This study investigates how plants adopted a cellular strategy for defense against microbial pathogens by recruiting a plasmodesmata-localized protein to regulate cell-to-cell communication and augment innate immune responses.
Abstract
Plasmodesmata (PD) are thought to play a fundamental role in almost every aspect of plant life, including normal growth, physiology, and developmental responses. However, how specific signaling pathways integrate PD-mediated cell-to-cell communication is not well understood. Here, we present experimental evidence showing that the Arabidopsis thaliana plasmodesmata-located protein 5 (PDLP5; also known as HOPW1-1-INDUCED GENE1) mediates crosstalk between PD regulation and salicylic acid–dependent defense responses. PDLP5 was found to localize at the central region of PD channels and associate with PD pit fields, acting as an inhibitor to PD trafficking, potentially through its capacity to modulate PD callose deposition. As a regulator of PD, PDLP5 was also essential for conferring enhanced innate immunity against bacterial pathogens in a salicylic acid–dependent manner. Based on these findings, a model is proposed illustrating that the regulation of PD closure mediated by PDLP5 constitutes a crucial part of coordinated control of cell-to-cell communication and defense signaling.
INTRODUCTION
In plants, plasmodesmata (PD) establish symplastic conduits through which small molecules such as ions, metabolites, and hormones can diffuse from one cell to another, thereby allowing the intercellular coordination of biochemical and physiological processes (Roberts and Oparka, 2003; Lucas and Lee, 2004; Maule, 2008; Benitez-Alfonso et al., 2010; Lee et al., 2010; Burch-Smith et al., 2011). Each plasmodesma forms a discrete cytoplasmic channel that is delimited by the plasma membrane externally and the endoplasmic reticulum membrane internally. In addition to this unique structural feature, PDs are fundamentally different from intercellular communication channels found in animals, such as gap junctions, in that PDs have the capacity to facilitate cell-to-cell trafficking of proteins, RNAs, and protein/RNA complexes (Zambryski and Crawford, 2000; Haywood et al., 2002; Heinlein and Epel, 2004; Oparka, 2004; Ding, 2009).
A significant body of evidence supports the idea that PDs play a crucial role in cell fate determination and epigenetic modification by facilitating cell-to-cell movement of specific transcription factors (Lucas et al., 1995; Nakajima et al., 2001; Kurata et al., 2005) and mobile small RNAs (Carlsbecker et al., 2010; Dunoyer et al., 2010; Molnar et al., 2010; Olmedo-Monfil et al., 2010), respectively. However, this fundamental intercellular trafficking machinery is exploited by opportunistic microbial pathogens, such as plant viruses and obligate biotrophic parasites (Boevink and Oparka, 2005; Lucas, 2006; Hofmann et al., 2007; Ding, 2009; Benitez-Alfonso et al., 2010). Notably, recent findings suggested that a hemibiotrophic fungal pathogen might also use PDs to spread infectious hyphae and fungal effectors (Kankanala et al., 2007; Khang et al., 2010). If these microbial pathogens have evolved mechanisms to recognize PD as easy cellular gateways, it is reasonable to think that plants must have also developed counteracting strategies. However, how PD function is integrated into plant immunity and defense signaling is not yet clear (Lee and Lu, 2011).
Isolation of purified, intact PD is extremely difficult because PDs are embedded in rigid cell walls and constitute only a minute subcellular fraction. Moreover, mutations in PD components are thought to be either detrimental or pleiotropic, making it difficult to screen for PD mutants. However, efforts to overcome these hurdles are producing exciting and promising results (Maule, 2008; Lee et al., 2010). For example, various genetic screens to identify genes that modulate intercellular trafficking have led to the isolation of increased size exclusion limit 1 and 2, mutants of RNA helicases (Kim et al., 2002; Kobayashi et al., 2007; Stonebloom et al., 2009; Burch-Smith and Zambryski, 2010); and green fluorescent protein arrested trafficking1, a mutant of m-type thioredoxin (Benitez-Alfonso and Jackson, 2009). These mutants were shown to differently affect PD structure, morphology, and/or function. In addition, the characterization of a guard cell–patterning mutant, chorus, has led to a fortuitous identification of a putative callose synthase implicated in intercellular trafficking (Guseman et al., 2010). Moreover, biochemical and molecular studies undertaken to isolate proteins that are directly associated with PDs have contributed to the identification of multiple proteins that affect PD permeability. These include class 1 reversibly glycosylated polypeptides (C1RGPs) (Sagi et al., 2005), plasmodesmata-associated β-1,3-glucanase (At-BG_ppap) (Levy et al., 2007), PD-callose binding proteins (PDCBs) (Simpson et al., 2009), and PD-located protein 1 (PDLP1) (Thomas et al., 2008).
Members of the PDLP family have been identified based on sequence homology to PDLP1, the isoform first discovered by surveying cell wall proteomics from Arabidopsis thaliana suspension cultured cells (Thomas et al., 2008). A further analysis of this proteome identified additional proteins that are partially associated with PDs (Fernandez-Calvino et al., 2011). PDLPs range from 30 to 35 kD in predicted size and are composed of two conserved Cys-rich repeats containing DUF26 domains at the N terminus, followed by a transmembrane domain (TMD) and a very short cytoplasmic tail at the C terminus. The DUF26 domain, a plant-specific protein module, is characterized by conserved Cys residues and is found in a plant protein superfamily including Cys-rich receptor-like kinases (CRKs) and Cys-rich secretory proteins (Chen, 2001). The eight PDLP members constitute an intermediate form that contains only DUF26 domain and TMD, which thus is anchored to the membrane but lacks the cytosolic kinase domain. Thomas et al. (2008) used fluorescent tags to show that all eight members of the PDLP family localize to punctate structures at the cell periphery reminiscent of PD. In addition, a comparative analysis of cell-to-cell trafficking by employing green fluorescent protein (GFP) as a probe in transgenic plants that either overexpress or have a knockout of PDLP1 demonstrated that increasing levels of PDLP1 decrease PD permeability (Thomas et al., 2008). A recent study, in which fluorescently labeled movement protein (MP) of the tubule-forming virus Grapevine fanleaf virus (GFLV) and each of eight PDLP members were coexpressed, reported that the MP interacts with PDLPs at the PD (Amari et al., 2010). Surprisingly, the movement of GFLV as well as its PD association were inhibited in the pdlp1 pdlp2 pdlp3 mutant, suggesting that this virus exploits these PDLPs as necessary endogenous factors to infect Arabidopsis.
Meta-analysis of public transcriptome data (http://www.genevestigator.com; http://www.weigelworld.org/resources/microarray/AtGenExpress) suggested that the PDLP family members have diverse expression patterns, but the specific biological function of each PDLP member has not yet been established. Through a proteomics analysis of a cell wall fraction prepared from Arabidopsis seedlings, we identified two members of the PDLP family, PDLP3 (At2g33330) and PDLP5 (At1g70690) (see Supplemental Figure 1 online). PDLP3 and PDLP5 show ~50 and 30% amino acid sequence identities, respectively, with PDLP1 (see Supplemental Figure 2 online). Interestingly, in an independent study, Lee et al. (2008) identified the PDLP5 gene, which was alternatively named HOPW1-1-INDUCED GENE1 (HWI1) based on its inducible expression upon infection by Pseudomonas syringae, as an effective marker for plant responses to P. syringae carrying an effector, hopW1-1 (see Supplemental Table 1 online). Whether PDLP5/HWI1 has an active role in defense responses and what impact it might have on pathogen susceptibility was not tested. However, the circumstantial evidence, inferred from the public metatranscriptome analysis, RT-PCR data shown by Lee et al. (2008), and sequence similarity of PDLP5 to PDLP1 led us to hypothesize that PDLP5 might have a unique function, linking PD trafficking and bacterial pathogenesis.
Here, we provide new experimental evidence that supports the role of PDLP5 in PD regulation and innate immunity. The molecular and genetic analyses revealed that overexpression of PDLP5 leads to spontaneous cell death through a feedback regulation of salicylic acid (SA). By employing an ultrastructual approach combining correlative light electron microscopy and immunogold labeling, the location of PDLP5 was mapped to the central region of PDs. Assays performed by introducing various fluorescent probes revealed that PDLP5 inhibits PD permeability, and this effect was correlated with enhanced callose deposition at PD. Consistent with the role of PDLP5 in basal defense responses, an Arabidopsis mutant with a partial loss-of-function mutation in PDLP5 exhibited a greater susceptibility to virulent P. syringae. In accordance with these data, a model is proposed that illustrates potential mechanisms by which PD-mediated cell-to-cell communication is coordinated with the SA signaling pathway through PDLP5.
RESULTS
Overexpression of PDLP5 Induces Spontaneous Cell Death and Chlorosis
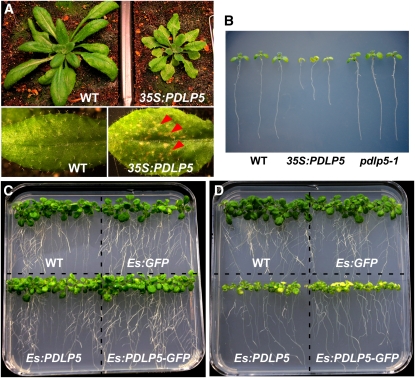
To gain insight into the biological function of PDLP5, we examined the morphological phenotypes brought about by modulating its expression level. For 35S:PDLP5, which expresses PDLP5 under the control of the Cauliflower mosaic virus 35S promoter, over 50 independent transgenic lines were produced and analyzed, and ~40% of the T1 lines exhibited visible growth and morphological phenotypes. In a search for a potential knockout mutant from a public database, we identified one T-DNA insertion line, SAIL_46_E06, which is predicted to contain T-DNA in the first intron of PDLP5. Genomic and transcriptional analyses confirmed the presence of the insertion and showed that the expression of PDLP5 is severely knocked down in this mutant, which was named pdlp5-1 (see Supplemental Figure 3 online). The pdlp5-1 plants did not exhibit any obvious growth defect compared with the wild type, but constitutive expression of PDLP5 in 35S:PDLP5 plants led to growth inhibition and chlorosis (Figure 1A). In addition, spontaneous lesions that resemble the hypersensitive response (HR), cell death induced by avirulent pathogens, were often observed in mature leaves of these plants, implying a potential role for PDLP5 in a cell death program. The chlorotic and inhibited growth caused by overexpression of PDLP5 was fully evident even at the seedling stage (Figure 1B).
Figure 1.
Constitutive or Induced Overexpression of PDLP5 Produces Necrotic Lesions and Chlorosis.
(A) Wild-type (WT) and a 35S:PDLP5 line with a strong chlorotic and cell death phenotype at the 3-week stage. Spontaneous lesion formation (arrowheads) observed in the leaf of the 35S:PDLP5 plant.
(B) Seedlings grown on MS agar 6 d after germination.
(C) and (D) Wild-type and estradiol-inducible transgenic lines expressing GFP, PDLP5, or PDLP5-GFP grown on MS agar for 10 d in the absence (C) and presence (D) of 5 μM estradiol.
[See online article for color version of this figure.]
