Abstract
Double-strand breaks (DSBs), arising from exposure to exogenous clastogens or as a by-product of endogenous cellular metabolism, pose grave threats to genome integrity. DSBs can sever whole chromosomes, leading to chromosomal instability, a hallmark of cancer. Healing broken DNA takes time, and it is therefore essential to temporarily halt cell division while DSB repair is underway. The seminal discovery of cyclin-dependent kinases as master regulators of the cell cycle unleashed a series of studies aimed at defining how the DNA damage response network delays cell division. These efforts culminated with the identification of Cdc25, the protein phosphatase that activates Cdc2/Cdk1, as a critical target of the checkpoint kinase Chk1. However, regulation works both ways, as recent studies have revealed that Cdc2 activity and cell cycle position determine whether DSBs are repaired by non-homologous end-joining or homologous recombination (HR). Central to this regulation are the proteins that initiate the processing of DNA ends for HR repair, Mre11–Rad50–Nbs1 protein complex and Ctp1/Sae2/CtIP, and the checkpoint kinases Tel1/ATM and Rad3/ATR. Here, we review recent findings and provide insight on how proteins that regulate cell cycle progression affect DSB repair, and, conversely how proteins that repair DSBs affect cell cycle progression.
Keywords: MRN complex, Cdc2, non-homologous end-joining, homologous recombination, Ataxia-telangiectasia-mutated, Ataxia-telangiectasia and Rad 3-related
1. Introduction
Preservation of the genome is crucial for the survival and well-being of all organisms. As genomes are under constant assault from exogenous DNA-damaging agents, such as UV light or natural radiation, and endogenous DNA-damaging agents, such as free radicals generated by oxidative metabolism, cells of all organisms are equipped with multiple pathways to recognize and repair DNA damage. One of the most harmful forms of DNA damage is the double-strand break (DSB). A DSB can be induced directly by exposure to, for instance, ionizing irradiation, or indirectly through chemical modifications of the DNA that cause replication fork stalling and collapse in actively cycling cells. In addition, DSBs are deliberately generated in meiotic cells and in lymphocytes during V(D)J recombination and class switch recombination [1]. When left unrepaired, DSBs can cause a plethora of chromosomal aberrations that often result in cell death or mutations that can lead to cancer phenotypes [2].
There are two major pathways that repair DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR). Key NHEJ proteins are conserved from yeast to humans, and include the Ku70–Ku80 heterodimer that binds DNA ends with high affinity, as well as XRCC4-like factor (XLF)/Cernunnos and DNA ligase IV [3,4]. During NHEJ, DNA ends are recognized, captured and brought together by Ku70–Ku80. The Ku heterodimer recruits nucleases (Artemis with DNA-dependent protein kinase catalytic subunit, DNA-PKcs), polymerases (µ and λ) and the ligase complex (XLF with DNA ligase IV). After little to no end processing by the nucleases and polymerases, the ends are directly ligated [5]. While NHEJ is highly efficient, its imprecise nature makes it prone to mutations. NHEJ is active throughout the entire cell cycle, but is the preferred mode of repair during G0, G1 and early S phase [5]. HR, on the other hand, is generally restricted to the late S and G2 phases of the cell cycle, as it usually uses the intact sister chromatid as template for synthesis-dependent repair in mitotic cells [6,7]. HR- or homology-directed repair (HDR) initiates when the Mre11–Rad50–Nbs1/Xrs2 (MRN) complex recognizes and binds the DNA end. The DNA ends on both sides of the DSB undergo 5′ to 3′ resection. Replication protein A (RPA) complex binds the resulting 3′ single-stranded DNA (ssDNA) overhangs. In yeast, Rad52 catalyses the assembly of Rad51 on the ssDNA, thereby displacing RPA [8–10]. The Rad51-covered filament initiates the homology search and catalyses strand exchange to allow the priming of DNA replication to repair the DSB [11,12]. HR is considered to be an error-free pathway, as it mainly uses the homologous sequence of the sister chromatid as a template for repair. Interestingly, DSBs in vertebrate cells are predominantly repaired by NHEJ rather than HR. While it has been postulated that chromosome condensation may make the search for homology extremely difficult, thereby resulting in the preferential use of NHEJ for DSB repair [13], chromosome condensation is not a feature of interphase chromatin. Rather, interphase chromatin is wrapped around histones, which has been thought to act as a barrier for the successful recognition and capture of homology by an invading Rad51-covered filament. Several studies performed in the last 5 years have however shown that wrapping of DNA around histones does not interfere with HR, both in vitro and in vivo [14–16].
Intriguingly, HR is essential for recombination between homologous chromosomes in meiosis, which is required for proper chromosome segregation and generation of genetic diversity. A multitude of human disease syndromes have been traced to NHEJ and HR defects, including those characterized by neurological, immunological and developmental disorders as well as radiation sensitivity, premature ageing diseases and cancer, stressing the importance of NHEJ and HR in maintaining genome stability [17–22].
The preference for NHEJ in G1 phase and HR in S and G2 phases implies that the modes of DSB repair are regulated during the cell cycle. At the same time, cell cycle checkpoints play a critical role in delaying the onset of mitosis until DSB repair is complete, otherwise chromosome fragments that are distal to the DSB would be lost during nuclear division. Here, we describe recent findings and provide insights on how proteins that regulate cell cycle progression affect DSB repair, and, conversely, how proteins that repair DSBs affect cell cycle progression.
2. Regulation of cell cycle progression
In the early 1950s, the first experimental evidence was published showing that plant and animal cells synthesize DNA within a certain limited period during cell division [23–26]. This led to the subdivision of cell division into four distinct phases: the mitotic phase or M, the first gap phase or G1, the DNA synthesis phase or S and the second gap phase or G2. The discovery of cyclin-dependent kinases (CDKs) provided the first clues on how the transitions from initiation of DNA replication and the entry into mitosis are regulated. The identification of Cdc25 in Schizosaccharomyces pombe, the phosphatase that activates Cdc2/Cdk1, propelled the understanding on cell cycle regulation further forward [27]. CDKs are protein Ser/Thr kinases that bind a cyclin to form an active heterodimer. Cyclin–CDK complexes are however kept in an inactive state through inhibitory phosphorylation by Wee1 and Myt1 [28]. Cdc25 dephosphorylates Cdc2/Cdk1 within the activation loop of the kinase domain to achieve full activity of the cyclin–CDK complex [28]. In S. pombe and Saccharomyces cerevisiae a single CDK, Cdc2 and Cdc28, respectively, triggers both the G1 to S and G2 to M transition. While many CDKs exist in mammalian cells, it appears that the Cdc2 homologue Cdk1, which is necessary for the onset of mitosis and can interact with all cyclins, can solely drive the essential mammalian cell cycle in culture [29]. The lack of Cdk1 results in embryonic lethality in mice, indicating that the ability of other CDKs to compensate for Cdk1 is incomplete [29]. The reverse is also true, as triple Cdk2 Cdk4 Cdk6 mutants lacking all interphase CDKs show embryonic lethality [29].
