Abstract
The molecular networks regulating the G1–S transition in budding yeast and mammals are strikingly similar in network structure. However, many of the individual proteins performing similar network roles appear to have unrelated amino acid sequences, suggesting either extremely rapid sequence evolution, or true polyphyly of proteins carrying out identical network roles. A yeast/mammal comparison suggests that network topology, and its associated dynamic properties, rather than regulatory proteins themselves may be the most important elements conserved through evolution. However, recent deep phylogenetic studies show that fungal and animal lineages are relatively closely related in the opisthokont branch of eukaryotes. The presence in plants of cell cycle regulators such as Rb, E2F and cyclins A and D, that appear lost in yeast, suggests cell cycle control in the last common ancestor of the eukaryotes was implemented with this set of regulatory proteins. Forward genetics in non-opisthokonts, such as plants or their green algal relatives, will provide direct information on cell cycle control in these organisms, and may elucidate the potentially more complex cell cycle control network of the last common eukaryotic ancestor.
Keywords: cell cycle, eukaryotes, evolution, opisthokonts, plants
1. Introduction
Progress in understanding eukaryotic cell cycle control over the past 40 years was largely initiated from two very different approaches. A biochemical line of inquiry was carried out in the embryos of marine invertebrates and Xenopus frogs. Independently, researchers applied microbial genetics to ascomycete fungi, especially Saccharomyces cerevisiae and Schizosaccharomyces pombe (budding and fission yeast, respectively), to identify genes with essential functions attributed specifically to cell cycle control or execution. These lines of research converged in the late 1980s. Cyclins, first identified in marine invertebrates, were found to periodically activate cyclin-dependent protein kinases (Cdks), identified as central to cell cycle regulation in yeast screens. Cdk–cyclin complexes and oscillation of Cdk kinase activity were subsequently found to be at the core of all eukaryotic cell cycle control.
Further work showed that other cell cycle regulators identified in yeast screens were present and functioning in highly similar ways in animal systems. Indeed, conservation of structure and function was frequently sufficient to allow for cross-kingdom genetic complementation [1]. Cross-species methods based on complementation in budding yeast were also used to identify many different fission yeast and mammalian cyclins [2–6]. This body of work also placed a molecular interpretation on the cell fusion experiments carried out decades earlier that suggested cis- and trans-acting regulators of cell cycle progression in cultured animal cells [7].
In addition to cyclin–Cdk complexes, the anaphase-promoting complex (APC; cyclosome) is another central and highly conserved controlling element that was independently identified by both biochemical and yeast genetic approaches. Active APC is an E3 ubiquitin ligase that targets many cyclins for degradation, so that in general, high APC activity correlates with low cyclin–Cdk activity. Conversely, high cyclin–Cdk is thought to promote initial activation of the APC driven by the Cdc20 co-activator. Later, after removal of cyclin by Cdc20–APC-driven degradation, APC activity is maintained by the Cdh1 co-activator. Reciprocally, Cdh1–APC is antagonized by cyclin–Cdk activity at the G1–S transition, so that cyclin–Cdk and APC activity oscillate, approximately out of phase with each other. The high Cdk/low APC condition approximately correlates with the conventional S, G2 and early M phases (i.e. cells in the process of cell duplication), while the converse condition correlates with late M–G1 (i.e. newborn cells). Studies across diverse organisms suggest that both molecular and dynamic functional aspects of this reciprocal Cdk–APC regulation may be conserved.
The conservation of molecular functions strongly argued for a conserved core of eukaryotic cell cycle control machinery. However, as more details emerged in animals and yeast, it became clear that some core regulatory proteins were frequently reapportioned for different uses. For example, budding yeast, fission yeast and animals share a highly similar checkpoint surveillance system monitoring DNA replication and integrity, but this system impinges on cell cycle control by entirely different mechanisms in budding yeast compared with other organisms [8]. As another example, re-initiation of DNA replication within a single S phase is blocked because cyclin–Cdk activity antagonizes loading of the ‘pre-replication complex’ including the Mcm replicative helicase at origins of replication. However, different organisms exhibit strikingly different mechanisms for this inhibition [9].
The central cyclin–CDK interaction has remained intact, despite extensive growth of eukaryotic cyclin and Cdk families through gene duplication. Functional overlap of gene duplicates runs the gamut from largely, if not completely, indistinguishable biological roles to almost complete separation of function. This rampant gene duplication results in ambiguity with respect to monophyly (unique line of descent) of some key cell cycle functions. For example, while cyclins E and A promote initiation of DNA replication in animals, they are absent in yeasts, where instead, B-type cyclins promote DNA replication [10]. A plausible interpretation of this observation is that control of replication was acquired by B cyclins in the yeast lineage after loss of cyclins E and A.
Other molecules central to mammalian G1 regulation, such as the retinoblastoma protein and Cip–Kip inhibitors, have no obvious sequence homologues in yeast [11]. Thus, important aspects of cell cycle control are not conserved between yeast and animals, and in some cases differ even within the fungal lineage. Here, we examine conserved and diverged features of eukaryotic cell cycle regulation. Our analysis leads to new ideas about cell cycle regulation in the last common ancestor (LCA) of eukaryotes. We propose that in some cases, network topology, and its associated dynamic properties, rather than regulatory proteins themselves may be the most important elements conserved through evolution.
2. The G1–S regulatory network in budding yeast and animals has a remarkably similar topology, but little sequence similarity among network components
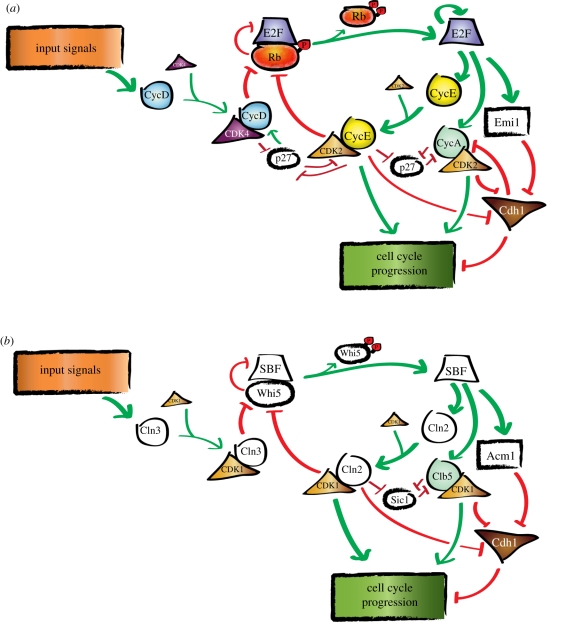
The molecular networks regulating the G1–S transition in budding yeast and animals, shown schematically in figure 1, exhibit a diverse set of molecular interactions arranged in remarkably similar network structures [11,12]. In both cases, the most likely model requires Cdk activity to reach a threshold at which an inhibitor of cell cycle progression is inactivated. Then, a positive feedback mechanism is activated to ensure switch-like, irreversible commitment to the next phase of the cell cycle (DNA replication, ultimately followed by cell division). However, many of the individual proteins performing network-similar or identical network roles appear to have unrelated sequences (table 1), suggesting either monophyly followed by extremely accelerated sequence evolution, or true polyphyly (i.e. convergent evolution) of unrelated proteins carrying out identical network roles.
Figure 1.
Schematic of (a) mammalian and (b) budding yeast G1–S control circuits indicates a common feedback-driven regulatory architecture. Shapes correspond to the type of protein (e.g. upward triangles denote cyclin-dependent kinases). Colour implies that the G1–S regulator has high sequence similarity (indicating homology, table 1) to the same regulator in another kingdom (animal, fungi, plant). The G1–S circuit in mammals is colourful (compared with budding yeast) because there are many identifiable sequence homologues between plants and animals.