There is always some concern that expressing a gene under a constitutively strong promoter that is active from the earliest stages of growth could lead to indirect sickness and growth inhibition of the transgenic plants. Thus, we examined whether overexpression of PDLP5 using inducible constructs under the control of the non-plant steroid estradiol, Es:PDLP5 and Es:PDLP5-GFP (Zuo et al., 2000), specifically causes the same symptoms observed in the 35S:PDLP5 line. All seedlings grew normally in the absence of estradiol treatment (Figure 1C); however, in the presence of 5 μM estradiol, both Es:PDLP5 and Es:PDLP5-GFP seedlings were severely affected in aerial tissue growth and became chlorotic, while the growth of wild-type or Es:GFP control seedlings remained unaffected (Figure 1D). Collectively, our results demonstrated that the observed plant phenotype is caused by the ectopic induction of PDLP5 and supports its negative impact on plant fitness.
Punctate Localization of PDLP5-GFP Correlates with PD Channels
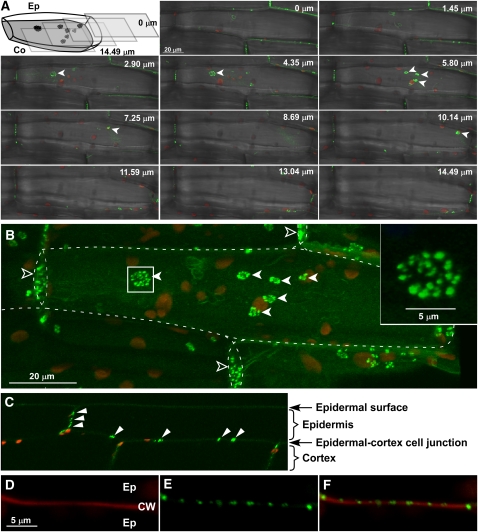
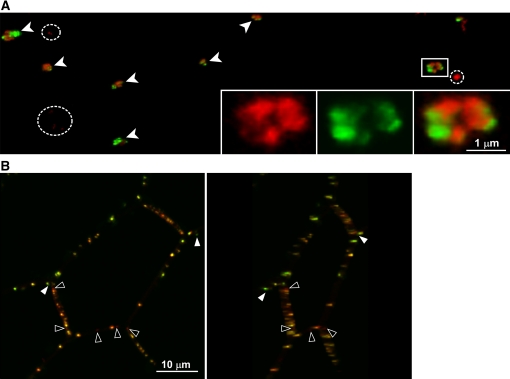
Next we confirmed the localization of PDLP5 by examining transgenic 35S:PDLP5-GFP seedlings. Here, we note that because overexpression of PDLP5 caused a growth phenotype (Figure 1), we selected transgenic lines with a relatively low expression level for the localization studies. Also, the growth phenotype was independent of whether PDLP5 was expressed as the native form or as a GFP fusion, indicating that PDLP5-GFP likely forms a functional construct that faithfully reports the intrinsic subcellular localization of PDLP5. Fluorescent signals produced by PDLP5-GFP were detected as punctate particles at the cellular boundaries of leaf epidermal cells (see Supplemental Figure 4 and Supplemental Movie 1 online), consistent with the previous report on its homolog, PDLP1 (Thomas et al., 2008). Serial z-section images (see Supplemental Figure 4A online) revealed that the punctate signals were only found at the cross-wall boundaries, supporting that the signals were likely associated with PD. This localization pattern was consistent in both the constitutive 35S:PDLP5-GFP and inducible Es:PDLP5-GFP lines. Within expanding lateral wall junctions between epidermal and cortex cells of the hypocotyl, PDLP5-GFP signals were detected as large, discrete structures reminiscent of PD pit fields (Seagull, 1983; Faulkner et al., 2008a) (Figures 2A and 2B). Each pit field–like fluorescent area varied in diameter and was composed of rosettes of smaller fluorescent foci (Figure 2B). A similar spatial organization of PDLP5-GFP signals also was detected at the cell wall junctions between leaf epidermal and mesophyll cells (see Supplemental Figure 5 and Supplemental Movie 1 online). By contrast, only smaller and more dispersed fluorescent foci were detected at the cross walls between epidermal or cortex cells within the hypocotyl (Figures 2A to 2C). Consistent with the punctate PDLP5-GFP signals that were associated with PD, they also were detected only at the cross-walls in hypocotyl cells (Figures 2C to 2F).
Figure 2.
PDLP5-GFP Localizes to PD Pit Fields.
Confocal images of PDLP5-GFP showing its association with PD pit fields. Closed arrowheads, PD pit fields; open arrowheads, cross-walls between epidermal cells.
(A) Confocal images shown in z-series optical sections, which span through a 14.49-μm-thick region of hypocotyl tissue across lateral wall junctions between epidermal (Ep) and cortex (Co) cells. Images show merged green, red, and transmitted channels. Illustration is included to help visualize the orientation of z-sections.
(B) A 3D maximum intensity projection reconstructed from the z-series displayed in (A). Images show merged green and red channels. Dashed lines, contour of epidermal cells; inset, high magnification of the boxed region.
(C) A reconstructed confocal image illustrating a longitudinal view of the hypocotyl tissue. Punctate PDLP5-GFP signals were only found in cross-walls. Images show merged green and red channels. Arrowheads, PD pit fields labeled by PDLP5-GFP.
(D) to (F) Confocal images showing punctate PDLP5-GFP signals within propidium iodide–stained cross-wall between epidermal hypocotyl cells, presented in red (D), green (E), and merged (F) channels. CW, cell wall.
[See online article for color version of this figure.]
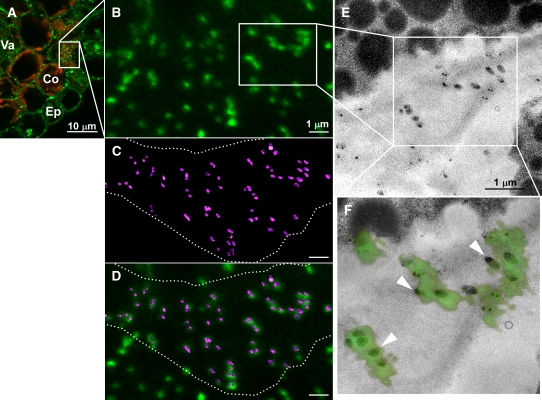
Next, to validate the punctate PDLP5-GFP signals as PD at an ultrastructural resolution, we applied the correlative light electron microscopy technique (van Rijnsoever et al., 2008) (Figure 3). A cross-wall segment containing PDLP5-GFP signals (Figure 3A) was selected for serial ultrathin sectioning to visualize the same wall region under a transmission electron microscope (TEM) as a reconstructed three-dimensional image (Figures 3B to 3D). The clusters of PD revealed in this three-dimensional (3D) TEM image completely correlated with the fluorescent signals (Figures 3B to 3F), providing compelling ultrastructural evidence that the punctate signals produced by PDLP5-GFP were indeed associated with PD.
Figure 3.
Correlative Light Electron Microscopy Analysis Showing That PDLP5-GFP Signals Overlap with PD Structures.
(A) Confocal image of a fixed, 50-μm-thick cryosection from hypocotyl tissue exposing the face-on view (boxed area) of dispersed PD at the cross-wall between cortex cells. Red, chlorophyll autofluorescence; green, PDLP5-GFP signals. Co, cortex; Ep, epidermis; Va, vasculature.
(B) to (F) Correlation between confocal image of PDLP5-GFP and electron microscopy images. PDs were false-colored in magenta for contrast. Dotted lines, boundary of cell wall area; arrowheads, correlated small clusters of PDs.
(B) High magnification of punctate PDLP5-GFP signals on a cross-wall from the rotated, boxed region in (A).
(C) A 3D rendering of nine serial ultrathin TEM sections of the imaged cell wall junction, corresponding to the same region in (B).
(D) Overlay of (B) and (D).
(E) Single section TEM image of boxed region in (B).
(F) Overlay of the PDLP5-GFP (boxed region in [B]) and TEM image in (E).
PDLP5 Localizes to the Central Region of PD Similar to Viral MPs
Immunogold studies of Tobacco mosaic virus (TMV) MP or PDCB have previously shown that these proteins localize to specific subdomains of PD (i.e., the median cavity) (Ding et al., 1992) or neck region (Simpson et al., 2009), respectively. To determine the PD subdomain that PDLP5 occupies, we first tried employing peptide antibodies specific to PDLP5 for conventional immunogold labeling. However, the results were not satisfactory in terms of resolving the location of PDLP5 within PD. As an alternative approach in improving the ultrastructural resolution of the immunogold labeling, we applied an antigen recovery method (Stirling and Graff, 1995), in which the tissue samples were processed according to a chemical fixation and epoxy resin embedding method, followed by chemical removal of the resin from ultrathin sections.
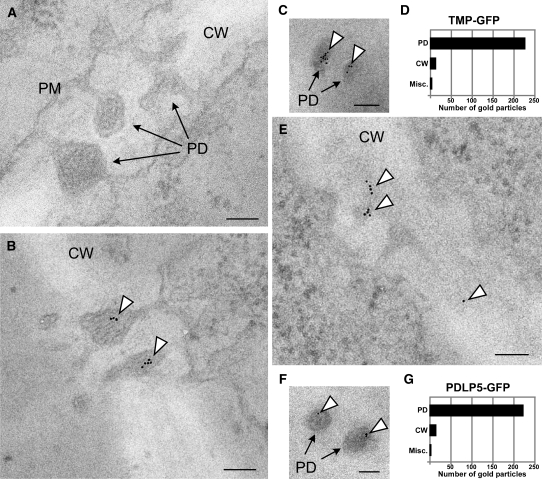
To validate the suitability of this approach for our experimental goal, we performed two control experiments. First, wild-type sections processed for the antigen retrieval were incubated with gold-conjugated secondary α-rabbit alone, without exposure to primary antibody. This experiment confirmed that the morphology of intracellular compartments, including PD, was well preserved and that nonspecific labeling of the secondary antibody was absent (Figure 4A). A second control experiment was performed on transgenic Arabidopsis expressing GFP-tagged TMV-MP (TMP-GFP) under the control of the 35S promoter. Here, we used high-titer, affinity-purified α-GFP prepared in-house as the primary antibody and the gold-conjugated α-rabbit as the secondary antibody. Examination of both longitudinal and cross sections (Figures 4B and 4C) revealed specific targeting of TMP-GFP to the median cavity of PD at a reasonably high resolution of PD morphology (see Supplemental Figure 6 online). Importantly, this localization was comparable and consistent with the previous report performed on transgenic tobacco by employing high-pressure freezing and freeze substitution (Ding et al., 1992), validating our experimental approach.
Figure 4.
PDLP5-GFP Specifically Localizes within PD.
(A) Representative immunogold TEM image of secondary antibody control.
(B) to (G) Representative immunogold TEM images performed using α-GFP as primary antibody and quantitative analysis of gold particles. Longitudinal (B) and cross sections (C) of PD revealing the localization of TMP-GFP. Detection of PDLP5-GFP at the longitudinal (E) and cross sections (F) of PD. Quantification of gold particles ([D] and [G]).