3. How to stall the cell cycle after DNA damage
Cells are under constant attack by DNA-damaging agents that interfere with the faithful transmission of genetic information when a cell divides. As it takes time to repair broken or damaged DNA, it is essential that cycle progression can be temporarily stalled. In the late 1980s, surveillance mechanisms that are capable of delaying the cell cycle in the presence of DNA damage were identified [30–32]. These mechanisms are now referred to as checkpoints. Mammalian cells have three major DNA repair checkpoints: G1/S, intra-S and G2/M [33], whereas fission yeast appears to have only two: the intra-S and G2/M checkpoints. In a simplified model of checkpoints, four groups of proteins can be identified: damage sensors, signal mediators, signal transducers and effectors. In the case of DSBs, it appears that the MRN complex serves as the main sensor, as it recognizes and locates to the DSB during all stages of the cell cycle [34,35]. MRN subsequently recruits the Ataxia-telangiectasia-mutated (ATM, or Tel1 in yeast) checkpoint kinase through binding to the Nbs1 subunit [36–38]. ATM belongs to the phosphoinositide-3 kinase-related protein kinase (PIKK) family of kinases and activates checkpoint signalling by phosphorylating downstream targets. Despite the fact that the MRN complex is required for recruitment of ATM in all species, the function of ATM at DSBs is not conserved. In yeast, ATM is not required for DSB repair but is primarily involved in telomere maintenance [39]. Two other members of the PIKK family kinases that play a role in checkpoint signalling are ATM- and Rad3-related (ATR, or Rad3 in S. pombe) and DNA–PKcs. Rad3 is recruited to sites of damage by binding of its interaction partner Rad26/ATRIP (ATR-interacting protein) to RPA-coated ssDNA [40]. In yeast, it is Rad3 rather than Tel1 that is responsible for the DNA damage signalling upon DSB detection [41,42]. DNA–PKcs, on the other hand, for which no homologue is identified in yeast, interacts with the Ku heterodimer, another DNA damage sensor that recognizes and binds to DSBs [43,44]. Activation of ATM, ATR and DNA–PKcs depends on their recruitment to sites of damage that is mediated through a conserved motif found in Nbs1, ATRIP and Ku80, respectively [37].
The PIKK kinases serve as transducers of the damage signal, ultimately phosphorylating and activating the downstream effector kinases: checkpoint kinases 1 and 2 (Chk1 and Cds1 in S. pombe). The relay of the signal from transducer to effector kinases is facilitated and enhanced by mediator proteins [45,46]. In S. pombe, a critical mediator for Chk1 activation is Crb2, as Crb2-deficient cells are unable to activate the G2/M checkpoint [47,48]. Two types of histone modifications ensure that Crb2 localizes to sites of damage: a tandem BRCA1 carboxyl terminal (BRCT) domain in Crb2 binds to C-terminal phosphorylated histone H2A (γ-H2A) [49,50], while a tandem Tudor domain binds to dimethylated lysine 20 on histone H4 (H4-K20me2) [51,52]. Most mediator proteins do not strictly depend on γ-H2A/H2AX for their recruitment to DSBs as they have additional interactions with proteins that localize at DSBs, rather, binding to γ-H2A/H2AX is required for the large scale and prolonged presence of these proteins at the break [53]. Interestingly, the transducer kinases ATM, ATR and DNA-PK (consisting of DNA-PKcs and the Ku heterodimer) are responsible for phosphorylation of H2A/H2AX [49,54,55]. A second factor required for Rad3-dependent activation of Chk1 is a protein complex consisting of Rad9–Rad1–Hus1 (the 9-1-1- complex or DNA damage checkpoint clamp) [56–59]. Interestingly, the loading of this protein complex at DNA lesions is enhanced by an interaction with RPA-coated ssDNA, similar to Rad3–Rad26 [60,61].
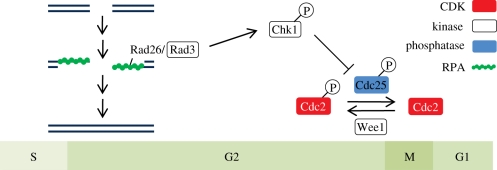
The actual cell cycle arrest is imposed by the effector kinases Chk1 and Cds1. In fission yeast, Chk1 is activated in G2, whereas Cds1 is activated in response to stalled replication forks during the S phase [33]. Studies in fission yeast have identified Cdc25 as a key target for Chk1 and Cds1 [62]. Phosphorylation of active Cdc25 by Chk1 and Cds1 was shown to inhibit Cdc25 activity [63]. In the absence of Cdc25, the inhibitory phosphorylation on Cdc2 is not removed and the cell cycle arrests. A representation of the interactions between RPA, PIKK and checkpoint kinases, Cdc25 and Cdc2 in S. pombe is shown in figure 1.
Figure 1.
Homologous recombination (HR) repair activates the DNA damage checkpoint. Double-strand breaks formed in S or G2 phases are processed for HR repair. Resection by nucleases generates 3′ ssDNA overhangs that are bound by RPA, which recruits the Rad3–Rad26 checkpoint kinase in Schizosaccharomyces pombe. Rad3 phosphorylates and thereby activates the checkpoint kinase Chk1. This event requires the Rad9–Rad1–Hus1 checkpoint clamp and Crb2 mediator proteins (not shown in the figure). Chk1 phosphorylates and thereby inactivates the mitotic inducer Cdc25, which is a protein phosphatase that activates Cdc2. Cdc2 is kept in an inactive state by Wee1 protein kinase. Inhibition of Cdc25 by Chk1 delays the onset of mitosis, which provides time to complete DSB repair. See main text for references.