Table 1.
Quantifying the sequence similarity and homology between known regulators of G1–S control. List of functionally similar G1–S regulators (figure 1) in animals (Homo sapiens) and fungi (Saccharomyces cerevisiae). For comparison, we also include putative G1–S regulators from plants (Arabidopsis thaliana) [13]. Two proteins are classified as homologues (i.e. descent from a common ancestral protein) if they have high-sequence similarity (NCBI BLASTP E-value lower than 1E-5; bold) [14]. Note that two proteins can be true homologues, yet not have any detectable sequence similarity.
| G1–S regulator | animal gene E-value (fungi and plant) | fungal gene E-value (animal and plant) | plant gene E-value (animal and fungi) |
|---|---|---|---|
| G1 cyclin | CycD1 (2E-02, 5E-18) | Cln3 (>1, >1) | CycD4 (5E-13, >1) |
| G1 Cdk | Cdk4/6 (2E-59, 3E-66) | Cdk1 (3E-58, 1E-105) | CdkA (3E-65, 2E-106) |
| G1–S cyclin | CycE1 (>1, 2E-10) | Cln2 (>1, >1) | CycD4 (2E-10, >1) |
| G1–S Cdk | Cdk2 (2E-109, 1E-117) | Cdk1 (1E-108, 1E-105) | CdkA (1E-117, 2E-106) |
| S cyclin | CycA1 (1E-27, 8E-09) | Clb5 (4E-27, 2E-08) | CycD4 (6E-11, 2E-09) |
| G1–S TF | E2F1 (>1, 8E-31) | Swi4 (>1, >1) | E2FB (2E-30, >1) |
| G1–S TF inhibitor | Rb1 (>1, 3E-45) | Whi5 (>1, >1) | Rbr1 (9E-45, >1) |
| Cdk inhibitor | p27/Kip1 (>1, 2E-03) | Sic1 (>1, >1) | Krp1/Ick1 (1E-04, >1) |
| Cdh1 inhibitor | Emi1 (>1, >1) | Acm1 (>1, >1) | |
| Cdh1 | Cdh1/Frz1 (3E-95, 8E-133) | Cdh1 (2E-94, 7E-91) | Fzr1 (3E-135, 4E-95) |
Globally, the cell cycle ‘step’ controlled by the budding yeast network in figure 1 is the ‘Start’ transition of cell cycle commitment [15]. Prior to Start, a cell integrates various internal and external signals, such as those arising from cell size, nutrients and mating pheromone, to produce an all-or-none decision to enter the mitotic cell cycle. G1 regulation proceeds in two steps separated by a rapid positive feedback-driven transition: the first step, dependent on the upstream cyclin Cln3, yields cell size control and initial inactivation of Whi5, while the second, cell size-independent step pertains to the activation of the cell division machinery by the rapidly increasing CDK activity arising from a transcriptional positive feedback loop in which Cln1,2 complete Whi5 inactivation and yield full and coherent expression of the entire regulon activated by the SBF (Swi4–Swi6) and MBF (Mbp1–Swi6) transcription factors [16–19]. The transition point (i.e. Start) between these two steps corresponds to cell cycle commitment arising from the abrupt feedback-driven increase in G1 cyclin transcription rate [19,20].
The animal cell transition illustrated in figure 1 has been called the ‘restriction point’ (R) since it corresponds to transition to independence from extracellular growth factors. Insightful time-lapse microscopy analysis [21] demonstrated that passage of R occurred in a sharp time-window about 3 h after division. Later events in G1 required a variable (0–10 h) period, but were growth factor-independent. Although the precise alignment of R with the events diagrammed in figure 1 remains somewhat unclear, it is possible that R consists primarily of induction of cyclin D and initial inactivation of Rb, with the growth factor-independent part of this transition corresponding to cyclin E–E2F-positive feedback leading ultimately to progression to DNA replication (reviewed by Foster et al. [22]).
These observations suggest a hypothetical alignment of R in animal cells and at least the first step of Start in yeast, with subsequent growth factor- or cell size-independent events in the two systems occurring after the induction of transcriptional-positive feedback. Whether this alignment reflects conserved network functions is difficult to determine at this time as cell size control in animal cells has been a contentious subject [23–25].
We begin by reviewing the similarities in G1–S regulatory network topology and molecular function of its constituent parts [12]. Cell cycle entry in mammalian cells is initiated by cyclin D (three variants D1/2/3) targeting CDK4 or CDK6 to phosphorylate the transcriptional-inhibitor pRb, the retinoblastoma protein. Phosphorylation of pRb inhibits its pocket domain from binding the transactivating domain of an activating E2F transcription factor. In budding yeast, the Cln3 cyclin phosphorylates the Whi5 transcriptional inhibitor preventing Whi5 from binding and inactivating the SBF transcription factor. Both pRb and Whi5 recruit HDAC (histone deacetylases) proteins to the promoter that inhibits transcription [26–28], and both proteins have multiple (approx. 10) CDK phosphorylation sites, suggesting that multi-site phosphorylation is a conserved or convergent feature of their inhibition [29–31]. However, the sequences of pRb and Whi5 are completely different.
Activity of the upstream cyclin–CDK complex that initiates the cell cycle is regulated through transcription, translation and localization [32–35]. Both Cln3 and cyclin D are unstable, which serves to make this network structure rapidly responsive to changes in environmental conditions [36–39].
In budding yeast and mammals, the upstream cyclin initiates multiple transcriptional-positive feedback loops. Cyclin D initiates the transcription of cyclin E (two variants E1/2), which also target pRb for phosphorylation and inhibition [40–42]. In mitotic cell cycles, cyclin E transcription and p27 degradation correlate with increased phosphorylation of pRb, consistent with this feedback-driven transition [31,43]. A similar process occurs in yeast, where Cln3 initiates a transcriptional-positive feedback loop of two downstream G1 cyclins, CLN1 and CLN2, which complete the inactivation of Whi5 [18]. This network structure probably results in bistability and hysteresis, both in animals and in yeast [42,44].
A complexity in both budding yeast and animals is that the activating transcription factors SBF and E2F represent distinct proteins. ‘SBF’ as illustrated in figure 1 actually is a stand-in for two related transcription factors, SBF and MBF, owing to a relatively recent gene duplication event in the budding yeast lineage, while E2F is a stand-in for E2F1, E2F2 and E2F3. The yeast factors are largely overlapping in function and can substitute for each other to a large extent [45], but they are distinct both with respect to upstream regulators and downstream targets [46]. For simplicity, however, we will treat them as one entity here.
As well as cyclins D and E, cyclin A is capable of phosphorylating and inactivating pRb [47]. Since cyclin A is activated later in the cell cycle, this network motif results in an ‘initiation versus maintenance’ control structure. Budding yeast lack cyclin E and A homologues; however, Whi5 inactivation is maintained by phosphorylation by B-type cyclin–Cdk complexes that are activated later in the cell cycle. So again, network structure is similar without evidence of monophyly of the controlling proteins.
A major role in the animal and the yeast systems is relief of inhibition of cyclin B–Cdk complexes (as well as of cyclin A in animal systems). These complexes are inhibited because of APC–Cdh1 activity and because of accumulation of stoichiometric inhibitors (Sic1 in budding yeast, Kip–Cip in animals). Cdh1 is inhibited in all systems by cyclin–Cdk phosphorylation, and is also inhibited by stoichiometric-binding proteins (Emi1 and Acm1). In animals, Emi1 is an E2F transcriptional target, which binds and inhibits Cdh1 [48]; in yeast, Acm1 similarly binds and inhibits Cdh1 [49] and is expressed specifically at G1–S, probably under SBF control [50]. However, Acm1 and Emi1 lack detectable sequence similarity [51]. Sic1 and Kip–Cip are both tight-binding stoichiometric inhibitors of cyclin–Cdk complexes, and Cdk1 phosphorylation targets both Sic1 and Kip for degradation via the SCF complexes ubiquitin ligase. Despite these biochemical similarities, there is no detectable sequence similarity between Sic1 and Kip–Cip, although similar secondary structure over a region of about 20 amino acids was suggested based on modelling [52]. Similarly, no obvious sequence similarity exists between SBF and E2F components.