CW, cell wall; Misc., miscellaneous subcellular compartments; PM, plasma membrane. Arrowheads, gold particles found within PD. Bars = 100 nm.
Next, we processed 35S:PDLP5-GFP plants using the same fixation, antigen retrieval, and immunogold labeling method. This experiment showed that PDLP5 was localized inside the PD channels similar to TMP-GFP (Figures 4E and 4F; see Supplemental Figure 6 online). A quantitative analysis of the gold labels for TMP and PDLP5 showed that the labeling was highly specific in terms of its association with PD (Figures 4D and 4G): a total of 227 out of 242 gold particles from 34 TEM images and 218 out of 235 from 59 images were found within PD of TMP-GFP and PDLP5-GFP sections, respectively.
The finding that both TMP-GFP and PDLP5-GFP localized to the central region of the PD raised the possibility that they might occupy the same PD subdomain. In an attempt to test this notion by colocalizing TMP and PDLP5, we produced PDLP5-specific peptide antibodies and tested for immunogold labeling because the TMP-specific antibody that was proven to be suitable for immunogold analysis (Ding et al., 1992) was not available. However, the peptide antibodies were not compatible with etched epoxy sections. As an alternative approach, we used high-resolution confocal microscopy of PDs colabeled by TMP-GFP and PDLP5-mRFP in transgenic Arabidopsis. We examined the cell wall interface between epidermal and cortex cells of hypocotyl tissue as this junction allowed for optimal visualization of the cross sections of PD. Fluorescent signals produced by TMP-GFP and PDLP5-mRFP partially overlapped within the clusters of PD (Figure 5A). A careful examination of the PD at the epidermal cell junctions by serial z-sections consistently showed that not all fluorescent speckles were evenly marked by both TMP-GFP and PDLP5-mRFP but that some spots were exclusively labeled by one or the other signal (Figure 5B). Although these results did not resolve whether the two proteins occupied the same subdomains of PD, we speculated that these proteins might either compete for the same plasmodesmal strand and become mutually exclusive or have a slightly different preference for a specific type of PD (see Discussion).
Figure 5.
PDLP5 Localization Pattern at PDs Does Not Completely Overlap with TMP.
(A) Merged confocal image of coexpressed TMP-GFP (green) and PDLP5-mRFP (red) at face-on view of cross-walls between epidermal and cortex cells of the hypocotyl. Arrowheads, a small cluster of PDs; boxed region, a larger cluster of PD; broken circles, speckles exhibiting only red signals from PDLP5-mRFP; insets, higher magnification of the boxed region (split into red and green channels and merged).
(B) Confocal images of coexpressed TMP-GFP (green) and PDLP5-mRFP (red) at the side view of cross-walls between epidermal cells of the hypocotyl. Left panel: serial z-sections capturing longitudinal PDs through cell wall junctions were reconstructed as a 3D maximum intensity projection and viewed in merged channel. Right panel, a z-axis merged view of the 3D maximum intensity projection. Closed arrowheads, nonoverlapping TMP-GFP signals; open arrowheads, nonoverlapping PDLP5-mRFP signals.
PDLP5 Modulates Both Basal and Induced PD Permeability
Experiments using GFP as a reporter for cell-to-cell diffusion have shown that PDLP1 negatively regulates PD permeability (Thomas et al., 2008). Based on the conserved domain structure, one can speculate that PDLP5 might also decrease PD permeability. However, given the low amino acid sequence identity between PDLP1 and PDLP5 (30%), it is possible that their functional characteristics differ. To gain insight into the potential role of PDLP5 at PD, we introduced three different types of reporters for cell-to-cell movement: a 560-D fluorescent probe for basal PD permeability, GFP for macromolecular diffusion, and viral MPs for active movement.
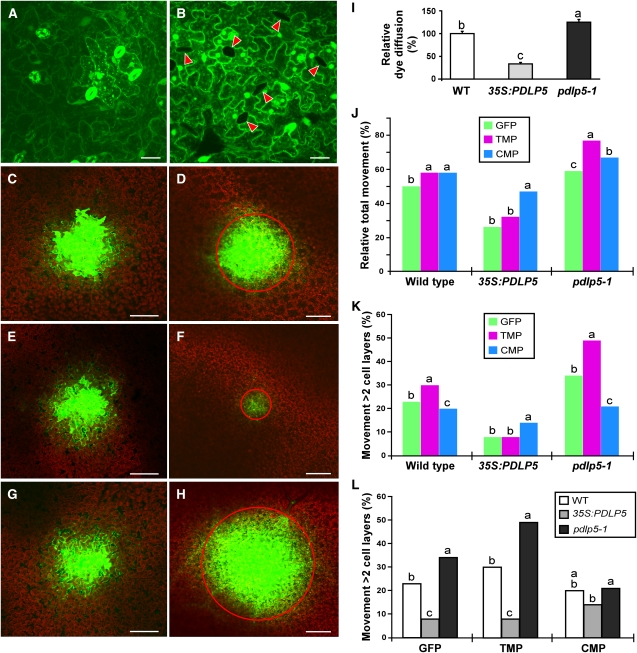
To assay basal PD permeability, we redesigned the dye-loading assay employing carboxy-fluorescine diacetate (CFDA), which is used as a symplastic tracer (Wright and Oparka, 1996). The nonmembrane-permeable fluorescent form of the dye, CF, is released from membrane-permeable, nonfluorescent CFDA by cellular esterases within the loaded cells, ensuring that any intercellular spread of CF in plants is through symplastic connections provided by PD. Our assay, which we named Drop-ANd-See (DANS) dye loading, used a noninvasive dye application method: a drop of CFDA was released onto the adaxial epidermal surface of an intact plant leaf and the cell-to-cell diffusion of cleaved CF was observed in the abaxial epidermis. The extent of fluorescent dye movement was quantified by measuring the diameter of the fluorescent area in the abaxial epidermis after 5 min of loading. As expected for a membrane-permeable dye, both guard cells and pavement cells within the adaxial epidermis, on which CFDA was directly loaded, showed fluorescent signals of released CF (Figure 6A). By contrast, no fluorescent signals were detected in mature guard cells on the abaxial side of the leaf epidermis, demonstrating that CF diffusion into these cells was prevented because they lack functional PD; hence, they were symplastically isolated (Figure 6B).
Figure 6.
PDLP5 Acts as a Negative Regulator for PD Permeability.
(A) to (I) PD permeabilities examined by employing CFDA-based DANS dye loading assays.
(A) and (B) Close-up confocal images of adaxial surface loaded with CFDA (A) and abaxial surface showing exclusion of fluorescent signals from symplasmically isolated guard cells (arrowheads) (B).
(C) to (H) Confocal images showing CFDA loaded onto adaxial surfaces ([C], [E], and [G]) of intact leaf and CF movement observed in abaxial surfaces ([D], [F], and [H]). The wild type ([C] and [D]), 35S:PDLP5 ([E] and [F]), and pdlp5 ([G] and [H]]. Red circle is used to illustrate the extent of dye diffusion. Bars = 20 μm in (A) and (B) and 200 μm in (C) to (H).
(I) Quantification of CF movement in the three genotypes. At least 10 plants were used per assay and more than three repeats were performed. The extent of dye diffusion was quantified by measuring diameters of diffusion area with respect to the fluorescent intensity distribution. Levels not connected by same letter are significantly different at the α = 0.05 level according to the LSD test following one-way ANOVA. Bars indicate se. WT, wild type.
(J) to (L) Quantification of cell-to-cell protein trafficking of GFP, TMP-GFP, or CMP-GFP in the wild type, 35S:PDLP5, and pdlp5. This assay used a biolistic DNA delivery-mediated transient expression system. More than three biological repeats per genotype were performed to collect a total of 969, 2146, and 1321 cell transformation events for GFP, CMP-GFP, and TMP-GFP, respectively (three to four leaves were collected from a single plant, and at least five plants from each genotype were used for each repeat). Protein movements out of each transformed cell are differentially illustrated by scoring total movements (percentage of the total number of transformed cells showing protein movement) regardless of the extent of the movement (J) and by counting only extensive movement beyond two cell layers ([K] and [L]). Levels not connected by same letter are significantly different at the α = 0.05 level according to the LSD test following one-way ANOVA.
[See online article for color version of this figure.]
After establishing that the DANS dye loading assay was reliable and effective in quantifying PD permeability by performing control experiments using wild-type plants, we applied the technique to address how the basal level of PD permeability might be affected in plants with altered levels of PDLP5. To this end, 35S:PDLP5 and pdlp5-1 were used. Compared with wild-type plants, CF movement was reduced by 70% in 35S:PDLP5 but enhanced by 25% in pdlp5-1 plants (Figures 6C to 6I). Consistent with the dye diffusion data, nongated trafficking of GFP between epidermal cells, which was assayed by employing a biolistic DNA delivery method, also was affected by PDLP5; overexpression of PDLP5 restricted GFP movement, and reduction led to an increased intercellular spread (Figures 6J to 6L; see Supplemental Figure 7 online). These results revealed an inverse relationship between the level of PDLP5 expression and the PD permeability, similar to the effects of altered PDLP1 levels as reported by Thomas et al. (2008).
It is notable that downregulation of PDLP5 alone was sufficient to enhance PD permeability; this is in contrast with PDLP1, which apparently shows a functional redundancy with other closely related isoforms (Thomas et al., 2008). In addition, Thomas et al. (2008) did not address whether PDLP1 or its close homologs could interfere with gated protein movement. Therefore, to test the effect of PDLP5 expression on gated macromolecular trafficking, two different viral MPs, TMP and Cucumber mosaic virus MP (CMP), along with a GFP control, were employed in a transient expression system using biolistic DNA delivery. Compared with the wild-type control, 35S:PDLP5 plants showed an overall 10 to 30% reduction in trafficking of both MPs, but pdlp5-1 had a 10 to 20% enhancement (Figure 6J; see Supplemental Figure 7 online). However, the extent to which the trafficking of these two MPs was affected by PDLP5 was quite dissimilar, especially when their extensive movement (e.g., protein trafficking into two or more cell layers from a single target cell) was analyzed (Figures 6K and 6L). A statistical analysis using Fisher’s LSD method showed that whereas CMP movement was minimally altered in 35S:PDLP5 or pdlp5-1 background (approximately ±5% variation relative to wild-type control), TMP trafficking was influenced significantly (approximately ±20%) (Figure 6L).