4. Ku and Mre11–Rad50–Nbs1: more than signalling
The first protein complexes that sense or recognize DSBs are the Ku heterodimer and the MRN complex. Besides initiating the activation of the DNA damage checkpoint, these protein complexes are required for the actual repair of DSBs by NHEJ and HR, respectively. As mentioned earlier, Ku recruits DNA–PKcs in mammalian cells to form DNA–PK and initiate checkpoint signalling. In addition, Ku recruits the XRCC4/DNA ligase IV complex that ligates the ends together [64–67]. This step is stimulated by XLF/Cernunnos [68,69]. Ku and the homologues of DNA ligase IV and XLF are required for DSB repair by NHEJ in all species [5,70–73], suggesting a high conservation of the mode of action of NHEJ repair. Interestingly, DNA–PKcs not only activates checkpoint signalling but is also thought to function as a bridging factor by bringing the two DNA ends of a DSB to close proximity [74]. Yeast cells lack DNA–PKcs, suggesting that the DNA end-bridging function is performed by another protein. The Mre11–Rad50–Xrs2 (MRX) complex may perform this function in budding yeast [75]. Curiously, MRN is not required for NHEJ in S. pombe [71], leaving open the bridging factor that brings the DNA ends together for NHEJ.
The first essential step in HDR of DSBs is the 5′ to 3′ resection of the DNA ends. Current models, which are largely based on studies performed with S. cerevisiae, suggest that resection is a two-step process that can be divided into resection initiation and resection extension. The MRN complex is required for the first step: initiation of resection [76,77]. While Mre11 has ssDNA endonuclease and 3′ to 5′ dsDNA exonuclease activities in vitro [78], these activities are not actually required for resection in budding yeast unless the exonuclease 1 (Exo1) and Sgs1–Dna2 proteins required for extended resection are absent (see below; [79–81]). Mre11 nuclease mutants in budding yeast are modestly sensitive to DNA-damaging agents such as γ-irradiation and methyl methanesulphonate, whereas the analogous nuclease-deficient mutant in fission yeast is highly sensitive to the same DNA-damaging agents [82–84], although in neither species are they as sensitive as Mre11 null mutants. This is in contrast to a study performed in mouse embryonic fibroblasts (MEFs), which indicated that Mre11 nuclease mutants phenocopy Mre11 deficiency [85]. While studies of Mre11 nuclease mutants in S. pombe have revealed a role for Mre11 nuclease activity in the processing of DNA ends that are covalently bound by protein [86–88], the function of Mre11 nuclease activity in the repair of DSBs arising from γ-irradiation remains a mystery.
The initiation of resection also involves the Sae2 protein in budding yeast [76,77]. However, as seen for mutants defective in Mre11 nuclease activities, the resection and DSB defects of sae2Δ mutants are modest in comparison with mrxΔ mutants, which lack one of the subunits of the MRX complex [80]. The contribution of Sae2 to resection becomes clear when the Exo1- and Sgs1-dependent activities required for extended resection are eliminated. In support of its role in resection, Sae2 was shown to have nuclease functions in vitro [89]. Schizosaccharomyces pombe Ctp1 and mammalian CtBP interacting protein (CtIP) share sequence similarities to Sae2 and are presumed to be Sae2 orthologues, although it is unknown whether they share the nuclease activities detected with Sae2. In contrast to sae2Δ mutants, which are only weakly sensitive to most DNA-damaging agents, S. pombe cpt1Δ cells are acutely sensitive to DNA-damaging agents, showing phenotypes equivalent to mrnΔ mutants [90–92]. The reasons for these species differences in the requirements for Sae2 versus Ctp1 are unknown, but they suggest that Ctp1 may be critical for resection in an otherwise wild-type background. This prediction is supported by chromatin immunoprecipitation (ChIP) studies showing that RPA localization at a site-specific DSB is strongly diminished in cpt1Δ cells [90]. It will be interesting to quantitatively measure resection of DNA ends in vivo in fission yeast to determine the effects of Ctp1 and MRN deletion on resection. Interestingly, siRNA experiments in mammalian cells showed that the formation of RPA foci is diminished when CtIP is knocked down [93], which indicates that CtIP may be required for efficient resection. From these studies, it appears that both Ctp1/CtIP may be critical for processing of DSBs in fission yeast and mammals, whereas Sae2 is less crucial in budding yeast, perhaps because alternative activities can more effectively substitute for Sae2. Curiously, Ctp1 and CtIP are recruited to DSBs through interaction with the Nbs1 subunit of the MRN complex [90,93], while Sae2 does not require the MRX protein complex to localize to DSBs [34]. CtIP localization at DSBs also requires an interaction with the tumour suppressor protein breast cancer 1 (BRCA1), which does not exist in budding or fission yeasts [94,95].
The second phase of resection that extends ssDNA formation several kilobases from the break involves the exonuclease Exo1 or the DNA helicase Sgs1 acting with the Dna2 nuclease [76,77]. Recently, several laboratories reconstituted the resection process in vitro, using either budding yeast Sgs1/Dna2/MRX or Exo1/MRX/Sae2 or mammalian Bloom's syndrome protein (BLM)/EXO1/MRN [96–98].
After resection, the newly generated ssDNA is coated by RPA to protect from degradation and allow the exchange of RPA with Rad51. The formation of RPA-coated ssDNA can be visualized by fluorescent tagging of RPA and monitoring the appearance of repair foci. This method is commonly used in mammalian cells, where it has so far not been possible to quantitatively measure resection. Using this technique, it was shown that depletion of CtIP, BLM (mammalian homologue of Sgs1) or EXO1 results in decreased RPA foci formation [93,99,100], supporting the idea that the three resection activities defined in budding yeast are conserved in mammals. Decreased RPA foci formation and reduced Chk1 phosphorylation after γ-irradiation was also observed in Mre11 nuclease-deficient MEFs [85]. This result is intriguing, as budding yeast nuclease-deficient Mre11 mutants do not show reduced ssDNA formation at DSBs [79]. These data suggest that Mre11 nuclease activity may be required for efficient resection of ionizing radiation (IR)-induced DSBs in mammalian cells.
