), LHP1 (At5g17690), SMZ (At3g54990), SVP (At2g22540), TEM1 (At1g25560), TEM2 (At1g68840), CLF (At2g23380), SWN (At4g02020), VIN3 (At5g57380), STM (At1g62360), RBSC1A (At1g67090), GSE20256 (Polycomb mutant seedlings microarrays), and E-MTAB-749 (H3K27me3 ChIP-chip data).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Representative Example of Hierarchical Clustering Based on Expression Data of a Subset of 500 Randomly Selected H3K27me3 Genes.
Supplemental Figure 2. Analysis of SVP, TEM1, TEM2, and SMZ Expression in the Wild Type, clf, and swn clf.
Supplemental Figure 3. LHP1:HA Binding and H3K27me1 Changes at AG and H3K4me3 Changes at FT and AG in the Wild Type, clf, and swn clf.
Supplemental Figure 4. The Vascular Tissue Loses Its Reticulated Pattern in swn clf.
Supplemental Figure 5. CO Protein Is Stable in Hormone-Supplemented Medium.
Supplemental Figure 6. Biological Repeat of Expression Data Presented in Figure 2.
Supplemental Figure 7. Biological Repeat of ChIP Data Presented in Figure 4.
Supplemental Table 1. List of Primers.
Supplemental Data Set 1. Expression Clusters of H3K27me3-Enriched Genes by K-Means Clustering.
Acknowledgments
We thank Daniel Schubert for providing the PcG mutant lines, Valentina Strizhova and Jesús López-Corrales for excellent technical help, and George Coupland, Wim Soppe, and José C. Reyes for critical reading of the manuscript. We gratefully acknowledge financial support from the Deutsche Forschungsgemeinschaft and the Max Planck Society.
AUTHOR CONTRIBUTIONS
S.F., F.L.T., J.G., and F.T. designed research; S.F., F.L.T., J.A., and L.S.-K. performed research; S.F., F.L.T., J.E., and X.D. analyzed data; and S.F. and F.T. wrote the article.
References
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero J.M., González-Bayón R., del Pozo J.C., Ponce M.R., Micol J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19: 2822–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F., López-Torrejón G., Koch M., Del Pozo J.C., Calonje M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Buzas D.M., Robertson M., Finnegan E.J., Helliwell C.A. (2011). Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant J. 65: 872–881 [DOI] [PubMed] [Google Scholar]
- Calonje M., Sanchez R., Chen L., Sung Z.R. (2008). EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos E.I., Reinberg D. (2009). Histones: Annotating chromatin. Annu. Rev. Genet. 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Carles C.C., Fletcher J.C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 23: 2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Amasino R.M. (2009). A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Aichinger E., Shu H., Wildhaber T., Alfarano P., Caflisch A., Gruissem W., Köhler C., Hennig L. (2009). The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4: e5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Coupland G., Turck F. (2008). The impact of chromatin regulation on the floral transition. Semin. Cell Dev. Biol. 19: 560–573 [DOI] [PubMed] [Google Scholar]
- Fisher C.L., Fisher A.G. (2011). Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Genet. Dev. 21: 140–146 [DOI] [PubMed] [Google Scholar]
- Göbel U., Reimer J., Turck F. (2010). Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part B: ChIP-chip data analysis. Methods Mol. Biol. 631: 161–184 [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Ha M., Ng D.W., Li W.H., Chen Z.J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21: 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51: 1694–1706 [DOI] [PubMed] [Google Scholar]
- Hu Y., Bao F., Li J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24: 693–701 [DOI] [PubMed] [Google Scholar]
- Imaizumi T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.H., Song H.R., Ko J.H., Jeong Y.M., Kwon Y.E., Seol J.H., Amasino R.M., Noh B., Noh Y.S. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L. (2010). Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 20: 541–547 [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Lee D., Choi G., Chung W.I. (2009). Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 60: 112–121 [DOI] [PubMed] [Google Scholar]
- Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., Tomita Y., Dohi K., Mori M., Araki T. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Ogas J., Cheng J.C., Sung Z.R., Somerville C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Reimer J.J., Turck F. (2010). Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part A: ChIP-chip molecular methods. Methods Mol. Biol. 631: 139–160 [DOI] [PubMed] [Google Scholar]
- Rizzardi K., Landberg K., Nilsson L., Ljung K., Sundås-Larsson A. (2011). TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J. 65: 897–906 [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., Trajanoski Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Takada S., Goto K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Toedling J., Skylar O., Krueger T., Fischer J.J., Sperling S., Huber W. (2007). Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C.L., Hester Q., Mattsson J. (2008). Identification of genes expressed in vascular tissues using NPA-induced vascular overgrowth in Arabidopsis. Plant Cell Physiol. 49: 457–468 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]