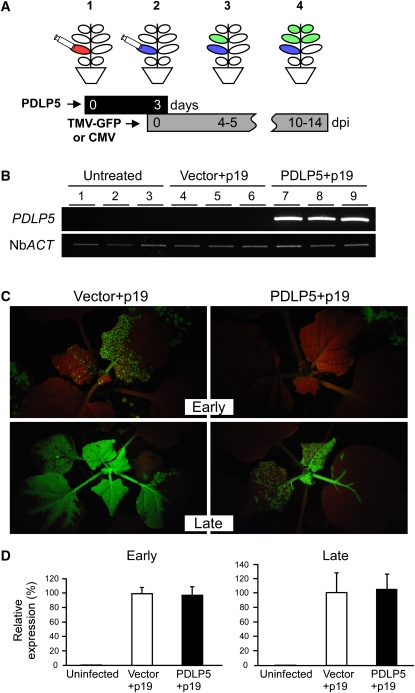
The finding that TMP movement was affected by PDLP5 expression led us to ask whether PDLP5 may also impact TMV infection. We first attempted to address this question by infecting Arabidopsis (Columbia-0) with GFP-tagged TMV (TMV-GFP) (Liu et al., 2002). However, no fluorescent infection foci were detectable in either infected or systemic leaves. Therefore, as an alternative approach, we examined specifically whether overexpression of PDLP5 could impact systemic viral infection by employing a heterologous system, Nicotiana benthamiana, in which TMV-GFP was previously shown to effectively spread (Liu et al., 2002) (Figure 7). To this end, leaves of N. benthamiana plants were coinfiltrated with two strains of agrobacteria: one transformed with the binary vector pMLBart carrying 35S:PDLP5 and the other transformed with a vector carrying p19 (Voinnet et al., 2003) to boost PDLP5 expression over the experimental time frame. Control plants were treated the same except that the empty vector (pMLBart) was substituted for 35S:PDLP5. Three days later, the plants were infected with TMV-GFP via infiltration of agrobacteria carrying 35S:TMV-GFP (Figure 7A).
Figure 7.
Ectopic Expression of PDLP5 Delays Systemic Movement of TMV-GFP in N. benthamiana.
(A) A schematic illustration showing the experimental setup for agro-mediated PDLP5 expression and viral infection: (1) agroinfiltration of PDLP5 or empty vector control at experimental day 0 into 4th and 5th leaves (red); (2) agroinfection of the leaves (blue with TMV-GFP [or CMV inoculation] 3 d after primary agroinfiltration; (3) movement of TMV-GFP (or CMV) in systemic leaves (green) at early time points (4 to 5 DAI [dpi]); (4) systemic movement of TMV-GFP (or CMV) at late time points (10 to 14 DAI). Black and gray bars indicate the time frames for PDLP5 expression and TMV-GFP infection, respectively.
(B) RT-PCR analysis confirming the expression of PDLP5 in the leaves agroinfiltrated with GV3101 mixture carrying 35S:PDLP5 and p19 at the time of viral infection. PCR products were electrophoresed in ethidium bromide–containing 0.8% agarose gel and visualized under UV. Lanes 1 to 3, three independent plant samples that were untreated; lanes 4 to 6, three independent plant samples that were infiltrated with GV3101 mixture carrying empty vector and p19; lanes 7 to 9, three independent plant samples that were infiltrated with GV3101 mixture carrying 35S:PDLP5, and p19. N. benthamiana ACTIN (Nb ACT) was used as control for RT-PCR.
(C) Systemic TMV-GFP infection detected by GFP fluorescence under UV illumination using a Black Ray UV lamp. Representative images are shown from at least three independent repeats using 10 plants for each treatment.
(D) Systemic CMV movement detected by RT-PCR. The expression level of CMV was normalized to Nb ACT. No statistical difference in CMV detection was found between vector- and PDLP5-infiltrated samples at either early or late infection time points. Bars indicate se.
[See online article for color version of this figure.]
To confirm that PDLP5 expression was persistent in the agroinfiltrated leaves at the time of the viral infection, RT-PCR analysis using PDLP5-specific primers was performed 3 d after agroinfiltration (Figure 7B). This experiment validated the ectopic expression of PDLP5 at the time of viral infection. Next, to examine the impact of PDLP5 on systemic viral spread, TMV-GFP–infected plants were monitored at early and late time points (Figure 7C). At early time points (4 to 5 d after infection [DAI]), TMV-GFP spread to systemic leaves was clearly detectable only in vector control and not in PDLP5-overexpressing plants. However, at late time points (10 to 14 DAI), systemic infection of TMV-GFP also became evident in PDLP5-overexpressing samples. By contrast, PDLP5 ectopic expression in N. benthamiana did not affect the systemic movement of Cucumber mosaic virus (CMV) either at early or late time points (Figure 7D). These results were consistent with the trafficking data that TMP movement, but not CMP movement, was affected by PDLP5 (Figure 6). Collectively, our data demonstrated that the changes in PDLP5 expression were sufficient to alter both basal PD permeability and gated movement of MPs, which suggests that PDLP5 has more potent effects on PD regulation than some other previously characterized isoforms (see Discussion).
PDLP5 Alters Callose Deposition at PD
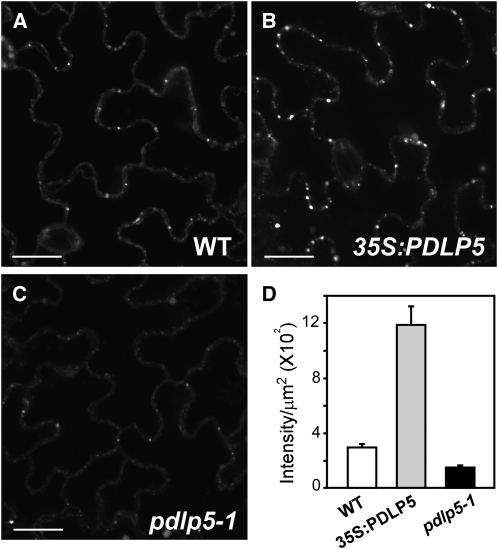
One of the mechanisms thought to regulate PD gating involves reversible callose deposition around PDs (Radford et al., 1998; Zavaliev et al., 2011). Correlations between the level of PD callose and the effect on PD gating have been made for other PD-associated proteins, such as C1RGP2 (Sagi et al., 2005), At-BG_ppap (Levy et al., 2007), and PDCB1 (Simpson et al., 2009). However, it is not known whether PDLP1 or other PDLP members also affect callose accumulation at PD. Thus, to gain new insight into how the change in PD permeability was brought about in pdlp5-1 and 35S:PDLP5 plants, we compared the level of PD callose in these plants.
We postulated that the physiological stresses from the chlorotic and lesion-forming phenotype observed in 35S:PDLP5 (Figure 1) could induce accumulation of callose as an indirect effect. If this was the case, pdlp5-1, having similar overall morphological phenotype as the wild type, would show no difference in callose deposition from the wild-type control. To test this, we performed aniline blue staining and quantified PD callose based on more than 80 images collected from each genetic background (Figure 8). This experiment showed that 35S:PDLP5-expressing leaf tissues contained approximately fourfold higher PD callose level than the wild type (Figures 8A, 8B, and 8D), but pdlp5-1 plants contained a twofold lower callose accumulation than the wild type (Figures 8A, 8C, and 8D). These results revealed an inverse relationship between the level of PDLP5 and callose deposition. Therefore, we postulated that PDLP5 has the capacity to modulate the accumulation of PD callose, which is likely necessary for its function in controlling PD permeability.
Figure 8.
PDLP5 Modulates Callose Accumulation.
(A) to (C) Representative images of callose staining in the wild type (WT) (A), 35S:PDLP5 (B), and pdlp5 (C). Bars = 20 μm.
(D) Quantitative comparison of fluorescence foci intensity per unit area in the three genotypes (two-tailed P values < 0.0001). Bars indicate se.
Overexpression of PDLP5 Triggers Cell Death through Hyperaccumulation of SA
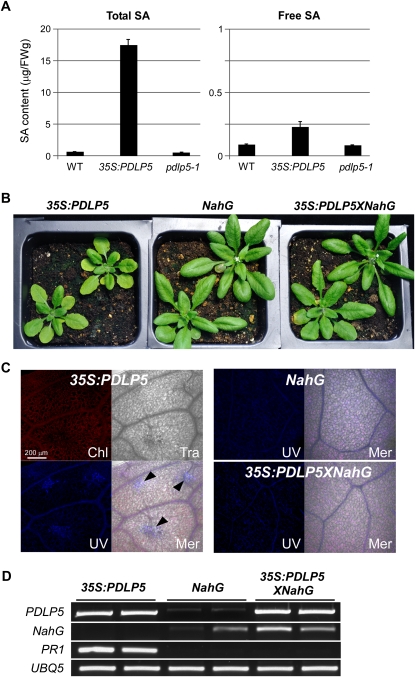
The formation of HR-like cell death or lesions is known to involve genes that are activated by a hyperaccumulation of SA (Glazebrook, 2005; Vlot et al., 2009). To test whether the lesions and chlorosis induced by PDLP5 overexpression are linked to an induced SA production, we measured the level of SA in 35S:PDLP5 and pdlp5-1 plants. The SA content in pdlp5-1 plants was similar to the wild-type control. However, 35S:PDLP5 plants accumulated 15-fold higher total SA (both free active SA and an inactive conjugated form, SAG) compared with the wild-type control; the total free SA alone was threefold higher (Figure 9A). This result suggested that the necrotic cell death caused by PDLP5 overexpression could be attributed to the high level of SA accumulation in these plants. To test this possibility, we introduced NahG, which encodes the SA-degrading bacterial enzyme salicylate hydroxylase (Friedrich et al., 1995), into 35S:PDLP5 plants by genetic cross. As anticipated, the cell death phenotype associated with overexpression of PDLP5 was suppressed in F2 progenies that expressed both NahG and PDLP5 (Figures 9B and 9C). Moreover, a marker gene for SA accumulation, PR1, was highly induced in 35S:PDLP5 and was suppressed by coexpression of NahG (Figure 9D). These results supported a positive role for PDLP5 in SA accumulation and spontaneous cell death.
Figure 9.
Growth Inhibition and Cell Death in 35S:PDLP5 Are Induced by a Hyperaccumulation of SA.
(A) Measurement of total and free SA content in the wild type (WT), 35S:PDLP5, and pdlp5. Bars indicate se. FW, fresh weight.
(B) to (D) NahG suppresses the growth (B), cell death lesion formation (C), and molecular phenotypes (D) observed in 35S:PDLP5. Growth comparison of 4-week-old 35S:PDLP5 and 35S:NahG with F2 progenies (35S:PDLP5 × 35S:NahG) carrying both ectopic PDLP5 and NahG from the parental lines (B). Confocal images of the leaves showing microlesions, which were visualized using autofluorescence under UV, detected only in 35S:PDLP5 collected from 35S:PDLP5, but not from 35S:NahG or 35S:PDLP5 X 35S:NahG plants (C). Chl, chlorophyll channel; Tra, transmitted light channel; UV, UV channel; and Mer, merged image. Arrowheads indicate cell death lesions. Molecular phenotyping using RT-PCR showing loss of PR1 induction in 35S:PDLP5 by introducing NahG (D).