5. Non-homologous end-joining or homologous recombination: competing interests?
It is clear that there are two protein complexes that can recognize DNA ends and each of them initiates a different mode of DSB repair. How do cells decide which pathway to use? Do NHEJ and HR proteins compete with each other for the same DNA end? Several studies have shown that Ku indeed interferes with repair of DSBs by HR [90,101–105]. More specifically, Ku inhibits Exo1-dependent resection in budding yeast, which is most apparent in the absence of MRN or Sae2 [80,81]. It would therefore seem a good idea to be able to remove Ku from DNA ends to allow resection to activate repair by HR. ChIP experiments in S. cerevisiae showing increased presence of Ku at DSBs in the absence of the MRX complex suggest that MRX displaces Ku from the DNA end [66,81,106]. While these results can be explained by competition between Ku and MRX for DNA ends, MRX is required for efficient NHEJ in budding yeast, arguing against this idea [72,107]. It therefore appears that Ku and MRN are not merely competing for DNA ends, but that MRN can actively release Ku from DNA ends when HR is the favoured mode of DSB repair.
Curiously, despite the fact that deletion of the Ku heterodimer in Mre11 nuclease-deficient budding yeast can improve cell survival upon high doses of IR, Mre11 nuclease activity is not required to remove Ku from DSBs [66,81]. In comparison with S. cerevisiae, Mre11 nuclease activity is more critical for survival of IR-induced DSBs in fission yeast, but this defect can be very effectively suppressed by eliminating Ku [80,84]. It will therefore be interesting to investigate the interplay between MRN and Ku in fission yeast, as it may reveal the mysterious function of Mre11 nuclease activity.
6. Cell cycle regulation of homologous recombination
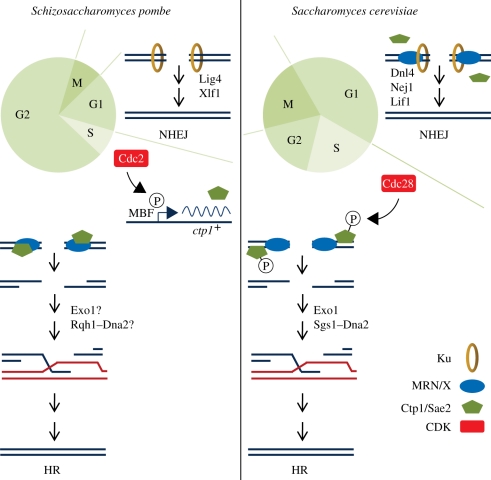
While it seems sensible to use HR activities to prevent NHEJ when a template is available for HDR, it increases the complexity of DSB repair. If the MRN complex is constantly available to bind DSBs and displace or actively release Ku from DNA ends, then NHEJ would never get a chance to repair DSBs. This suggests that the binding of the MRN complex to DNA ends is insufficient to promote HR at the expense of NHEJ. As HR repair is most favoured during S and G2 phases, it is likely that cell cycle-regulating proteins are involved in this decision-making step. One way to control MRN and Ctp1 activity during the cell cycle would be to regulate their abundance à la B-type cyclins. The MRN/X protein complex is however assembled and abundant throughout the cell cycle in all organisms that have been studied. On the other hand, the abundance of fission yeast Ctp1 and its human homologue CtIP are cell cycle-regulated, with no (Ctp1) to low (CtIP) protein in G1 and the highest amount observed during S and G2 phases [90,108]. Transcription of ctp1+ mRNA is regulated by MluI binding factor (MBF), a transcription factor responsible for the periodic expression of a large set of genes that are required for DNA replication and other cell cycle-regulated events [79]. MBF activity is regulated by CDKs, thus the same activity that drives the transition from G1 to S phase is also responsible for the expression of ctp1+ mRNA (figure 2). In view of the critical roles that Ctp1 and CtIP have in promoting HDR, it is likely that the onset of their expression in late G1 phase is a major determining factor in the switch from NHEJ to HR as the preferred mode of DSB repair at that stage of the cell cycle.
Figure 2.
Cell cycle regulation of double-strand break (DSB) repair. Schizosaccharomyces pombe and Saccharomyces cerevisiae use different mechanisms to control the modes of DSB repair during the cell cycle. In both organisms, the Ku heterodimer binds DSBs and promotes NHEJ repair in G1 phase. Other NHEJ factors include Lig4 and Xlf1 in S. pombe, and Dnl4, Nej1 and Lif1 in S. cerevisiae. The MRX protein complex stimulates NHEJ in S. cerevisiae, whereas MRN is not required for NHEJ in S. pombe. It is unknown whether the MRN complex binds DSBs during G1 in S. pombe. Sae2 is present during G1 phase in S. cerevisiae, but it is unknown whether it associates with DSBs prior to the onset of S phase CDK activity triggers the onset of S phase. In S. pombe, Cdc2 activates the MBF transcription factor in late S phase, which leads to expression of Ctp1. Ctp1 is recruited to DSBs through its interaction with the forkhead-associated domain of Nbs1. Genetic studies, replication protein A chromatin immunoprecipitation assays and checkpoint assays indicate that MRN and Ctp1 are essential for efficient resection which is required for HR repair of DSBs in fission yeast. In S. cerevisiae, Sae2 is expressed throughout the cell cycle, but its activity depends on phosphorylation by Cdc28. Both MRX and Sae2 locate to the DSB, but in contrast to fission yeast, Sae2 binds to DSBs independently of MRX. Sae2 and MRX initiate resection, with extended resection performed by Exo1 or Sgs1–Dna2. See main text for references.