The positive feedback in the network has been demonstrated in budding yeast to result in an irreversible transition [44]. In budding yeast and in fission yeast, activation of B-type cyclins resets the system by inactivating the SBF-related G1–S-specific transcription factors, in collaboration with the Nrm1 repressor [46,53,54]. Nrm1 lacks known animal homologues; however, a parallel negative control of E2F by cyclin A–Cdk complexes does occur in animal cells [55]. As noted above, cyclin A appears to carry out multiple roles carried out by B-type cyclins in yeast, including initiation of DNA replication and inactivation of Cdh1, as well as inactivation of G1–S transcription.
Taken together, these data indicate an extraordinarily similar network topology and mechanism in yeast and mammals, often without detectable sequence similarity, or (as in the cyclin A/cyclin B divergences noted above) with plausibly polyphyletic acquisition of network roles by distinct members of the cyclin superfamily.
3. Mammalian G1–S regulation is more complex
When there are significant differences between the yeast and mammalian G1 control network, it is typically owing to additional regulation in mammals [12]. Examples include microRNA regulation of CDK inhibitors in mammals [56] and a G1 role for the Cdc25 phosphatase [57].
Notably, the animal system contains additional positive feedback loops affecting G1 control. The animal F-box protein Skp2 targets p27 for degradation. Skp2 is a direct E2F transcription target and its stability (required for timely S phase entry) is regulated by the APCCdh1. Thus, as CDK activity increases through G1, Skp2 is both transcribed and stabilized, which is required to avoid premature S phase [58]. A second transcriptional positive feedback loop that is unique to mammalian cells is the regulation of E2F1 by E2F1–3 [59]. This animal-specific regulatory loop clearly reinforces the same regulatory themes provided by the network structure described above.
In mammals, multiple cyclin-dependent kinases play complementary roles in cell cycle progression. The most notable are CDK4 and CDK6, which preferentially bind cyclin D, while CDK2 and CDK1 preferentially bind the downstream cyclins E, A and B [60]. Nevertheless, there is significant cross-binding of cyclins and CDKs, as most strikingly evidenced by the fact that Cdk2–/– Cdk4–/– Cdk6–/– embryos undergo organogenesis and develop to mid-gestation [61]. Additionally, the E2F (1–8) and the pocket protein (pRb, p107 and p130) families are greatly expanded in mammals. These animal-specific regulatory expansions clearly reinforce the same regulatory themes provided by the network structures described above. This might be expected to avoid novel regulatory mechanisms operating at odds with an established system. An interesting consequence, developed below, is that if the novel regulatory mechanisms become sufficiently strong, they could ultimately replace the original components without disrupting the network structure.
Perhaps similarly, budding yeast contain an additional Cdk in addition to the major Cdk1/Cdc28, called Pho85. Pho85 is activated by the Pcl class of cyclins, which appear specific to the yeast lineage. At least some Pcl cyclins overlap with G1 cyclins Cln1 and Cln2 in function and transcriptional regulation [62,63]. Intriguingly, animals contain a possible Pho85 homologue, Cdk5, which appears to have neuronal, non-cell cycle roles, and is activated by proteins (p25 and p35) with no evident sequence relationship to Pcls or to other cyclins; strikingly, though, Cdk5 and Pho85 cross-complement and are cross-activated by Pcls and p25/p35 [64].
4. G1–S control in yeast and animals: diverged sequence with conserved function, or independent convergence onto the identical network?
As discussed above, the very similar G1–S network structures in mammals and budding yeast are surprising when sequence similarity of the key proteins is considered. Rb and Whi5, E2F and SBF, Emi1 and Acm1, p27-Kip1 and Sic1 all have no detectable sequence homology. Cln3 and cyclin D both share core structural features of the cyclin superfamily, as do Cln1, 2 and cyclins E, A, but detailed phylogenetic analysis (N. E. Buchler et al. 2011, unpublished data) has so far not demonstrated close affinity of any yeast G1 cyclins to any specific animal G1 cyclins. Similarly, the B-type cyclin Clb5 in budding yeast carries out a very similar network role to cyclin A in animals (see above), but Clb5 has greater sequence similarity to animal B-type cyclins than to cyclin A, and yeasts lack any clear cyclin A homologue.
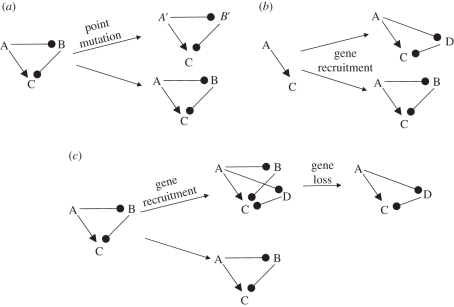
We can propose three possible general explanations for this apparent lack of molecular conservation (schematized for a simple network element in figure 2). (i) The LCA had the mammalian/yeast network structure. Highly accelerated sequence evolution along one or both lineages resulted in enough sequence divergence to make homology unrecognizable. (ii) The LCA had a different (possibly simpler) network structure, which convergently evolved to the structure shown in figure 1, independently in fungal and animal lineages. (iii) The LCA of budding yeast and animals had the mammalian/yeast network structure comprising either the yeast or the animal versions of the proteins. Then, other proteins, originally with different functions, evolved functional overlap with some original network components. As is noted above, selection for consistent regulatory function in novel evolved circuits could ultimately result in redundancy of the original components. This would then allow loss of the original components without loss of the network.
Figure 2.
Schematic of possibilities for network evolution. (a) A simple network is conserved in two lineages, but along one or both lineages, highly accelerated sequence evolution results in loss of detectable homology in modern descendants (e.g. A′ and A are direct sequence descendants of A in the ancestor, and have carried out the same network role throughout evolution, but A and A′ are no longer sequence-alignable). In this case, both network and sequences are monophyletic: from a single origin, the network has retained the same topology, and all sequences have kept the same network role. (b) A simple network independently recruits new elements to elaborate the network (note that the ‘sense’ of the network remains the same, with A still activating the downstream C). In this case, the enhanced network is polyphyletic, as are the new sequences B and D. (c) Along one lineage, the network acquires an independent loop redundant with the B loop, allowing subsequent loss of B along this lineage, without losing network function at any step. In such a case, we consider the network monophyletic, even though the sequences are polyphyletic. Note that in the case of recruitment, B and D could be ancient relatives. Provided D did not carry out the indicated network role in precursor organisms, this still constitutes sequence polyphyly for this network.
These possibilities are difficult to evaluate at present. Discovery of a ‘missing link’ organism with hybrid controls would be revealing, as would be in-depth analysis of G1–S control in other organisms among yeasts or animals, to determine the degree of similarity of network topology and physiological function.
5. Conservation of sequence does not imply conservation of network architecture
We argued above that network topology can be strikingly similar despite lack of detectable sequence similarity of most of the controlling components, and suggested three potential mechanisms by which this could have come about. We note that it is well documented that network topology can change while the underlying molecules are conserved. Very similar, almost certainly monophyletic molecules can be found in diverse networks playing even more diverse biological roles.