PDLP5 Is Restricted to Vasculature during Normal Development but Is Highly Induced during Senescence and by Exogenous SA
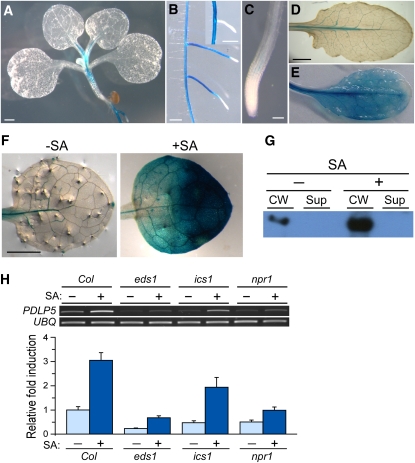
To further elucidate the relationship between PDLP5 and SA, we investigated how PDLP5 expression was connected to the SA defense pathway by performing histochemical analysis. First, we analyzed the expression pattern of PDLP5 under normal developmental conditions using PDLP5pro:GUS (for β-glucuronidase) plants. This experiment showed that the GUS activity was mostly associated, though at a low level, with vascular tissues of petioles and the hypocotyl of seedlings (Figure 10A). GUS staining was stronger overall in roots but was almost excluded from the meristematic and elongation zones of primary and lateral roots (Figure 10B). Only a very low level of GUS staining associated with the provasculature was detectable in root tips (Figure 10C). A basal level of vascular expression revealed by GUS staining also was observed in the leaves of seedlings and mature plants (Figure 10D).
Figure 10.
Control of PDLP5 Expression by SA Pathway.
(A) to (E) GUS-stained PDLP5pro:GUS under normal growth conditions. Arial tissue of 6-d-old seedling (A); primary root and elongated secondary roots (B); primary root tip (C); and young (D) and senescing rosette leaves (E).
(F) and (G) Histochemical and immunoblot analyses showing PDLP5 induction by exogenous SA. Mock and 100 μM SA-treated PDLP5pro:GUS leaves (F). Immunoblot analysis with α-PDLP5 showing the PDLP5 protein induction in wild-type leaves by SA treatment (G). CW, cell wall; Sup, supernatant fractions.
(H) PDLP5 expression in various SA pathway mutants in the presence and absence of exogenous SA application. RT-PCR products were electrophoresed in ethidium bromide–containing 0.8% agarose gel and visualized under UV (top). Quantification of PDLP5 transcript level was normalized to the corresponding UBQ level using ImageJ software (bottom). The data set between treatments is significantly different at P < 0.0001. Bars indicate se. Col, Columbia-0.
[See online article for color version of this figure.]
Contrary to the low, vascular expression of PDLP5 in seedlings and mature leaves, its expression pattern and level were altered in senescing leaves as exhibited by a strong GUS staining throughout the whole leaf tissue (Figure 10E). This was in agreement with public transcriptome data predicting a relatively high expression of PDLP5 in senescing tissues (http://www.genevestigator.com). As SA signaling is known to be involved in senescence (Vlot et al., 2009), we then tested whether an exogenous application of SA could induce PDLP5 expression. A strong GUS staining in SA-treated PDLP5pro:GUS leaves compared with mock-treated leaves demonstrated that PDLP5 expression was highly upregulated by SA (Figure 10F). Consistent with this result, immunoblot analysis using PDLP5-specifc antibodies revealed that SA treatment of wild-type Arabidopsis resulted in an approximately fivefold upregulation of PDLP5 protein (Figure 10G). Collectively, these results suggested the possibility that a positive feedback regulatory loop exists between PDLP5 expression and accumulation of SA.
Next, to further demonstrate SA-dependent PDLP5 induction, RT-PCR analysis was performed by employing several known SA mutants (Figure 10H). These include genes encoding an SA biosynthetic enzyme, isochorismate synthase1 (Wildermuth et al., 2001); a lipase-like protein required for accumulation of SA, enhanced disease susceptibility1 (Falk et al., 1999); and a central regulator of SA signal transduction, nonexpressor of PR1 (Cao et al., 1994). PDLP5 transcript was downregulated in all three tested SA mutants even with exogenous SA application, showing that induction of PDLP5 was dependent on both SA biosynthetic and signaling components.
PDLP5 Is Linked to Innate Immunity against Bacterial Pathogens
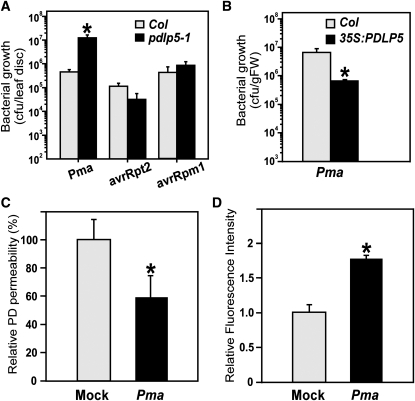
The findings that SA induced PDLP5 expression and that overexpression of PDLP5 conferred a SA-dependent spontaneous HR-like cell death suggested that PDLP5 might play a positive role in defense responses. For a proof of concept, we chose P. syringae pv maculicola ES4326 (Pma), a virulent bacterial pathogen, along with isogenic avirulent strains Pma avrRpt2 and Pma avrRpm1 (Dong et al., 1991; Guttman and Greenberg, 2001), for infection of pdlp5-1 and analyzed the pathogen susceptibility of the plants. Upon bacterial infection, PDLP5 was highly induced (see Supplemental Figure 8 online), which was consistent with the published data (Lee et al., 2008). Pma bacterial growth was increased in pdlp5-1 plants by over 10-fold at 3 DAI, revealing a greater susceptibility in pdlp5-1 compared with the wild-type control (Figure 11A). By contrast, the avirulent Pma strains did not show much difference in their growth between pdlp5-1 and the wild type. To further examine the role of PDLP5 in innate immunity, we then investigated whether overexpression of PDLP5-conferred resistance against bacterial infection by examining the bacterial growth in 35S:PDLP5. Compared with the wild-type control, Pma growth in 35S:PDLP5 plants was reduced by 10-fold (Figure 11B), indicating that PDLP5 overexpression brought about an enhanced basal defense in Arabidopsis.
Figure 11.
PDLP5 Is Required for Basal Immunity against Bacterial Pathogen.
(A) Susceptibility of pdlp5 to bacterial infection with virulent Pma and its avirulent derivatives. Asterisk indicates a significant difference (P < 0.005; n = 6) between two samples infected with the same pathogen. Bars indicate se. Col, Columbia-0.
(B) Comparison of Pma growth in 35S:PDLP5 and wild-type Arabidopsis. Asterisk indicates a significant difference (P < 0.001; n = 4). Bars indicate se. FW, fresh weight.
(C) DANS assay showing a reduction in PD permeability upon Pma infection. Three-week-old Arabidopsis wild-type plants were infected with Pma, and DANS assay was performed on systemic leaves at 48 h after inoculation. Two systemic leaves from at least five different plants were used for DANS assay per repeat. The data set between treatments is significantly different at P < 0.0001. Bars indicate se.
(D) Aniline blue staining showing an increased PD callose accumulation upon Pma infection. Three-week-old Arabidopsis wild-type plants were infected with Pma, and systemic leaves were stained at 48 h after inoculation for callose quantification. Two to three systemic leaves from at least five different plants were used for callose staining per repeat. The data set between treatments is significantly different at P < 0.0001. Bars indicate se.
Our findings that PDLP5 was induced by bacterial infection and played a role in controlling PD permeability while affecting the susceptibility to Pma suggested that the regulation of PD constitutes a part of the innate immune response. Moreover, this model predicted that bacterial infection would induce a phenotype similar to 35S:PDLP5 in terms of aberrant PD closure and callose deposition. To test this notion, DANS dye loading assay and callose staining were performed on systemic leaves of 3-week-old wild-type Arabidopsis plants at 24 h after Pma infection (Figures 11C and 11D). The result of this experiment revealed that Pma infection caused changes in PD permeability as well as PD callose deposition. Collectively, our data support the idea that the control of PD permeability is integrated into innate immune response and that this process is mediated by PDLP5.
DISCUSSION
Localization of PDLP5 within the Central Region of PD
It was predicted that PDLP5, as a transmembrane protein, may distribute along the endoplasmic reticulum or plasma membrane of the PD. To our surprise, PDLP5 localizes to the central part of the PD channels, similar to TMP (Figure 4). Intriguingly, however, fluorescently tagged PDLP5-mRFP and TMP-GFP did not show a complete overlap of their signals when examining their colocalization pattern in either the face-on view of PD pit fields at the epidermal and cortex cellular junctions or the PD at the epidermal junctions in hypocotyl tissue (Figure 5).
We speculate that multiple factors might be responsible for this apparent discrepancy. For example, even though both PDLP5 and TMP associate with PD, characteristics determining their positioning at the ultrastructural level may be unique, so that they occupy different subdomains of PD. Simple PD can undergo structural modifications over time, forming complex PD with various morphologies (Ehlers and Kollmann, 2001; Faulkner et al., 2008b; Burch-Smith et al., 2011), providing potentially heterogeneous binding sites. With regard to this point, it is also possible that the lack of complete overlap has to do with specific types of PD at different cell junctions that are differentially preferred by PDLP5 or TMP whether they target the same or different subdomain(s). An alternative possibility is that PDLP5 and TMP simply compete for the same sites such that one is excluded from the site if the other gets to that site first.
Given the similarity between PDLP5 and TMP in terms of their PD localization, it is tantalizing to speculate as to whether PDLP5 also moves between cells like TMP. Trafficking of PDLP5 might provide a mechanism to induce closure of PD or to enhance innate immune responses in neighboring cells. Likewise, one could reason that the range of PDLP5 activity might be controlled by confining PDLP5 to a specific subdomain of PD, limiting its function to the local tissues where its expression is induced. However, our examination performed by a biolistic DNA delivery, in which over 100 transformed Arabidopsis epidermal cells were analyzed, showed that PDLP5 does not have the capacity to traffic cell to cell (see Supplemental Figure 9 online).
If PDLP5 and TMP indeed occupy the same PD subdomain, this would raise other interesting questions: What is the molecular basis that supports association of PDLP5, a transmembrane protein (unlike TMP) to a particular subdomain of PD? Do all PDLP members localize to the same site within PD? What is special about the median cavity area? Addressing these questions would help substantiate the functional significance of the association of PDLP5 with the central region of PD. Future investigation aided either by dual immunogold labeling or superresolution fluorescent microscopy techniques, which have recently been developed to allow nanoscale imaging of cells (Bates et al., 2008; Chi, 2009; Lippincott-Schwartz and Patterson, 2009), might be helpful to discern between those possibilities.
Regulation of PD Permeability by PDLP5
Our findings that PDLP5 impacts both gated and nongated PD trafficking with correlative callose accumulation suggest that PDLP5 could control PD permeability via the recruitment of callose. This interpretation is consistent with the aniline blue staining and dye-loading results of the pdlp5-1 mutant, in which reduced PDLP5 lowered the callose deposition and promoted PD trafficking activity. It could be that PDLP5 accumulation at PD elicits a change in the microenvironment at the plasma membrane lining or extracellular matrix, an event that then generates secondary messengers or chemicals to directly affect the activities of callose synthases or hydrolases (Brownfield et al., 2009; Zavaliev et al., 2011). It is also possible that the defense or cell death signaling induced by PDLP5 overexpression could have reinforced the callose deposition by way of SA amplification; in fact, multiple isoforms of putative callose synthases have been shown to be upregulated by SA (Dong et al., 2008).