Another way to potentially control the mode of DSB repair is to regulate protein activity through post-translational modification, such as phosphorylation by protein kinases. CDKs are the obvious candidates, as the increase in CDK activity that triggers the G1–S transition coincides with the switch from NHEJ to HR as the preferred mode of DSB repair. Indeed, studies of S. cerevisiae in the mid-2000s showed that resection is decreased in the absence of CDK1 (Cdc28) activity [109–111]. This was followed by the discovery that Cdc28 phosphorylates Sae2 on Ser267 in a region of sequence similarity with CtIP ([112]; figure 2). Interestingly, this C-terminal domain is absent in S. pombe, plants and in some other species, indicating that it was lost from these organisms during evolution. Mutating Ser267 in Sae2 to abolish phosphorylation reduces processing of DSBs and increases sensitivity to DSB-inducing agents [112]. Later studies with CtIP identified a corresponding phosphorylation at Thr847, which is part of a CDK-consensus motif [113]. Mutation of this residue impaired CtIP function. CtIP is phosphorylated on a second site, Ser327, enabling the interaction of CtIP with BRCA1 that is necessary to recruit CtIP to DSBs [94,114]. Curiously, while Ctp1 is phosphorylated both basally and upon DNA damage, only basal phosphorylation is partially CDK-dependent and even then does not appear to affect the DNA repair-associated activities of Ctp1 [115]. Rather, DNA damage-induced phosphorylation of Ctp1 is suggested to be required for localization of Ctp1 to DSBs through the interaction with Nbs1 [116–118].
7. Concluding remarks
Years of inspired research have culminated in a good understanding of the processes of DSB repair. Most or all of the proteins involved in HR are known, and more specifically, we have a good working knowledge on the nucleases and helicases responsible for the resection during HR in S. cerevisiae. The proteins required for NHEJ have also been characterized. Additionally, much is understood about how checkpoints detect DNA damage and regulate the cell cycle, and conversely, how cell cycle status influences the mode of DSB repair used by the cell. One of the next steps is likely to be in the direction of how resection is switched off. This may involve proteins belonging to the checkpoint-signalling pathway. In fact, it appears that the mediator proteins Rad9 in budding yeast (Crb2 in S. pombe) and 53BP1 in mammalian cells inhibit resection of DNA ends [119–121]. Interestingly, all three homologues are phosphorylated by CDKs [122–124]. The effect of this phosphorylation remains to be elucidated.
8. Note added in proof
Two late-breaking studies address important topics of this review. Cdk1, which activates Sae2/CtIP, was found to also activate Dna2, the partner of Sgs1 in budding yeast [125]. Ctp1, the S. pombe Sae2/CtIP orthologue, was shown to be crucial for resection initiation and release of Ku from DNA ends [126]. Mre11 endonuclease activity was found to be dispensable for resection while essential for efficient release of Ku and accumulation of RPA on resected DNA ends [126]. These results correlate with the strong radiosensitive phenotypes of mutants lacking Mre11 endonuclease activity or Ctp1 [84,90].
Acknowledgements
DNA damage research in the Russell laboratory was funded by NIH grants GM59447, CA77325 and CA117638. P.L. received financial support from the Netherlands Organization for Scientific Research (NWO).
References
- 1.Wyman C., Kanaar R. 2006. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383 10.1146/annurev.genet.40.110405.090451 (doi:10.1146/annurev.genet.40.110405.090451) [DOI] [PubMed] [Google Scholar]
- 2.Aguilera A., Gomez-Gonzalez B. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217 10.1038/nrg2268 (doi:10.1038/nrg2268) [DOI] [PubMed] [Google Scholar]
- 3.Pitcher R. S., Wilson T. E., Doherty A. J. 2005. New insights into NHEJ repair processes in prokaryotes. Cell Cycle 4, 675–678 10.4161/cc.4.5.1676 (doi:10.4161/cc.4.5.1676) [DOI] [PubMed] [Google Scholar]
- 4.Lees-Miller S. P., Meek K. 2003. Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85, 1161–1173 10.1016/j.biochi.2003.10.011 (doi:10.1016/j.biochi.2003.10.011) [DOI] [PubMed] [Google Scholar]
- 5.Lieber M. R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 10.1074/jbc.R700039200 (doi:10.1074/jbc.R700039200) [DOI] [PubMed] [Google Scholar]
- 6.Kadyk L. C., Hartwell L. H. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonoda E., Takata M., Yamashita Y. M., Morrison C., Takeda S. 2001. Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA 98, 8388–8394 10.1073/pnas.111006398 (doi:10.1073/pnas.111006398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28 194–28 197 10.1074/jbc.272.45.28194 (doi:10.1074/jbc.272.45.28194) [DOI] [PubMed] [Google Scholar]
- 9.New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 10.1038/34950 (doi:10.1038/34950) [DOI] [PubMed] [Google Scholar]
- 10.Shinohara A., Ogawa T. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–407 10.1038/34943 (doi:10.1038/34943) [DOI] [PubMed] [Google Scholar]
- 11.Sung P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 10.1126/science.8066464 (doi:10.1126/science.8066464) [DOI] [PubMed] [Google Scholar]
- 12.Ogawa T., Yu X., Shinohara A., Egelman E. H. 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259, 1896–1899 10.1126/science.8456314 (doi:10.1126/science.8456314) [DOI] [PubMed] [Google Scholar]
- 13.Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. 2006. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029 10.1016/j.dnarep.2006.05.022 (doi:10.1016/j.dnarep.2006.05.022) [DOI] [PubMed] [Google Scholar]
- 14.Sinha M., Peterson C. L. 2008. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol. Cell 30, 803–810 10.1016/j.molcel.2008.04.015 (doi:10.1016/j.molcel.2008.04.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P. A. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 10.1016/j.molcel.2008.05.017 (doi:10.1016/j.molcel.2008.05.017) [DOI] [PubMed] [Google Scholar]
- 16.Noon A. T., Shibata A., Rief N., Lobrich M., Stewart G. S., Jeggo P. A., Goodarzi A. A. 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12, 177–184 10.1038/ncb2017 (doi:10.1038/ncb2017) [DOI] [PubMed] [Google Scholar]
- 17.O'Driscoll M., Gennery A. R., Seidel J., Concannon P., Jeggo P. A. 2004. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair 3, 1227–1235 10.1016/j.dnarep.2004.03.025 (doi:10.1016/j.dnarep.2004.03.025) [DOI] [PubMed] [Google Scholar]
- 18.Sharpless N. E., et al. 2001. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol. Cell 8, 1187–1196 10.1016/S1097-2765(01)00425-7 (doi:10.1016/S1097-2765(01)00425-7) [DOI] [PubMed] [Google Scholar]