Although close sequence homologues frequently have conserved nearest-neighbour interacting partners, the likelihood of continued conservation of interacting proteins falls off steeply as a function of the number of degrees of separation. Examples of neighbour conservation include the Cdk activation by cyclin binding and the Ras–GEF–GAP module [65]. A clear example of network rearrangement is found in DNA damage response. In budding yeast and fission yeasts, a similar machinery from the point of damage to the activation of downstream kinases (upstream: 9-1-1 complex, MRN complex; downstream: Chk1, Chk2) is followed by different interfaces with the core cell cycle. In fission yeast, Cdc25 is targeted to block mitosis, while in budding yeast, the primary target is likely Pds1, resulting primarily in a block to cohesin cleavage despite high cyclin–Cdk activity levels) [8]. Similarly, the Cdc14 phosphatase drives the high-to-low CDK transition (M–G1) in budding yeast, yet has a much less central role in other organisms [66].
6. Most analysis of eukaryotic cell cycle control has been carried out in the opisthokont branch
In the 1970s and 1980s, when cell cycle research in yeast model organisms provided key insights into widely conserved mechanisms, broad eukaryotic phylogenetics was not well resolved, leading to the concept of yeast as a ‘universal cell’ [67], exhibiting a ‘universal M-phase control mechanism’ [68]. Budding yeast and fission yeast appeared to be very highly diverged from each other, and both appeared very highly diverged from animals. Since by standard evolutionary logic, a trait shared by two distantly related organisms is likely to have been present in their LCA, it was hoped that common findings among these organisms would triangulate to include essentially all eukaryotes.
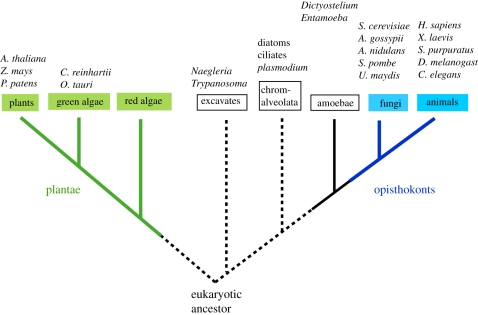
However, it has become clear that budding yeast and fission yeast are much more closely related to each other than either is to animals [69]. The apparently similar distance between the two yeasts and between yeast and animals in earlier sequence comparisons is likely due to rapid evolution along the fungal branches (N. E. Buchler et al. unpublished data). Further, recent work shows that fungi and metazoans (collectively, the ‘opisthokonts’) share a more recent LCA relative to all other eukaryotic groups (figure 3). This consensus view is derived from analysis of slowly evolving phylogenetic characters including 18S rRNA, proteins encoded in mitochondrial genomes, ‘RGC-CAMs’ (rare genomic changes after conserved amino acids multiple substitutions), and an ensemble of highly conserved, slowly evolving proteins (EF-1a, actin, b-tubulin and HSP70) [70–73]. While these studies differ on relative divergence of eukaryotes other than metazoans and fungi, the conclusion that metazoans and fungi diverged most recently is uniformly supported by these analyses.
Figure 3.
A current phylogeny of the eukaryotic supergroups, adapted from Rogozin et al. [69]. Branch lengths are not drawn to scale. Phylogenetic data support the idea that plants, fungi and animals are monophyletic (solid lines), that fungi and animals are part of a larger group known as the opisthokonts, and that plants diverged from the opisthokonts at an early point in eukaryotic evolution. However, the root of the eukaryotic tree and the relative placement of other supergroups (excavates, chromalveolata) with respect to each other, and the eukaryotic ancestor is still under debate.
The proximal fungal/animal evolutionary relationship indicates that the massive enterprise to understand cell cycle control in yeasts and animals emphatically does not provide triangulation for all eukaryotes, because analysis is restricted to two branches within the opisthokonts: the fungal branch (represented primarily by two ascomycete twigs, budding and fission yeast), and the metazoan branch. Although this information is highly informative for opisthokonts, the unsettling implication is that cell cycle control could be significantly different in other eukaryotes. These other eukaryotes represent most eukaryotic genomic diversity. Among them are many medically significant parasites, and also the land plants, which form the basis of all terrestrial ecosystems and provide us with food and oxygen. Thus, this systematic gap in knowledge about cell cycle control provides grounds for academic and practical concern.
7. Cell cycle analysis in plants and protists: evolutionary perspective beyond opisthokonts
The availability of genome sequences for a broad range of eukaryotes shows that many molecular aspects of cell cycle control may be conserved beyond the opisthokont lineage. All eukaryotic genomes contain sequences for core elements of the opisthokont cell cycle programme, including cyclins, Cdks and the APC, which are uniformly missing from archaeal and bacterial genomes. Thus, at least some key functional relationships are likely to be conserved, such as the requirement for cyclins to activate Cdk enzymatic activity, and the role of the APC in degrading cyclins.
Outside of the opisthokonts, a significant amount of cell cycle research has focused on plants. Based on sequence comparisons, plants appear to contain most ‘standard’ opisthokont cell cycle regulatory components. In fact, considering the overall evolutionary relationships (figure 3), plant genomes contain a surprising number of very likely homologues for many of the proteins shown in figure 1 that are present in animals but missing in yeast. These include Rb, E2F, cyclin A, cyclin D and p27-related proteins (Kip-related protein; KRP) [74]. This suggests that animal cell cycle regulators were present in the LCA of animals and plants, but were either replaced or subjected to rapid sequence evolution (to unrecognizability) in fungi. Sequence conservation of cell cycle regulators between plants and animals is especially remarkable considering that a recent study suggested that the divergence of plants concomitant with chloroplast acquisition was the most ancient divergence in the eukaryotic tree [70].
8. Looking where the light is in the plant cell cycle
Despite the reassuring presence of familiar sequences, the frequent discovery of strong sequence conservation embedded in very distinct circuits, as well as the converse case (see above), means that, in principle, rather little can be assumed from sequence conservation alone. Direct experimental analysis is required.
Much experimental analysis of the cell cycle in plants relies significantly on prior discovery of opisthokont cell cycle control genes for which similar sequences (likely homologues) were then identified in plants, for example, generation of antibodies and transcriptional reporters to determine if the basic behaviour is similar (e.g. B-type cyclins are degraded in mitosis in plants [75]). Functionally, overexpression of the homologues can show whether roughly similar biological function is noted (e.g. overexpression of the Kip–CIP-like KRP family of Cdk inhibitors inhibiting mitosis [76]). Loss-of-function analysis (by dominant-negatives, by siRNA, or by identification and testing of T-DNA insertion null alleles) of the homologues provides complementary functional information (e.g. requirement for cdka;1, the most likely Cdc2 homologue in Arabidopsis, for early cell cycles in gametogenesis [77]).
While such studies do provide insight into the plant cell cycle, and provide support for the validity of the opisthokont model in plants, significant discrepancies have been noted. Some of these may be due to incomplete knowledge, while others may reflect real differences. For example, plants contain clear homologues of the ATM and ATR kinases. In yeast and animals, these kinases are essential for responses to DNA damage and to replicative stress, respectively. In Arabidopsis, as in opisthokonts, ATM is required for DNA damage response, while ATR is required for response to replicative stress (hydroxyurea treatment) [13]. However, the well-documented mechanism for ATM and ATR activity absolutely requires the ‘downstream’ kinases Chk1 and Chk2 [8], which are missing in plant genomes [13]. Chk1 or Chk2 candidate homologues in plant genomes identified by BLAST (F. Cross 2011, unpublished data) appear to be selected on the basis of protein kinase similarity, rather than by any of the well-defined domain features of Chk1 and Chk2 [78]. If plants genuinely lack Chk1/2 equivalents, then the downstream interface of ATM/ATR with the cell cycle must be significantly different.