Apart from the role of callose, we speculate that the specific localization of PDLP5 within the central region of PD could provide an important clue as to how PDLP5 controls PD gating. A simple model can be drawn on the basis of a physical clogging, which predicts that accumulation of PDLP5 will cause obstruction of PD channels. With this model, however, the specificity is an issue because not all proteins that accumulate at PD have a negative effect on PD permeability. Given the targeting of TMP to the seemingly similar location and the opposite effects of PDLP5 and TMP on PD permeability, it is tempting to speculate whether this region may provide a structural platform that acts in modulation of PD size exclusion limits. For example, we could hypothesize that the central site might serve as a diaphragm of the PD. In this model, the capacity to open or close the diaphragm is attributed to the characteristics of specific proteins that associate with the site to bring about a desired effect.
The overall pattern and characteristics of PDLP5 expression seem to indicate that it is intended as a PD-gating control mechanism meant to close PD during a defense response. However, a reduction in PD permeability stimulated by the PDLP5 accumulation and accompanying PD callose deposition may not lead to a complete blockage of PD. We argue that two lines of circumstantial evidence provide support for the latter notion. One is that the overexpression of PDLP5 led to a reduction in PD permeability but not a full inhibition (Figure 6; see Supplemental Figure 7 online). Second, the effect of PDLP5 on viral MPs was not uniformly potent. When the movements of CMP and TMP beyond two cell layers were compared, there was ±20% difference in pdlp5-1 or 35S:PDLP5 from the wild type for TMP versus ±5% for CMP (Figure 6L). Furthermore, our analyses clearly indicated that the influence on TMP movement was statistically significant but that on CMP movement was not. Besides, our study points out that trafficking capacities or characteristics of CMP and TMP were distinct even in the wild-type Arabidopsis background: CMP movement was predominantly limited to a single cell layer and showed the least capacity among the three proteins tested (i.e., GFP, TMP, and CMP) to traffic two or more cell layers. By contrast, TMP exhibited the highest percentage of extensive movement in wild-type plants. Differential trafficking behavior of these two MPs, however, is consistent with other observations from independent studies (Cooper and Dodds, 1995; Cooper et al., 1996; Kragler et al., 2000; Murphy and Carr, 2002; Lee et al., 2003; Su et al., 2010). Therefore, we speculate that the differential effect of PDLP5 on TMP and CMP likely stems from their intrinsically different molecular characteristics and mechanisms to pass through PD.
Our results showing that the movement of TMP, but not CMP, was affected by PDLP5 indicated also that these viral proteins might have a distinct capacity to interfere with or reverse the effect of PDLP5 on PD. In our study, we found that the systemic accumulation of TMV was decreased by overexpression of PDLP5 in N. benthamiana. This impact on TMV systemic movement could be interpreted as the result of the inhibitory effect of PDLP5 on TMP movement. Alternatively, it is possible that TMV spread was affected indirectly by PDLP5-induced accumulation of SA. SA treatment of tobacco has been shown to inhibit TMV spread (Murphy and Carr, 2002). Here again, these authors found that CMV infection was not affected by SA. A recent study by Amari et al. (2010) demonstrated that the viral tubule formation by GFLV MP at PD is dependent on PDLPs. Moreover, the infection of the tubule-forming GFLV was unexpectedly aided by the interaction between GFLV MP and PDLPs, whereas the infection of Oil-seed rape mosaic virus, a nontubule virus and close relative of TMV, was not affected in pdlp1/2/3 triple mutant (Amari et al., 2010). Perhaps different plant viruses exploit the same endogenous component to different ends within a plant host and, thus, PDLP members, for example, have varying effects on infection hindrance or spread, depending on the virus.
Role of DUF26 Proteins in Biotic and Abiotic Stresses
The DUF26 domain is unique to plant systems and constitutes a large gene family: ~100 and over 250 DUF26-containing genes are encoded by Arabidopsis and rice (Oryza sativa), respectively (Chen, 2001). The findings presented in our study together with previously published data (Czernic et al., 1999; Du and Chen, 2000; Ohtake et al., 2000; Chen et al., 2003; Acharya et al., 2007; Sawano et al., 2007; Zhang et al., 2009) support the possibility that the DUF26-containing proteins might have evolved to cope with various biotic and abiotic stresses in plants. For example, several DUF26-containing CRKs have been implicated in SA-dependent defense pathways, oxidative stress, and resistance to bacterial pathogens in Arabidopsis (Czernic et al., 1999; Du and Chen, 2000; Ohtake et al., 2000; Chen et al., 2003; Acharya et al., 2007). Overexpression of these CRK isoforms was shown to induce accumulation of SA and HR-like cell death (Chen et al., 2003; Acharya et al., 2007) similar to PDLP5. A secreted DUF26 domain–containing Cys-rich secretory protein isolated from the endosperm of Ginko seeds, ginkbilobin-2 (Gnk2), has been reported to exhibit a novel antifungal effect (Sawano et al., 2007). Another secreted DUF26 protein has been implicated in salt stress response in rice (Zhang et al., 2009), extending the role of the DUF26 domain to abiotic stresses.
Although molecular mechanisms by which PDLP5 and CRKs bring about defense responses are not yet known, important structural and functional insights into the DUF26 domains were recently provided (Miyakawa et al., 2009). The crystal structure of Gnk2 revealed that the core DUF26 together with the N-terminal extension formed compact single-domain architecture with an α+β fold, which provided two surface areas: two α-helices on one side and one antiparallel β-sheet on the opposite side. This fold was apparently stabilized by three intramolecular disulfide bridges involving six conserved Cys residues. In terms of the structure and function relationship of Gnk2, the authors postulated that positively charged α-helix on one surface could associate with negatively charged fungal cell surface for antifungal activity. They also proposed to define the Gnk2 homologous sequence as a new functional protein module. Consistent with this idea, the sequence analysis of PDLP5 showed that the extracellular domain, which constituted over 80% of the protein, contained two repeats of extended DUF26 sequences similar to Gnk2. Considering the tertiary structure of the Gnk2, it is tempting to speculate that the extracellular domain of PDLP5 might provide a potential interface for interacting with specific cell wall proteins. For example, it is conceivable that this domain might interact with a specific component of callose synthases or glucanases to affect their activities if PDLP5 has a direct effect on callose synthesis.
PDLP5, a Potential Molecular Link between PD Regulation and Innate Immunity
In this study, we presented experimental evidence showing that PDLP5 is localized within PD channels and modulates PD permeability and defense signaling. Similar to many known disease resistance genes, PDLP5 expression is highly inducible by bacterial infection and SA application. Likewise, overproduction of PDLP5 leads to spontaneous cell death and chlorosis via stimulation of SA production, compromising overall plant fitness.
It is quite intriguing how a PD-localized protein could bring about such defense signaling responses. Whether these phenotypes are direct or indirect consequences of PDLP5 accumulation at PD remains to be resolved. However, it is notable that the cellular and morphological phenotypes manifested by 35S:PDLP5 plants apparently require PD association of the protein (J.-Y. Lee, unpublished data). Thomas et al. (2008) showed that TMD is necessary for PD targeting of PDLP1, but whether this domain is essential for PDLP1 function was not reported. We found that although the TMD sequences of these two isoforms were not highly conserved (see Supplemental Figure 2B online), TMD was required for PD association of PDLP5, consistent with the role of this domain in PD association as shown by Thomas et al. (2008). However, overexpression of a mutant lacking TMD did not mimic the cellular and morphological phenotypes (e.g., growth retardation and spontaneous lesion formation) manifested by overexpression of full-length PDLP5, indicating that the targeting of PDLP5 to PD was important for those PDLP5 overexpression phenotypes (J.-Y. Lee, unpublished data). Nonetheless, a systemic mutagenesis and characterization will be necessary in future investigations to map the exact functional domains and/or motifs of PDLP5. These studies could help answer interesting questions about the significance of PD localization for PDLP5 to perform its biological function in defense signaling.
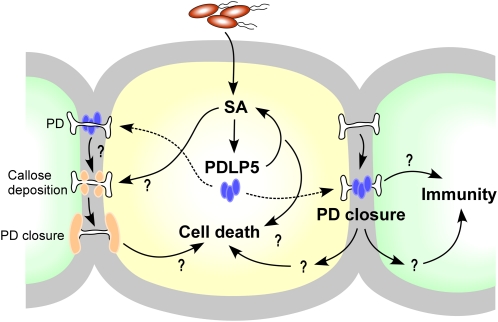
It has been shown by various analyses, including gene expression studies, that a shift from normal growth to defense mode alters both biochemical and physiological processes in an affected plant (Wang et al., 2005; Kwon et al., 2008). While undergoing such changes, a decision needs to be made at a tissue level that determines the fate of infected and healthy cells, killing some to deter the spread of pathogens and allowing the survival of others to prepare for transmitting systemic signals. Under this scenario, effective, local cell communications are inevitable. With this view, we propose a working model that the accumulation of PDLP5 at PD upon microbial infection recruits downstream molecules that feed into an SA amplification pathway, which in return will prepare the cells for death and defense responses (Figure 12). Concomitantly, PDLP5 accumulation at PD, either directly or indirectly recruiting callose, constricts PD, eventually leading to the loss of cell–cell coupling. Enhanced SA production also might indirectly help augment the callose level at PD. These responses might help the affected cells to isolate themselves from adjacent cells and safely execute death. In the neighboring healthy cells, signals that are transmitted from the infected cells prior to their isolation or generated by disconnection from the infected/dying cells via PDLP5 at PD might lead to immunity. The exact molecular mechanisms controlling how these responses are brought about by PDLP5 are under investigation.
Figure 12.
A Model Illustrating the PDLP5-Mediated Crosstalk between the Control of Cell Communication and Innate Immunity during Bacterial Infection.
PDLP5 is induced by bacterial infection in an SA-dependent manner. PDLP5 protein (blue ovals) is targeted to and accumulates at PD, which is indicated by broken arrows. The cell undergoing death owing to a hyper-SA production via the feedback amplification induced by PDLP5 is colored yellow to differentiate from healthy neighboring cells. Potential but unknown pathways or signals are denoted by question marks.
[See online article for color version of this figure.]
METHODS
Plants
Arabidopsis thaliana plants were grown in controlled environment growth chambers at 22°C under 16-h-light conditions. Seedling culture was performed under sterile conditions on agar plates supplemented with 0.5× Murashige and Skoog (MS) salts. Transgenic plants were produced by the floral dip method (Clough and Bent, 1998) in wild-type ecotype Columbia-0. T-DNA insertion line (SAIL_46_E06.v1) was obtained from the ABRC.