- 19.Difilippantonio M. J., Petersen S., Chen H. T., Johnson R., Jasin M., Kanaar R., Ried T., Nussenzweig A. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196, 469–480 10.1084/jem.20020851 (doi:10.1084/jem.20020851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Amours D., Jackson S. P. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3, 317–327 10.1038/nrm805 (doi:10.1038/nrm805) [DOI] [PubMed] [Google Scholar]
- 21.Hoeijmakers J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 10.1038/35077232 (doi:10.1038/35077232) [DOI] [PubMed] [Google Scholar]
- 22.Lamb N. E., Sherman S. L., Hassold T. J. 2005. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet. Genome Res. 111, 250–255 10.1159/000086896 (doi:10.1159/000086896) [DOI] [PubMed] [Google Scholar]
- 23.Lajtha L. G., Oliver R., Ellis F. 1954. Incorporation of 32P and adenine 14C into DNA by human bone marrow cells in vitro. Br. J. Cancer 8, 367–379 10.1038/bjc.1954.38 (doi:10.1038/bjc.1954.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swift H. 1950. The constancy of deoxyribose nucleic acid in plant nuclei. Proc. Natl Acad. Sci. USA 36, 643–654 10.1073/pnas.36.11.643 (doi:10.1073/pnas.36.11.643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swift H. H. 1950. The desoxyribose nucleic acid content of animal nuclei. Physiol. Zool. 23, 169–198 [DOI] [PubMed] [Google Scholar]
- 26.Howard A., Pelc S. R. 1953. Synthesis of deoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Heredity 6, 261–273 [Google Scholar]
- 27.Russell P., Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145–153 10.1016/0092-8674(86)90546-5 (doi:10.1016/0092-8674(86)90546-5) [DOI] [PubMed] [Google Scholar]
- 28.Coleman T. R., Dunphy W. G. 1994. Cdc2 regulatory factors. Curr. Opin. Cell Biol. 6, 877–882 10.1016/0955-0674(94)90060-4 (doi:10.1016/0955-0674(94)90060-4) [DOI] [PubMed] [Google Scholar]
- 29.Santamaria D., et al. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815 10.1038/nature06046 (doi:10.1038/nature06046) [DOI] [PubMed] [Google Scholar]
- 30.Weinert T. A., Hartwell L. H. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241, 317–322 10.1126/science.3291120 (doi:10.1126/science.3291120) [DOI] [PubMed] [Google Scholar]
- 31.Weinert T., Hartwell L. 1989. Control of G2 delay by the rad9 gene of Saccharomyces cerevisiae. J. Cell Sci. Suppl. 12, 145–148 [DOI] [PubMed] [Google Scholar]
- 32.Hartwell L. H., Weinert T. A. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 10.1126/science.2683079 (doi:10.1126/science.2683079) [DOI] [PubMed] [Google Scholar]
- 33.Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 10.1146/annurev.biochem.73.011303.073723 (doi:10.1146/annurev.biochem.73.011303.073723) [DOI] [PubMed] [Google Scholar]
- 34.Lisby M., Barlow J. H., Burgess R. C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 10.1016/j.cell.2004.08.015 (doi:10.1016/j.cell.2004.08.015) [DOI] [PubMed] [Google Scholar]
- 35.Kim J. S., Krasieva T. B., Kurumizaka H., Chen D. J., Taylor A. M., Yokomori K. 2005. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 170, 341–347 10.1083/jcb.200411083 (doi:10.1083/jcb.200411083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. H., Paull T. T. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308, 551–554 10.1126/science.1108297 (doi:10.1126/science.1108297) [DOI] [PubMed] [Google Scholar]
- 37.Falck J., Coates J., Jackson S. P. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 10.1038/nature03442 (doi:10.1038/nature03442) [DOI] [PubMed] [Google Scholar]
- 38.You Z., Chahwan C., Bailis J., Hunter T., Russell P. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25, 5363–5379 10.1128/MCB.25.13.5363-5379.2005 (doi:10.1128/MCB.25.13.5363-5379.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabourin M., Zakian V. A. 2008. ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol. 18, 337–346 10.1016/j.tcb.2008.04.004 (doi:10.1016/j.tcb.2008.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou L., Elledge S. J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 10.1126/science.1083430 (doi:10.1126/science.1083430) [DOI] [PubMed] [Google Scholar]
- 41.Harrison J. C., Haber J. E. 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 10.1146/annurev.genet.40.051206.105231 (doi:10.1146/annurev.genet.40.051206.105231) [DOI] [PubMed] [Google Scholar]
- 42.Rhind N., Russell P. 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113, 3889–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottlieb T. M., Jackson S. P. 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72, 131–142 10.1016/0092-8674(93)90057-W (doi:10.1016/0092-8674(93)90057-W) [DOI] [PubMed] [Google Scholar]
- 44.Dvir A., Peterson S. R., Knuth M. W., Lu H., Dynan W. S. 1992. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA 89, 11 920–11 924 10.1073/pnas.89.24.11920 (doi:10.1073/pnas.89.24.11920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Zou L. 2005. Sensing, signaling, and responding to DNA damage: organization of the checkpoint pathways in mammalian cells. J. Cell. Biochem. 94, 298–306 10.1002/jcb.20355 (doi:10.1002/jcb.20355) [DOI] [PubMed] [Google Scholar]
- 46.McGowan C. H., Russell P. 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16, 629–633 10.1016/j.ceb.2004.09.005 (doi:10.1016/j.ceb.2004.09.005) [DOI] [PubMed] [Google Scholar]
- 47.Willson J., Wilson S., Warr N., Watts F. Z. 1997. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 25, 2138–2146 10.1093/nar/25.11.2138 (doi:10.1093/nar/25.11.2138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saka Y., Esashi F., Matsusaka T., Mochida S., Yanagida M. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11, 3387–3400 10.1101/gad.11.24.3387 (doi:10.1101/gad.11.24.3387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura T. M., Du L. L., Redon C., Russell P. 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24, 6215–6230 10.1128/MCB.24.14.6215-6230.2004 (doi:10.1128/MCB.24.14.6215-6230.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilkenny M. L., Dore A. S., Roe S. M., Nestoras K., Ho J. C., Watts F. Z., Pearl L. H. 2008. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 22, 2034–2047 10.1101/gad.472808 (doi:10.1101/gad.472808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders S. L., Portoso M., Mata J., Bahler J., Allshire R. C., Kouzarides T. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 10.1016/j.cell.2004.11.009 (doi:10.1016/j.cell.2004.11.009) [DOI] [PubMed] [Google Scholar]
- 52.Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 10.1016/j.cell.2006.10.043 (doi:10.1016/j.cell.2006.10.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675–679 10.1038/ncb1004 (doi:10.1038/ncb1004) [DOI] [PubMed] [Google Scholar]
- 54.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 (doi:10.1074/jbc.273.10.5858) [DOI] [PubMed] [Google Scholar]