Similarly, in fission yeast and in animals (though not in budding yeast), ATM–ATR activation results in inhibition of Cdc2–cyclin B, by inhibition of the Cdc25 phosphatase and/or by activation of the Mik1 (and perhaps Wee1) kinases [8]. Wee1–Mik1 phosphorylate and inhibit Cdc2–cyclin B, while Cdc25 phosphatase removes this inhibitory phosphorylation. This mechanism ensures that cells will not enter mitosis with damaged or incompletely replicated DNA. Null alleles in Arabidopsis Wee1 are viable, but exhibit defective response to replicative stress [79]. However, this Wee1 functional activity was definitively shown to NOT require inhibitory phosphorylation of CDKA;1, the most likely cdc2 homologue in Arabidopsis [80]. Other potentially non-opisthokont-like observations from plants include suggestions that D-type cyclins might be directly involved in mitotic control [81]; and the plant-specific CDKB family, which is synthesized in G2 and degraded in mitosis [74].
9. Unbiased discovery required outside opisthokonts
The major discovery path to date for the plant cell cycle has involved examination of molecules already known from opisthokonts. Any aspect of cell cycle control that is prominent in plants, but cryptic or absent in opisthokonts cannot be identified by such approaches. This includes regulatory innovation along the plant branch, and mechanisms present in the LCA of plants and opisthokonts, but lost in the early opisthokont lineage. Thus, the further elucidation of cell cycle evolution requires a forward unbiased discovery strategy that can be applied outside the opisthokonts.
A central line of inquiry in the opisthokonts was loss-of-function genetics to identify cell cycle control molecules, initially in yeast and subsequently in Drosophila, Caenorhabditis elegans, and mice with the availability of siRNA and gene knock-out/knock-in methods. Although loss-of-function genetics has been central to plant biology, it can be highly challenging owing to the frequent presence of enormous numbers of gene duplicates (e.g. in Arabidopsis, there are eight cyclin A, 6 Cdc20, 5 Cdh1, 7 Kip–CIP-related (KRP) genes).
Even in the face of this challenge, some forward genetics has been successful. For example, the Siamese family of CDK inhibitors, which is only found in the plant kingdom, was discovered in a sensitive forward-genetic screen owing to a loss-of-function phenotype for SIA in one specific cell type (the Arabidopsis trichome), despite the presence of multiple Siamese-related, highly similar proteins [82]. As an alternative forward discovery method, recent work on protein interaction networks in the plant cell cycle [83] has identified a number of molecules not recognizable as having opisthokont homologues involved in cell cycle control.
Another means of forward discovery could revisit the screening techniques so successfully applied in yeast. The single-celled alga Chlamydomonas reinhardtii grows as a haploid, has well-established experimental genetics, and has a sequenced genome revealing two key features: a generally ‘plant-like’ suite of genes, but with most of these genes present in single copy [84]. These features extend specifically to the cell cycle control molecules known from opisthokonts [85]. It should, therefore, be possible to identify cell cycle control genes in an unbiased mutant hunt. Such an approach was taken in the past [86], but was not taken to completion, probably because of the great challenge of identifying the mutated genes. Advances in sequencing technology should minimize this difficulty, allowing a comprehensive approach.
Previous work has laid an interesting physiological foundation for thinking about the Chlamydomonas cell cycle as controlled by cell ‘sizers’ and ‘timers’ [87]. Mat3 null mutations led to an extreme disruption of ‘sizer’ control, with resulting extremely tiny cells [88]. The mat3 gene was shown to be a clear Rb homologue, including the LxCxE motif implicated in cyclin D interaction [88]. Remarkably, a small-cell phenotype is also observed in animal cells lacking all Rb family members [89,90]. Mutations in an E2F homologue as well as in a DP homologue (essential partner of E2F) suppressed the mat3 phenotype [91], exactly as would be expected from the canonical animal pathway in which Rb inhibits E2F-Dp. However, mat3 nulls showed no detectable change in cell cycle-regulated transcription of multiple plausible E2F target genes, suggesting that Rb function in the plant kingdom cannot be simply determined by an opisthokont overlay. Chlamydomonas Rb exhibits cell cycle-regulated phosphorylation-like animal Rb, but unlike the case in animals, no change in chromatin association could be detected through the cell cycle [92].
Arabidopsis Rb inactivation results in gametophyte lethality [93], and broad defects in differentiation and organ development. It is likely that analysis in the simpler Chlamydomonas system will provide testable hypotheses for Rb function in higher plants. A similar programme could be envisaged generally for a suitably comprehensive collection of sequence-identified Chlamydomonas cell cycle mutants.
10. Were Rb, E2F, Kip1, cyclins D and E present in the eukaryotic last common ancestor?
It has been proposed that the ancestral eukaryotic cell cycle consisted of a single B-type cyclin–Cdk complex, alternating in activity with the APC [94]. Strong justification for this view was the finding that these components and no others are common to yeast and animals, and at that time, the full eukaryotic phylogeny (figure 3) was unclear.
The current placement of the divergence of plants significantly before divergence of yeast and animals (figure 3) combined with the presence in plants of Rb, E2F and cyclins A and D, suggests instead that the eukaryotic LCA had cell cycle control carried out in some manner by all of these components, along with the universal eukaryotic APC–Cdc20–Cdh1 system. According to this view, many of these components were lost early in the fungal lineage, replaced in some still-mysterious way by other components playing identical network roles (figure 1).
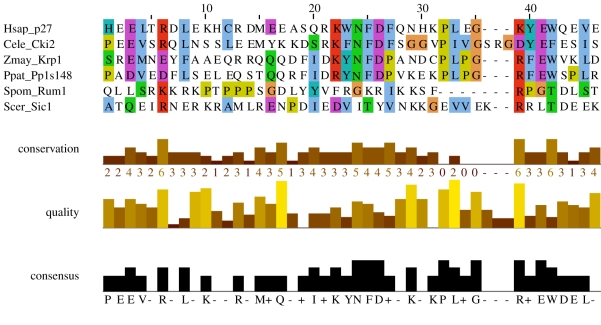
It is important to emphasize that Rb homology, for example, is unambiguous when looking at the plant or Chlamydomonas sequences aligned with the animal sequences [88]. Even more distantly related homologues, such as the KRP homologues of animal Kip–Cip Cdk inhibitors, show quite clear sequence similarity over key regions known from structural biology to be central to cyclin–Cdk inhibition by p27 and known from detailed mapping studies to be critical for Cdk inhibition by KRP [95–97]. Figure 4 shows sequence alignments of this critical region of KRP from maize [95] and moss with animal p27, along with a presumably best-case alignment of budding yeast Sic1 and the homologous Rum1 from fission yeast [99] to the same region, based on structural considerations and modelling [52]. The alignment between the plant and animal protein is obvious and almost sure to indicate monophyly; while Sic1 and Rum1 align with each other, they do not obviously align to the p27/KRP family any better than would be expected by chance (note that even the few possibly conserved residues between Sic1 and KRPs are not generally conserved in Rum1). While it is impossible to prove that any two proteins are truly unrelated (since in principle, sequential nucleotide substitution combined with deletion/insertion could transit between any two arbitrary sequences), it is logical and indeed frequently observed that sequence homology persists in a restricted ‘active site’ region. The KRP versus Sic1/Rum1 comparison is, therefore, a particularly clear case for arguing polyphyly because deletion mapping and structural studies restrict the key functional region to a very short stretch lacking any detectable alignment (figure 4).
Figure 4.
A sequence alignment of animal p27 from human (Homo sapiens) and C. elegans (nematode) [98], plant KRP1 [95] from Zea mays (maize) and P. patens (moss), along with a best-case alignment of Rum1 from fission yeast (Schizosaccharomyces pombe) and Sic1 from budding yeast (Saccharomyces cerevisiae) to the same region based on structural considerations and modelling [52,98,99]. The homology between the plant KRP1 and animal p27 is likely indicative of monophyly; it is not clear that the Sic1/Rum1 alignment to KRP1/p27 is any better than would be expected by chance.