Plasmid Constructs
Fluorescently tagged fusion constructs were produced using the pdFN(C) vector system (Ben-Nissan et al., 2008). To clone the PDLP5 open reading frame (ORF), cDNAs were synthesized from 5 μg total RNA prepared from 10-d-old Arabidopsis seedlings using M-MLV reverse transcriptase (New England Biolabs) and subsequently PCR amplified using a high-fidelity DNA polymerase, Phusion (New England Biolabs), and PDLP5 ORF primers containing restriction enzyme sites (see Supplemental Table 2 online). To produce PDLP5-GFP, PDLP5 ORF DNA fragment and the vector plasmid (pdGC) were ligated through XhoI and SalI sites. PDLP5-GFP was then subcloned into pMLBart binary vector plasmid using the NotI site. pPDLP5pro:GUS was constructed by subcloning the entire 5′ upstream sequence (3335bp) of PDLP5 ORF, which was PCR amplified using PDLP5pro Fw and Rv primers (see Supplemental Table 2 online) into pRITA-GUS. Estradiol inducible constructs were subcloned into pER8 vector using XhoI and blunt-ended SpeI.
Cell Wall Preparation from Arabidopsis Seedlings
Clean cell wall proteins were prepared from Arabidopsis seedlings grown in liquid culture media supplemented with Gamborg salt (Sigma-Aldrich) for 7 to 9 d. Seedlings were homogenized in cold H buffer containing 100 mM Tris-HCl, pH 8.0, 100 mM KCl, 10 mM EDTA, 10% glycerol, and 1× protease mix containing 5 μg/mL Leupeptin (USB), 10 μg/mL Aprotinin (Fisher Scientific), and 1 mM PMSF(Amresco). Samples were examined under a phase-contrast microscope for a desired purity.
Protein Identification by Mass Spectrometry Analysis
Cell wall proteins were extracted using Tris-buffered phenol, pH 8.0, precipitated with 0.1 M NH4OAC made in methanol, and washed with 80% acetone. Peptides were extracted, following reduction, alkylation, in-gel digestion with trypsin, from 12% SDS-PAGE gels (30 μg of cell wall proteins per lane) (Shevchenko et al., 2006). The peptide extracts resuspended in 15 to 20 μL 5% formic acid were analyzed by data-dependent tandem mass spectrometry using an online LC-LTQ-Orbitrap (Thermo Electron) (Olinares et al. 2010). Based on the mass spectrometry data, peak lists were generated and searched with Mascot v2.2 (Matrix Science) against The Arabidopsis Information Resource ATH database v8 as described (Fiala, 2005).
Antibody Production and Purification
PDLP5-specific peptide polyclonal antibodies were produced by immunizing rabbits (Covance) with the peptide derived from amino acid residues 289 to 297. IgGs were purified from the sera through an affinity column produced by sephadex beads conjugated with protein G (Pharmacia). GFP polyclonal antibodies were raised against 5 mg pure GFP produced in Escherichia coli. The sera bled from rabbits immunized with GFP were affinity purified against a GFP column prepared by cross-linking pure GFP, prepared according to Cristea et al. (2005), to UltraLink sepharose beads (Pharmacia).
Confocal Microscopy Imaging
For confocal microscopy, intact plant samples were placed in a single-welled Nalge Nunc chambered cover glass system and imaged on a Zeiss AxioObserver inverted light microscope using either the LM 780 or LSM 510 META scanhead on a Zeiss LSM 5 DUO confocal microscope. Targeted plant structures were documented with either a ×10 Plan-Apochromat (0.45 numerical aperture) or ×40 C-Apochromat (1.2 numerical aperture) water immersion objective lens with appropriate laser and filter combinations as follows: 488-nm laser line with a 500- to 550-nm band-pass emission filter (for enhanced GFP or CFDA); 636- to 754-nm emission wavelengths with META detector (for chloroplast autofluorescence); the 561-nm laser line with a 575-to 615-nm band-pass emission filter (for mRFP-Cherry); and 405-nm laser excitation and a 505-nm long-pass emission filter for aniline blue. All multilabeled images were acquired in fast-line switch mode to eliminate crosstalk. A series of optical sections were acquired as Z-stacks and rendered as 3D maximum intensity projections with Zeiss LSM 510 AIM software (Rel. 4.2).
Correlative Light Electron Microscopy
Arabidopsis seedlings were fixed in the dark in 4% paraformaldehyde and 0.1% glutaraldehyde in 0.2 M Sorensen’s phosphate buffer, pH 7.2, overnight at 4°C and washed and infiltrated with a gradient of 2.3 M sucrose in 0.1 M Sorensen’s phosphate buffer. Cryoprotected seedlings were embedded in TFM tissue freezing medium embedding matrix and frozen at −30°C in a Leica CM3050 S cryostat. Fifty-micron sections of hypocotyl were collected onto cold adhesive Permanox microscope slides using the CryoJane tape-transfer system. Following confocal mapping, samples were processed for TEM and embedded in Embed-812 resin, followed by serial sectioning on a Reichert-Jung Ultracut E ultramicrotome. Sections were poststained with Reynolds’ lead citrate and methanolic uranyl acetate, imaged with a Zeiss Libra 120 Plus TEM at 120 kV, and digital images were acquired with a Gatan Ultrascan 1000 2k × 2k CCD camera. A subset of serial section micrographs were aligned with Reconstruct (Fiala, 2005) and then modeled with IMOD (Kremer et al., 1996). Confocal-TEM and confocal-model overlays were generated in Adobe Photoshop CS3.
Immunogold Electron Microscopy
Arabidopsis seedlings were fixed and processed as described above for correlative fluorescence microscopy. Ultrathin sections were collected onto 200-mesh formvar-carbon coated nickel grids. Antigen retrieval treatment was performed basically as described (Stirling and Graff, 1995). Briefly, grids were incubated on a saturated aqueous solution of sodium metaperiodate for 1 h at room temperature, washed on eight drops of water, and then exposed to 10 mM sodium citrate buffer, pH 6.0, heated to 95 to 100°C for 10 min and washed on eight drops of water. Following antigen retrieval, samples were blocked for 1 h in filtered 5% nonfat milk buffer in 1× TBS and then incubated with primary antibody, polyclonal α-GFP (see the Antibody Production and Purification section of Methods) or a commercial α-GFP (catalog number ab6556; Abcam), diluted 1:200 in block for 2 h at room temperature. Grids were washed on eight drops of 1× TBS containing 0.05% Tween 20 (TTBS) and incubated in a 1:20 dilution of Aurion goat anti-rabbit 6-nm gold-conjugated secondary antibody in block for 1 h at room temperature. Samples were washed on eight drops of TTBS followed by 12 drops of water, air-dried, poststained with 0.5% aqueous uranyl acetate for 5 min, washed on five drops of water, dried, and imaged by TEM. Secondary antibody alone was included as a control. Specificity of α-GFP was tested on wild-type plant materials.
PD Permeability Assays
DANS dye loading and biolistic DNA delivery assays were performed on 3- or 4-week-old plants grown in 12-h-light conditions. DANS dye loading assay was performed by loading 1 μL drop of 1 mM CFDA (Sigma-Aldrich) on the adaxial side of intact rosette leaf and incubating for 5 min before withdrawal. Subsequently, the leaf was cut and mounted on a glass slide for a confocal imaging using the Fluar ×5/0.25 objective. The extent of dye diffusion was quantified by measuring diameters of diffusion area with respect to the fluorescent intensity distribution tool. Biolistic DNA delivery was performed as described elsewhere (Ben-Nissan et al., 2008). Statistical difference among samples was analyzed using analysis of variance (ANOVA) test in conjunction with Fisher’s LSD test.
Callose Staining and Quantification
Mature leaves of 3-week-old Arabidopsis were cleared with ethanol followed by staining in 0.1% aniline blue (Acros). Callose quantification was performed using Volocity 3.1.1 (Perkin-Elmer) by automatically extracting PD-associated puncta based on size and a background threshold and plotting fluorescence intensity per unit area.
RT-PCR and Quantification
Total RNA was isolated using Trizol (Invitrogen) from leaf tissues collected from 3- to 4-week-old Arabidopsis. For RT-PCR, cDNA was synthesized using M-MuLV reverse transcriptase (New England Biolabs) from 1 μg of RNA, followed by PCR amplification using DyNAzyme II DNA polymerase (DyNAzyme) for 20 cycles. Ethidium bromide–stained PCR products were visualized by Alpha Imager HP and quantified by densitometric analysis with Image J software. All samples were normalized against their respective UBQ or NbACT band intensities as a control. At least three biological plus three technical repeats were performed for each experiment. Statistical significance was analyzed with ANOVA test.
Pathogen Infection
For viral infection study, 3-week-old Nicotiana benthamiana plants were coagroinfiltrated with Agrobacterium tumefaciens GV3101 transformed with 35S:PDLP5 or empty vector plasmid mixed with GV3101 carrying p19 (Voinnet et al., 2003) adjusted to OD600 = 1.0. Transformed agrobacteria were cultured at 28°C and resuspended in infiltration buffer containing 10 mM MES, pH 5.7, 10 mM MgCl2, and 200 μM acetosyringone. After 3 d of incubation, the infiltrated leaves were either infected with TMV-GFP by performing a second agroinfiltration with GV3101 carrying TMV-GFP adjusted to OD600 = 0.02 or rub-inoculated with CMV containing extract (10 μL per leaf). To detect PDLP5 expression at the time of viral infection, total RNA of N. benthamiana leaves were extracted using Trizol reagent. RT was performed with 1 μg total RNA and M-MuLV reverse transcriptase (New England Biolabs). N. benthamiana ACTIN (Nb ACT) and PDLP5 DNA fragments were PCR amplified using Genscript Taq polymerase and the following gene-specific primer sets: 5′-ATGGCAGACGGTGAGGATATTCA-3′ and 5′-GCCTTTGCAATCCACATCTGTTG-3′ as forward and reverse primers, respectively, for Nb ACT (Zhang et al., 2008) and 5′-CCGCTACGCCAACTTCACAG-3′ and 5′-CTTCTCTCCATGACCAAAGT-3′ as forward and reverse primers, respectively, for full-length PDLP5. PCR reactions were performed using a Bio-Rad DNA Engine under the following conditions: 94°C for 5 min (one cycle); 94°C for 30 s, 59°C (for Nb ACT) or 55°C (for PDLP5) for 45 s, and 72°C for 90 s (22 cycles). CMV was prepared from infected pumpkin tissue by grinding the tissue in 10 mM phosphate buffer, pH 7.2. To detect CMV CP DNA fragment, RT and PCR were performed using CP gene-specific RV1 (5′-CACGACTGACCATTTTAGCC-3′) and a pair of FW1 (5′-GATAAGAAGCTTGTTTCGCGCA-3′) and RV2 (5′-AAGCTGGATGGACAACCCGTTC-3′) primers, respectively. RV1 and RV2 are nested reverse primers. For normalization of the RT-PCR, Nb ACT was PCR amplified from cDNAs synthesized in RT using oligo(dT). At least three biological and two technical repeats were performed for each time point. Student’s t test was employed to analyze statistical difference between samples.