- 55.Park E. J., Chan D. W., Park J. H., Oettinger M. A., Kwon J. 2003. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31, 6819–6827 10.1093/nar/gkg921 (doi:10.1093/nar/gkg921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carr A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1, 983–994 10.1016/S1568-7864(02)00165-9 (doi:10.1016/S1568-7864(02)00165-9) [DOI] [PubMed] [Google Scholar]
- 57.Zhou B. B., Elledge S. J. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 10.1038/35044005 (doi:10.1038/35044005) [DOI] [PubMed] [Google Scholar]
- 58.Majka J., Burgers P. M. 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 78, 227–260 10.1016/S0079-6603(04)78006-X (doi:10.1016/S0079-6603(04)78006-X) [DOI] [PubMed] [Google Scholar]
- 59.Rouse J., Jackson S. P. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551 10.1126/science.1074740 (doi:10.1126/science.1074740) [DOI] [PubMed] [Google Scholar]
- 60.Zou L., Liu D., Elledge S. J. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl Acad. Sci. USA 100, 13 827–13 832 10.1073/pnas.2336100100 (doi:10.1073/pnas.2336100100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majka J., Binz S. K., Wold M. S., Burgers P. M. 2006. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 281, 27 855–27 861 10.1074/jbc.M605176200 (doi:10.1074/jbc.M605176200) [DOI] [PubMed] [Google Scholar]
- 62.Furnari B., Rhind N., Russell P. 1997. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277, 1495–1497 10.1126/science.277.5331.1495 (doi:10.1126/science.277.5331.1495) [DOI] [PubMed] [Google Scholar]
- 63.Furnari B., Blasina A., Boddy M. N., McGowan C. H., Russell P. 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10, 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doherty A. J., Jackson S. P. 2001. DNA repair: how Ku makes ends meet. Curr. Biol. 11, R920–R924 10.1016/S0960-9822(01)00555-3 (doi:10.1016/S0960-9822(01)00555-3) [DOI] [PubMed] [Google Scholar]
- 65.Nick McElhinny S. A., Snowden C. M., McCarville J., Ramsden D. A. 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20, 2996–3003 10.1128/MCB.20.9.2996-3003.2000 (doi:10.1128/MCB.20.9.2996-3003.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu D., Topper L. M., Wilson T. E. 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178, 1237–1249 10.1534/genetics.107.083535 (doi:10.1534/genetics.107.083535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bryans M., Valenzano M. C., Stamato T. D. 1999. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res. 433, 53–58 [DOI] [PubMed] [Google Scholar]
- 68.Buck D., et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124, 287–299 10.1016/j.cell.2005.12.030 (doi:10.1016/j.cell.2005.12.030) [DOI] [PubMed] [Google Scholar]
- 69.Ahnesorg P., Smith P., Jackson S. P. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124, 301–313 10.1016/j.cell.2005.12.031 (doi:10.1016/j.cell.2005.12.031) [DOI] [PubMed] [Google Scholar]
- 70.Cavero S., Chahwan C., Russell P. 2007. Xlf1 is required for DNA repair by nonhomologous end joining in Schizosaccharomyces pombe. Genetics 175, 963–967 10.1534/genetics.106.067850 (doi:10.1534/genetics.106.067850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manolis K. G., Nimmo E. R., Hartsuiker E., Carr A. M., Jeggo P. A., Allshire R. C. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20, 210–221 10.1093/emboj/20.1.210 (doi:10.1093/emboj/20.1.210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daley J. M., Palmbos P. L., Wu D., Wilson T. E. 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39, 431–451 10.1146/annurev.genet.39.073003.113340 (doi:10.1146/annurev.genet.39.073003.113340) [DOI] [PubMed] [Google Scholar]
- 73.Critchlow S. E., Jackson S. P. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23, 394–398 10.1016/S0968-0004(98)01284-5 (doi:10.1016/S0968-0004(98)01284-5) [DOI] [PubMed] [Google Scholar]
- 74.Spagnolo L., Rivera-Calzada A., Pearl L. H., Llorca O. 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 10.1016/j.molcel.2006.04.013 (doi:10.1016/j.molcel.2006.04.013) [DOI] [PubMed] [Google Scholar]
- 75.Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A. E. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115 10.1016/S1097-2765(01)00388-4 (doi:10.1016/S1097-2765(01)00388-4) [DOI] [PubMed] [Google Scholar]
- 76.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 10.1016/j.cell.2008.08.037 (doi:10.1016/j.cell.2008.08.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mimitou E. P., Symington L. S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 10.1038/nature07312 (doi:10.1038/nature07312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paull T. T., Gellert M. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 10.1016/S1097-2765(00)80097-0 (doi:10.1016/S1097-2765(00)80097-0) [DOI] [PubMed] [Google Scholar]
- 79.Llorente B., Symington L. S. 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24, 9682–9694 10.1128/MCB.24.21.9682-9694.2004 (doi:10.1128/MCB.24.21.9682-9694.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mimitou E. P., Symington L. S. 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 10.1038/emboj.2010.193 (doi:10.1038/emboj.2010.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., Zhu Z., Paull T. T., Ira G., Lee S. E. 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 10.1038/emboj.2010.219 (doi:10.1038/emboj.2010.219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreau S., Ferguson J. R., Symington L. S. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., Ohta K. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17, 6412–6425 10.1093/emboj/17.21.6412 (doi:10.1093/emboj/17.21.6412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams R. S., et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109 10.1016/j.cell.2008.08.017 (doi:10.1016/j.cell.2008.08.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J. M., Chang S., Ferguson D. O. 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96 10.1016/j.cell.2008.08.015 (doi:10.1016/j.cell.2008.08.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hartsuiker E., Neale M. J., Carr A. M. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 10.1016/j.molcel.2008.11.021 (doi:10.1016/j.molcel.2008.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartsuiker E., Mizuno K., Molnar M., Kohli J., Ohta K., Carr A. M. 2009. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29, 1671–1681 10.1128/MCB.01182-08 (doi:10.1128/MCB.01182-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rothenberg M., Kohli J., Ludin K. 2009. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 5, e1000722. 10.1371/journal.pgen.1000722 (doi:10.1371/journal.pgen.1000722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651 10.1016/j.molcel.2007.11.001 (doi:10.1016/j.molcel.2007.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., Russell P. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 10.1016/j.molcel.2007.09.009 (doi:10.1016/j.molcel.2007.09.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prinz S., Amon A., Klein F. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rattray A. J., McGill C. B., Shafer B. K., Strathern J. N. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. 2007. Human CtIP promotes DNA end resection. Nature 450, 509–514 10.1038/nature06337 (doi:10.1038/nature06337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu X., Chen J. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24, 9478–9486 10.1128/MCB.24.21.9478-9486.2004 (doi:10.1128/MCB.24.21.9478-9486.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20, 34–46 10.1101/gad.1381306 (doi:10.1101/gad.1381306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 10.1101/gad.2003811 (doi:10.1101/gad.2003811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nicolette M. L., Lee K., Guo Z., Rani M., Chow J. M., Lee S. E., Paull T. T. 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17, 1478–1485 10.1038/nsmb.1957 (doi:10.1038/nsmb.1957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., Kowalczykowski S. C. 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 10.1038/nature09355 (doi:10.1038/nature09355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gravel S., Chapman J. R., Magill C., Jackson S. P. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 10.1101/gad.503108 (doi:10.1101/gad.503108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.You Z., et al. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell 36, 954–969 10.1016/j.molcel.2009.12.002 (doi:10.1016/j.molcel.2009.12.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tomita K., et al. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23, 5186–5197 10.1128/MCB.23.15.5186-5197.2003 (doi:10.1128/MCB.23.15.5186-5197.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., Haber J. E. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409 10.1016/S0092-8674(00)81482-8 (doi:10.1016/S0092-8674(00)81482-8) [DOI] [PubMed] [Google Scholar]
- 103.Pierce A. J., Hu P., Han M., Ellis N., Jasin M. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 10.1101/gad.946401 (doi:10.1101/gad.946401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wasko B. M., Holland C. L., Resnick M. A., Lewis L. K. 2009. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair 8, 162–169 10.1016/j.dnarep.2008.09.010 (doi:10.1016/j.dnarep.2008.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fukushima T., et al. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 276, 44 413–44 418 10.1074/jbc.M106295200 (doi:10.1074/jbc.M106295200) [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y., Hefferin M. L., Chen L., Shim E. Y., Tseng H. M., Kwon Y., Sung P., Lee S. E., Tomkinson A. E. 2007. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14, 639–646 10.1038/nsmb1261 (doi:10.1038/nsmb1261) [DOI] [PubMed] [Google Scholar]
- 107.Hefferin M. L., Tomkinson A. E. 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4, 639–648 10.1016/j.dnarep.2004.12.005 (doi:10.1016/j.dnarep.2004.12.005) [DOI] [PubMed] [Google Scholar]
- 108.Yu X., Baer R. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275, 18 541–18 549 10.1074/jbc.M909494199 (doi:10.1074/jbc.M909494199) [DOI] [PubMed] [Google Scholar]
- 109.Aylon Y., Liefshitz B., Kupiec M. 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 10.1038/sj.emboj.7600469 (doi:10.1038/sj.emboj.7600469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ira G., et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 10.1038/nature02964 (doi:10.1038/nature02964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zierhut C., Diffley J. F. 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27, 1875–1885 10.1038/emboj.2008.111 (doi:10.1038/emboj.2008.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huertas P., Cortes-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 10.1038/nature07215 (doi:10.1038/nature07215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huertas P., Jackson S. P. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 10.1074/jbc.M808906200 (doi:10.1074/jbc.M808906200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yun M. H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 10.1038/nature07955 (doi:10.1038/nature07955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Akamatsu Y., Murayama Y., Yamada T., Nakazaki T., Tsutsui Y., Ohta K., Iwasaki H. 2008. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 28, 3639–3651 10.1128/MCB.01828-07 (doi:10.1128/MCB.01828-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williams R. S., et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139, 87–99 10.1016/j.cell.2009.07.033 (doi:10.1016/j.cell.2009.07.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lloyd J., Chapman J. R., Clapperton J. A., Haire L. F., Hartsuiker E., Li J., Carr A. M., Jackson S. P., Smerdon S. J. 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139, 100–111 10.1016/j.cell.2009.07.043 (doi:10.1016/j.cell.2009.07.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dodson G. E., Limbo O., Nieto D., Russell P. 2010. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle 9, 1516–1522 10.4161/cc.9.8.11260 (doi:10.4161/cc.9.8.11260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lazzaro F., Sapountzi V., Granata M., Pellicioli A., Vaze M., Haber J. E., Plevani P., Lydall D., Muzi-Falconi M. 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27, 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bothmer A., Robbiani D. F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M. C. 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207, 855–865 10.1084/jem.20100244 (doi:10.1084/jem.20100244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bunting S. F., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 10.1016/j.cell.2010.03.012 (doi:10.1016/j.cell.2010.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakamura T. M., Moser B. A., Du L. L., Russell P. 2005. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol. Cell. Biol. 25, 10721–10730 10.1128/MCB.25.24.10721-10730.2005 (doi:10.1128/MCB.25.24.10721-10730.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Toh G. W., Lowndes N. F. 2003. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 31, 242–246 10.1042/BST0310242 (doi:10.1042/BST0310242) [DOI] [PubMed] [Google Scholar]
- 124.Linding R., et al. 2007. Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 10.1016/j.cell.2007.05.052 (doi:10.1016/j.cell.2007.05.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen X., Niu H., Chung W.-H., Zhu Z., Papusha A., Shim E. Y., Lee S. E., Sung P., Ira G. 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18, 1015–1019 (doi:10.1038/nsmb.2105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Langerak P., Mejia-Ramirez E., Limbo O., Russell P. 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271 (doi:10.1371/journal.pgen.1002271) [DOI] [PMC free article] [PubMed] [Google Scholar]