For animal E2F/Dp compared with yeast Swi4/Mbp1, the lack of detectable sequence alignment is reinforced by structural studies showing a significantly different fold in the DNA-binding domain between E2F/DP and yeast Mbp1 [100–102]. While structures of plant E2F are not available, sequence alignment to animal E2F is clear, and conservation is especially clear in residues involved in DNA binding. Thus, the ancestral transcription factor, as the CDK inhibitor, may have been replaced in the fungal lineage.
The surprising degree of conservation of the opisthokont G1/S network topology, taken together with sequence divergence implying possible replacement of central controlling molecules with polyphyletic substitutes, suggests network structure is the key conserved feature. This structure clearly has many favourable dynamical features that we have documented in recent work in the yeast system: cell size control [16], positive feedback-driven coherent cell cycle entry [18,19] and irreversibility [20,44]. Many of these features have been attributed to the animal system as well [40,42]. If similar analysis suggests the same network features in the plant superkingdom, or in other highly diverged eukaryotes, it is likely that the opisthokont network topology originated in the LCA of the eukaryotes, and persists in modern yeast despite the possible near-complete replacement of key proteins with polyphyletic substitutes.
Compared with G1–S control proteins, mitotic control proteins appear to show greater sequence conservation, as key elements such as cyclin B, Cdk, APC, Polo kinase, Cdc20 and Cdh1 are recognizable in most or all eukaryotic genomes. However, the network topology and its associated dynamic properties are less well understood and may be quite complex. Significant differences clearly exist even among relatively closely related organisms. For example, Wee1/Cdc25 is central to mitotic control in fission yeast but not in budding yeast. The nimA kinase is essential for mitosis in Aspergillus, but nimA relatives have more peripheral mitotic role in other eukaryotes [103]. Strikingly, despite a highly similar genome, there may be significant differences in mitotic control in Ashbya gossypii compared with budding yeast [104,105]. We expect that a properly grounded comparative phylogenetic approach will be central to extracting essential elements of mitotic control and their conservation or divergence through eukaryotic evolution.
Recent comparative genomics of protists implies that the LCA had a remarkably sophisticated and versatile signalling, metabolic, cytoskeletal repertoire [106]. Any novel and initially simple circuit will probably have undergone substantial evolution prior to its more elaborate implementation in the LCA of multiple descendant lines. Consistent with this idea, our analysis suggests that ancestral cell cycle control may not have been equivalent to the minimal essential circuit, but rather had complexities and redundancies similar to those found in modern organisms.
Acknowledgements
We thank Amanda Amodeo, Liam Holt and Danny Lew for reviewing the manuscript. We thank the following funding sources for their support: Burroughs Wellcome Fund (Career Award at the Scientific Interface to N.E.B. and J.M.S.), March of Dimes Foundation (Grant no. 5-FY11-70 to N.E.B.), NSF (CAREER Award no. 1054025 to J.M.S.), NIH (RO1 GM092925 to J.M.S., GM47238 and GM78153 to F.C.).
References
- 1.Lee M. G., Nurse P. 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327, 31–35 10.1038/327031a0 (doi:10.1038/327031a0) [DOI] [PubMed] [Google Scholar]
- 2.Forsburg S. L., Nurse P. 1991. Identification of a G1-type cyclin puc1+ in the fission yeast Schizosaccharomyces pombe. Nature 351, 245–248 10.1038/351245a0 (doi:10.1038/351245a0) [DOI] [PubMed] [Google Scholar]
- 3.Koff A., Cross F., Fisher A., Schumacher J., Leguellec K., Philippe M., Roberts J. M. 1991. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66, 1217–1228 10.1016/0092-8674(91)90044-Y (doi:10.1016/0092-8674(91)90044-Y) [DOI] [PubMed] [Google Scholar]
- 4.Léopold P., O'Farrell P. H. 1991. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell 66, 1207–1216 10.1016/0092-8674(91)90043-X (doi:10.1016/0092-8674(91)90043-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lew D. J., Dulić V., Reed S. I. 1991. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66, 1197–1206 10.1016/0092-8674(91)90042-W (doi:10.1016/0092-8674(91)90042-W) [DOI] [PubMed] [Google Scholar]
- 6.Xiong Y., Connolly T., Futcher B., Beach D. 1991. Human D-type cyclin. Cell 65, 691–699 10.1016/0092-8674(91)90100-D (doi:10.1016/0092-8674(91)90100-D) [DOI] [PubMed] [Google Scholar]
- 7.Rao P. N., Johnson R. T. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature 225, 159–164 10.1038/225159a0 (doi:10.1038/225159a0) [DOI] [PubMed] [Google Scholar]
- 8.Rhind N., Russell P. 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113, 3889–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearsey S. E., Cotterill S. 2003. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12, 1067–1075 10.1016/S1097-2765(03)00441-6 (doi:10.1016/S1097-2765(03)00441-6) [DOI] [PubMed] [Google Scholar]
- 10.Araki H. 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell. Biol. 22, 766–771 10.1016/j.ceb.2010.07.015 (doi:10.1016/j.ceb.2010.07.015) [DOI] [PubMed] [Google Scholar]
- 11.Cooper K. 2006. Rb, whi it's not just for metazoans anymore. Oncogene 25, 5228–5232 10.1038/sj.onc.1209630 (doi:10.1038/sj.onc.1209630) [DOI] [PubMed] [Google Scholar]
- 12.Sherr C. J., Roberts J. M. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18, 2699–2711 10.1101/gad.1256504 (doi:10.1101/gad.1256504) [DOI] [PubMed] [Google Scholar]
- 13.De Veylder L., Beeckman T., Inzé D. 2007. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell. Biol. 8, 655–665 10.1038/nrm2227 (doi:10.1038/nrm2227) [DOI] [PubMed] [Google Scholar]
- 14.Rost B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94 10.1093/protein/12.2.85 (doi:10.1093/protein/12.2.85) [DOI] [PubMed] [Google Scholar]
- 15.Pringle J. R., Hartwell L. H. 1981. The Saccharomyces cerevisiae cell cycle. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- 16.Di Talia S., Skotheim J. M., Bean J. M., Siggia E. D., Cross F. R. 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 10.1038/nature06072 (doi:10.1038/nature06072) [DOI] [PubMed] [Google Scholar]
- 17.Eser U., Falleur-Fettig M., Johnson A., Skotheim J. M. 2011. Commitment to a cellular transition precedes genome-wide tansriptional change. Mol. Cell. 43, 515–527 10.1016/j.molcel.2011.06.024 (doi:10.1016/j.molcel.2011.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skotheim J. M., Di Talia S., Siggia E. D., Cross F. R. 2008. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454, 291–296 10.1038/nature07118 (doi:10.1038/nature07118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eser U., Falleur-Fettig M., Johnson A., Skotheim J. M. 2011. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol. Cell. 43, 515–527 10.1016/j.molcel.2011.06.024 (doi:10.1016/j.molcel.2011.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doncic A., Falleur-Fettig M., Skotheim J. M. 2011. Distinct interactions select and maintain a specific cell fate. Mol. Cell. 43, 528–539 10.1016/j.molcel.2011.06.025 (doi:10.1016/j.molcel.2011.06.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zetterberg A., Larsson O. 1985. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl Acad. Sci. USA 82, 5365–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster D. A., Yellen P., Xu L., Saqcena M. 2010. Regulation of G1 cell cycle progression: distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 1, 1124–1131 10.1177/1947601910392989 (doi:10.1177/1947601910392989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon I., Raff M. 2003. Differences in the way a mammalian cell and yeast cells coordinate cell growth and cell-cycle progression. J. Biol. 2, 7. 10.1186/1475-4924-2-7 (doi:10.1186/1475-4924-2-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Killander D., Zetterberg A. 1965. A quantitative cytochemical investigation of the relationship between cell mass and initiation of DNA synthesis in mouse fibroblasts in vitro. Exp. Cell Res. 40, 12–20 10.1016/0014-4827(65)90285-5 (doi:10.1016/0014-4827(65)90285-5) [DOI] [PubMed] [Google Scholar]