Bacterial strains were derived from Pma ES4326 (Guttman and Greenberg, 2001). For bacterial growth assay, 25-d-old plants, grown in an Enconair growth chamber with 12-h-light/12-h-dark cycle were infected with Pma ES4326 (OD600 = 0.0001), Pma avrRpt2 (OD600 = 0.0004), or Pma avrRpm1 (OD600 = 0.0004) through infiltration and incubated for 3 d before bacterial measurement. Alternatively, 21-d-old plants, grown in a Conviron growth chamber with 16-h-light/8-h-dark cycle were infiltrated with Pma (OD600 = 0.001), and bacterial growth was measured at 2 DAI. Leaf discs from the infected fifth to seventh leaves were taken for bacterial growth measurement and analysis as described previously (Lu et al., 2003). Statistical difference between samples was analyzed with Student’s t test.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers AT1G70690, AT2G33330, AT2G14610, and AT3G62250.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MS/MS-Based Identification of At1g70690.1 and At2g33330.
Supplemental Figure 2. Sequence Comparison of PDLP1, PDLP3, and PDLP5.
Supplemental Figure 3. Confirmation of T-DNA Insertion and Near-Null Expression of PDLP5 in SAIL_46_E06 line (pdlp5-1).
Supplemental Figure 4. PDLP5 Localizes to Punctate Structures at the Cellular Boundaries.
Supplemental Figure 5. PDLP5 Localizes to Primary Pit Fields.
Supplemental Figure 6. Localization of TMP-GFP and PDLP5-GFP within PD.
Supplemental Figure 7. PD Permeability Assays Performed Using Fluorescently Tagged Viral Movement Proteins.
Supplemental Figure 8. Pma Infection Induces PDLP5.
Supplemental Figure 9. PDLP5-GFP Is Cell-Autonomous.
Supplemental Table 1. Members of the PDLP Family.
Supplemental Table 2. List of Additional Primers Used.
Supplemental Movie 1. Animated Optical Z-Sections Showing That PDLP5-GFP Punctate Signals Are Detected Only at the Cross-Walls.
Acknowledgments
We thank B.-C. Yoo for helpful discussions and comments on the manuscript, J. Sherrier for helpful suggestions on TEM techniques, A. Maule for sharing unpublished information and helpful comments, W. Park for cloning PDLP5-GFP, X. Dong for npr1 seeds, N. Chua for pER8 plasmid, J. Caplan for assisting with confocal imaging and providing GV3101 cells carrying TMV-GFP and p19, M. Gilbertson for providing CMV, and L. Crozier and S. Kitto for proofreading. Publically available Arabidopsis mutant lines were obtained from ABRC. The funds supporting this work were provided by the National Science Foundation (IOS 0954931), the National Institutes of Health (COBRE P20 RR15588), the University of Delaware Research Initiative Strategic Initiative Program to J.-Y.L., and the University of Maryland Baltimore County (a start-up fund) and National Science Foundation (RIG-0818651) to H.L.
AUTHOR CONTRIBUTIONS
J.-Y.L. designed research. J.-Y.L., X.W., W.C., R.S., S.M., K.C., B.Z., C.Z., and V.L. performed research. J.-Y.L., K.C., K.v.W., and H.L. analyzed data. J.-Y.L. wrote the article.
References
- Acharya B.R., Raina S., Maqbool S.B., Jagadeeswaran G., Mosher S.L., Appel H.M., Schultz J.C., Klessig D.F., Raina R. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50: 488–499 [DOI] [PubMed] [Google Scholar]
- Amari K., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 6: e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M., Huang B., Zhuang X. (2008). Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr. Opin. Chem. Biol. 12: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A.J. (2010). Plasmodesmata: Gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Jackson D. (2009). Redox homeostasis regulates plasmodesmal communication in Arabidopsis meristems. Plant Signal. Behav. 4: 655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nissan G., Cui W., Kim D.J., Yang Y., Yoo B.C., Lee J.Y. (2008). Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 148: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Oparka K.J. (2005). Virus-host interactions during movement processes. Plant Physiol. 138: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Doublin M., Fincher G., Bacic A. (2009). Biochemical and molecular properties of biosynthetic enzymes for (1,3)-β-glucans. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides, Bacic A., Fincher G.B., Stone B.A., (New York: Elsevier/Academic Press; ), pp. 283–326 [Google Scholar]
- Burch-Smith T.M., Stonebloom S., Xu M., Zambryski P.C. (2011). Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma 248: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Zambryski P.C. (2010). Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr. Biol. 20: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Du L., Chen Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53: 61–74 [DOI] [PubMed] [Google Scholar]
- Chen Z. (2001). A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 126: 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi K.R. (2009). Microscopy: Ever-increasing resolution. Nature 462: 675–678 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper B., Dodds J.A. (1995). Differences in the subcellular localization of tobacco mosaic virus and cucumber mosaic virus movement proteins in infected and transgenic plants. J. Gen. Virol. 76: 3217–3221 [DOI] [PubMed] [Google Scholar]
- Cooper B., Schmitz I., Rao A.L., Beachy R.N., Dodds J.A. (1996). Cell-to-cell transport of movement-defective cucumber mosaic and tobacco mosaic viruses in transgenic plants expressing heterologous movement protein genes. Virology 216: 208–213 [DOI] [PubMed] [Google Scholar]
- Cristea I.M., Williams R., Chait B.T., Rout M.P. (2005). Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4: 1933–1941 [DOI] [PubMed] [Google Scholar]
- Czernic P., Visser B., Sun W., Savouré A., Deslandes L., Marco Y., Van Montagu M., Verbruggen N. (1999). Characterization of an Arabidopsis thaliana receptor-like protein kinase gene activated by oxidative stress and pathogen attack. Plant J. 18: 321–327 [DOI] [PubMed] [Google Scholar]
- Ding B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47: 105–131 [DOI] [PubMed] [Google Scholar]
- Ding B., Haudenshield J.S., Hull R.J., Wolf S., Beachy R.N., Lucas W.J. (1992). Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hong Z., Chatterjee J., Kim S., Verma D.P. (2008). Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229: 87–98 [DOI] [PubMed] [Google Scholar]
- Dong X., Mindrinos M., Davis K.R., Ausubel F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Chen Z. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 24: 837–847 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916 [DOI] [PubMed] [Google Scholar]
- Ehlers K., Kollmann R. (2001). Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D., Daniels M.J., Parker J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C., Akman O., Bell K., Jeffree C., Oparka K. (2008b). Peeking into pit fields: A new model of secondary plasmodesmata formation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 150: S140–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C., Akman O.E., Bell K., Jeffree C., Oparka K. (2008a). Peeking into pit fields: A multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., Maule A. (2011). Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C. (2005). Reconstruct: A free editor for serial section microscopy. J. Microsc. 218: 52–61 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Vernooij B., Gaffney T., Morse A., Ryals J. (1995). Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 29: 959–968 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Guttman D.S., Greenberg J.T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant Microbe Interact. 14: 145–155 [DOI] [PubMed] [Google Scholar]
- Haywood V., Kragler F., Lucas W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.): S303–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M., Epel B.L. (2004). Macromolecular transport and signaling through plasmodesmata. Int. Rev. Cytol. 235: 93–164 [DOI] [PubMed] [Google Scholar]
- Hofmann C., Sambade A., Heinlein M. (2007). Plasmodesmata and intercellular transport of viral RNA. Biochem. Soc. Trans. 35: 142–145 [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Hempel F.D., Sha K., Pfluger J., Zambryski P.C. (2002). Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129: 1261–1272 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Otegui M.S., Krishnakumar S., Mindrinos M., Zambryski P. (2007). INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F., Monzer J., Xoconostle-Cázares B., Lucas W.J. (2000). Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 19: 2856–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Kwon C., Bednarek P., Schulze-Lefert P. (2008). Secretory pathways in plant immune responses. Plant Physiol. 147: 1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Cho S.K., Sager R. (2010). Plasmodesmata and non-cell-autonomous signaling in plants. In The Plant Plasma Membrane, Murphy A.S., Peer W., Schulz B., (Heidelberg, Germany: Springer; ), pp. 87–108 [Google Scholar]
- Lee J.Y., Lu H. (2011). Plasmodesmata: The battleground against intruders. Trends Plant Sci. 16: 201–210 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Yoo B.C., Rojas M.R., Gomez-Ospina N., Staehelin L.A., Lucas W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299: 392–396 [DOI] [PubMed] [Google Scholar]
- Lee M.W., Jelenska J., Greenberg J.T. (2008). Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J. 54: 452–465 [DOI] [PubMed] [Google Scholar]
- Levy A., Erlanger M., Rosenthal M., Epel B.L. (2007). A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Patterson G.H. (2009). Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 19: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lu H., Rate D.N., Song J.T., Greenberg J.T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Lee J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5: 712–726 [DOI] [PubMed] [Google Scholar]
- Maule A.J. (2008). Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Miyakawa T., Miyazono K., Sawano Y., Hatano K., Tanokura M. (2009). Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins 77: 247–251 [DOI] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Murphy A.M., Carr J.P. (2002). Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol. 128: 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Ohtake Y., Takahashi T., Komeda Y. (2000). Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 41: 1038–1044 [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., Demesa-Arévalo E., Autran D., Grimanelli D., Slotkin R.K., Martienssen R.A., Vielle-Calzada J.P. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9: 33–41 [DOI] [PubMed] [Google Scholar]
- Radford J.E., Vesk M., Overall R.L. (1998). Callose deposition at plasmodesmata. Protoplasma 201: 30–37 [Google Scholar]
- Roberts A.G., Oparka K.J. (2003). Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 26: 103–124 [Google Scholar]
- Sagi G., Katz A., Guenoune-Gelbart D., Epel B.L. (2005). Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17: 1788–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano Y., Miyakawa T., Yamazaki H., Tanokura M., Hatano K. (2007). Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 388: 273–280 [DOI] [PubMed] [Google Scholar]
- Seagull R.W. (1983). Differences in the frequency and disposition of plasmodesmata resulting from root cell elongation. Planta 159: 497–504 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling J.W., Graff P.S. (1995). Antigen unmasking for immunoelectron microscopy: Labeling is improved by treating with sodium ethoxide or sodium metaperiodate, then heating on retrieval medium. J. Histochem. Cytochem. 43: 115–123 [DOI] [PubMed] [Google Scholar]
- Stonebloom S., Burch-Smith T., Kim I., Meinke D., Mindrinos M., Zambryski P. (2009). Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Liu Z., Chen C., Zhang Y., Wang X., Zhu L., Miao L., Wang X.C., Yuan M. (2010). Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22: 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnsoever C., Oorschot V., Klumperman J. (2008). Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat. Methods 5: 973–980 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Oparka K.J. (1996). The fluorescent probe HPTS as a phloem-mobile, symplastic tracer: An evaluation using confocal laser scanning microscopy. J. Exp. Bot. 47: 439–445 [Google Scholar]
- Zambryski P., Crawford K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16: 393–421 [DOI] [PubMed] [Google Scholar]
- Zavaliev R., Ueki S., Epel B.L., Citovsky V. (2011). Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tian L.H., Zhao J.F., Song Y., Zhang C.J., Guo Y. (2009). Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 149: 916–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ma F., Wang Y., Yang B., Chen S. (2008). Expression of v-cath gene from HearNPV in tobacco confers an antifeedant effect against Helicoverpa armigera. J. Biotechnol. 138: 52–55 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]