- 25.Tzur A., Kafri R., LeBleu V. S., Lahav G., Kirschner M. W. 2009. Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–171 10.1126/science.1174294 (doi:10.1126/science.1174294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbour J., Luo R., Santi A., Postigo A., Dean D. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98, 859–869 10.1016/S0092-8674(00)81519-6 (doi:10.1016/S0092-8674(00)81519-6) [DOI] [PubMed] [Google Scholar]
- 27.Takahata S., Yu Y., Stillman D. 2009. The E2F functional analogue SBF recruits the Rpd3 (L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 28, 3378–3389 10.1038/emboj.2009.270 (doi:10.1038/emboj.2009.270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Carey L. B., Cai Y., Wijnen H., Futcher B. 2009. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 7, e1000189. 10.1371/journal.pbio.1000189 (doi:10.1371/journal.pbio.1000189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costanzo M., et al. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899–913 10.1016/j.cell.2004.05.024 (doi:10.1016/j.cell.2004.05.024) [DOI] [PubMed] [Google Scholar]
- 30.de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., 3rd, Wittenberg C. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117, 887–898 10.1016/j.cell.2004.05.025 (doi:10.1016/j.cell.2004.05.025) [DOI] [PubMed] [Google Scholar]
- 31.Weinberg R. 1995. The retinoblastoma protein and cell cycle control. Cell 81, 323–330 10.1016/0092-8674(95)90385-2 (doi:10.1016/0092-8674(95)90385-2) [DOI] [PubMed] [Google Scholar]
- 32.Di Talia S., Wang H., Skotheim J. M., Rosebrock A. P., Futcher B., Cross F. R. 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 7, e1000221. 10.1371/journal.pbio.1000221 (doi:10.1371/journal.pbio.1000221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall M. N., Raff M. C., Thomas G. 2004. Cell growth: control of cell size. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 34.Sherr C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60, 3689–3695 [PubMed] [Google Scholar]
- 35.Wang H., Gari E., Verges E., Gallego C., Aldea M. 2004. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin-Cdk complex to late G1. EMBO J. 23, 180–190 10.1038/sj.emboj.7600022 (doi:10.1038/sj.emboj.7600022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross F. R., Blake C. M. 1993. The yeast Cln3 protein is an unstable activator of Cdc28. Mol. Cell Biol. 13, 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehl J. A., Cheng M., Roussel M. F., Sherr C. J. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499. 10.1101/gad.12.22.3499 (doi:10.1101/gad.12.22.3499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanker S., Valdivieso M. H., Wittenberg C. 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271, 1597–1601 10.1126/science.271.5255.1597 (doi:10.1126/science.271.5255.1597) [DOI] [PubMed] [Google Scholar]
- 39.Tyers M., Tokiwa G., Futcher B. 1993. Comparison of the Saccharomyces-cerevisiae G1 cyclins—Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12, 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee T. J., Yao G., Bennett D. C., Nevins J. R., You L. 2010. Stochastic E2F activation and reconciliation of phenomenological cell-cycle models. PLoS Biol. 8, e1000488. 10.1371/journal.pbio.1000488 (doi:10.1371/journal.pbio.1000488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundberg A. S., Weinberg R. A. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao G., Lee T. J., Mori S., Nevins J. R., You L. 2008. A bistable Rb-E2F switch underlies the restriction point. Nat. Cell. Biol. 10, 476–482 10.1038/ncb1711 (doi:10.1038/ncb1711) [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa M., et al. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15, 7060–7069 [PMC free article] [PubMed] [Google Scholar]
- 44.Charvin G., Oikonomou C., Siggia E. D., Cross F. R. 2010. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 8, e1000284. 10.1371/journal.pbio.1000284 (doi:10.1371/journal.pbio.1000284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bean J. M., Siggia E. D., Cross F. R. 2005. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 171, 49–61 10.1534/genetics.105.044560 (doi:10.1534/genetics.105.044560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bruin R. A., Kalashnikova T. I., Chahwan C., McDonald W. H., Wohlschlegel J., Yates J., 3rd, Russell P., Wittenberg C. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23, 483–496 10.1016/j.molcel.2006.06.025 (doi:10.1016/j.molcel.2006.06.025) [DOI] [PubMed] [Google Scholar]
- 47.Hinds P., Mittnacht S., Dulic V., Arnold A., Reed S., Weinberg R. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70, 993–1006 10.1016/0092-8674(92)90249-C (doi:10.1016/0092-8674(92)90249-C) [DOI] [PubMed] [Google Scholar]
- 48.Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. 2002. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat. Cell Biol. 4, 358–366 10.1038/ncb785 (doi:10.1038/ncb785) [DOI] [PubMed] [Google Scholar]
- 49.Martinez J. S., Jeong D. E., Choi E., Billings B. M., Hall M. C. 2006. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol. Cell Biol. 26, 9162–9176 10.1128/MCB.00603-06 (doi:10.1128/MCB.00603-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrezuelo F., Colomina N., Futcher B., Aldea M. 2010. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 11, R67. 10.1186/gb-2010-11-6-r67 (doi:10.1186/gb-2010-11-6-r67) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostapenko D., Burton J. L., Wang R., Solomon M. J. 2008. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol. Cell Biol. 28, 4653–4664 10.1128/MCB.00055-08 (doi:10.1128/MCB.00055-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barberis M., De Gioia L., Ruzzene M., Sarno S., Coccetti P., Fantucci P., Vanoni M., Alberghina L. 2005. The yeast cyclin-dependent kinase inhibitor Sic1 and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain. Biochem. J. 387, 639–647 10.1042/BJ20041299 (doi:10.1042/BJ20041299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amon A., Tyers M., Futcher B., Nasmyth K. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74, 993–1007 10.1016/0092-8674(93)90722-3 (doi:10.1016/0092-8674(93)90722-3) [DOI] [PubMed] [Google Scholar]
- 54.Ayté J., Schweitzer C., Zarzov P., Nurse P., DeCaprio J. A. 2001. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat. Cell. Biol. 3, 1043–1050 10.1038/ncb1201-1043 (doi:10.1038/ncb1201-1043) [DOI] [PubMed] [Google Scholar]
- 55.Dynlacht B. D., Moberg K., Lees J. A., Harlow E., Zhu L. 1997. Specific regulation of E2F family members by cyclin-dependent kinases. Mol. Cell. Biol. 17, 3867–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivanovska I., et al. 2008. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28, 2167–2174 10.1128/MCB.01977-07 (doi:10.1128/MCB.01977-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blomberg I., Hoffmann I. 1999. Ectopic expression of Cdc25A accelerates the G1/S transition and leads to premature activation of cyclin E-and cyclin A-dependent kinases. Mol. Cell. Biol. 19, 6183–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yung Y., Walker J., Roberts J., Assoian R. 2007. A Skp2 autoinduction loop and restriction point control. J. Cell Biol. 178, 741–747 10.1083/jcb.200703034 (doi:10.1083/jcb.200703034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson D., Ohtani K., Nevins J. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8, 1514–1525 10.1101/gad.8.13.1514 (doi:10.1101/gad.8.13.1514) [DOI] [PubMed] [Google Scholar]
- 60.Morgan D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell. Dev. Biol. 13, 261–291 10.1146/annurev.cellbio.13.1.261 (doi:10.1146/annurev.cellbio.13.1.261) [DOI] [PubMed] [Google Scholar]
- 61.Santamaría D., et al. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815 10.1038/nature06046 (doi:10.1038/nature06046) [DOI] [PubMed] [Google Scholar]
- 62.Moffat J., Andrews B. 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell. Biol. 6, 59–66 10.1038/ncb1078 (doi:10.1038/ncb1078) [DOI] [PubMed] [Google Scholar]
- 63.Nishizawa M., Kawasumi M., Fujino M., Toh-e A. 1998. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell. 9, 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang D., Patrick G., Moffat J., Tsai L. H., Andrews B. 1999. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl Acad. Sci. USA 96, 14 445–14 450 10.1073/pnas.96.25.14445 (doi:10.1073/pnas.96.25.14445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rojas J., Santos E. 2006. Ras-gefs and Ras gaps. Proteins Cell Regul. 4, 15–43 10.1007/1-4020-4708-8_2 (doi:10.1007/1-4020-4708-8_2) [DOI] [Google Scholar]
- 66.Mocciaro A., Schiebel E. 2010. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 123, 2867–2876 10.1242/jcs.074815 (doi:10.1242/jcs.074815) [DOI] [PubMed] [Google Scholar]
- 67.Herskowitz I. 1985. A master regulatory locus that determines cell specialization in yeast. Harvey Lect. 81, 67–92 [PubMed] [Google Scholar]
- 68.Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344, 503–508 10.1038/344503a0 (doi:10.1038/344503a0) [DOI] [PubMed] [Google Scholar]
- 69.James T. Y., et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822 10.1038/nature05110 (doi:10.1038/nature05110) [DOI] [PubMed] [Google Scholar]
- 70.Rogozin I. B., Basu M. K., Csürös M., Koonin E. V. 2009. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1, 99–113 10.1093/gbe/evp011 (doi:10.1093/gbe/evp011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cavalier-Smith T., Chao E. E. 2003. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J. Mol. Evol. 56, 540–563 10.1007/s00239-002-2424-z (doi:10.1007/s00239-002-2424-z) [DOI] [PubMed] [Google Scholar]
- 72.Lang B. F., O'Kelly C., Nerad T., Gray M. W., Burger G. 2002. The closest unicellular relatives of animals. Curr. Biol. 12, 1773–1778 10.1016/S0960-9822(02)01187-9 (doi:10.1016/S0960-9822(02)01187-9) [DOI] [PubMed] [Google Scholar]
- 73.Steenkamp E. T., Wright J., Baldauf S. L. 2006. The protistan origins of animals and fungi. Mol. Biol. Evol. 23, 93–106 10.1093/molbev/msj011 (doi:10.1093/molbev/msj011) [DOI] [PubMed] [Google Scholar]
- 74.Dewitte W., Murray J. A. 2003. The plant cell cycle. Annu. Rev. Plant Biol. 54, 235–264 10.1146/annurev.arplant.54.031902.134836 (doi:10.1146/annurev.arplant.54.031902.134836) [DOI] [PubMed] [Google Scholar]
- 75.Criqui M. C., Parmentier Y., Derevier A., Shen W. H., Dong A., Genschik P. 2000. Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 24, 763–773 10.1111/j.1365-313X.2000.t01-1-.x (doi:10.1111/j.1365-313X.2000.t01-1-.x) [DOI] [PubMed] [Google Scholar]
- 76.Verkest A., et al. 2005. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736 10.1105/tpc.105.032383 (doi:10.1105/tpc.105.032383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dissmeyer N., Nowack M. K., Pusch S., Stals H., Inzé D., Grini P. E., Schnittger A. 2007. T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19, 972–985 10.1105/tpc.107.050401 (doi:10.1105/tpc.107.050401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stracker T. H., Usui T., Petrini J. H. 2009. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 8, 1047–1054 10.1016/j.dnarep.2009.04.012 (doi:10.1016/j.dnarep.2009.04.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Schutter K., et al. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19, 211–225 10.1105/tpc.106.045047 (doi:10.1105/tpc.106.045047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dissmeyer N., et al. 2009. Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654 10.1105/tpc.109.070417 (doi:10.1105/tpc.109.070417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sorrell D. A., Combettes B., Chaubet-Gigot N., Gigot C., Murray J. A. 1999. Distinct cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco bright yellow-2 cells. Plant Physiol. 119, 343–352 10.1104/pp.119.1.343 (doi:10.1104/pp.119.1.343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Churchman M. L., et al. 2006. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18, 3145–3157 10.1105/tpc.106.044834 (doi:10.1105/tpc.106.044834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Leene J., et al. 2010. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397. 10.1038/msb.2010.53 (doi:10.1038/msb.2010.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merchant S. S., et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 10.1126/science.1143609 (doi:10.1126/science.1143609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bisova K., Krylov D. M., Umen J. G. 2005. Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137, 475–491 10.1104/pp.104.054155 (doi:10.1104/pp.104.054155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harper J., Wu L., Sakuanrungsirikul S., John P. 1995. Isolation and partial characterization of conditional cell division cycle mutants in Chlamydomonas. Protoplasma 186, 149–162 10.1007/BF01281325 (doi:10.1007/BF01281325) [DOI] [Google Scholar]
- 87.Donnan L., John P. C. 1983. Cell cycle control by timer and sizer in Chlamydomonas. Nature 304, 630–633 10.1038/304630a0 (doi:10.1038/304630a0) [DOI] [PubMed] [Google Scholar]
- 88.Umen J. G., Goodenough U. W. 2001. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15, 1652–1661 10.1101/gad.892101 (doi:10.1101/gad.892101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dannenberg J. H., Van Rossum A., Schuijff L., Te Riele H. 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14, 3051–3064 10.1101/gad.847700 (doi:10.1101/gad.847700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sage J., Mulligan G. J., Attardi L. D., Miller A., Chen S., Williams B., Theodorou E., Jacks T. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14, 3037–3050 10.1101/gad.843200 (doi:10.1101/gad.843200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang S. C., de los Reyes C., Umen J. G. 2006. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2, e167. 10.1371/journal.pgen.0020167 (doi:10.1371/journal.pgen.0020167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olson B. J., et al. 2010. Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22, 3331–3347 10.1105/tpc.110.076067 (doi:10.1105/tpc.110.076067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebel C., Mariconti L., Gruissem W. 2004. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429, 776–780 10.1038/nature02637 (doi:10.1038/nature02637) [DOI] [PubMed] [Google Scholar]
- 94.Nasmyth K. 1995. Evolution of the cell cycle. Phil. Trans. R. Soc. Lond. B 349, 271–281 10.1098/rstb.1995.0113 (doi:10.1098/rstb.1995.0113) [DOI] [PubMed] [Google Scholar]
- 95.Coelho C. M., Dante R. A., Sabelli P. A., Sun Y., Dilkes B. P., Gordon-Kamm W. J., Larkins B. A. 2005. Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol. 138, 2323–2336 10.1104/pp.105.063917 (doi:10.1104/pp.105.063917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Russo A. A., Jeffrey P. D., Patten A. K., Massagué J., Pavletich N. P. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382, 325–331 10.1038/382325a0 (doi:10.1038/382325a0) [DOI] [PubMed] [Google Scholar]
- 97.Zhou Y., Li G., Brandizzi F., Fowke L. C., Wang H. 2003. The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J. 35, 476–489 10.1046/j.1365-313X.2003.01821.x (doi:10.1046/j.1365-313X.2003.01821.x) [DOI] [PubMed] [Google Scholar]
- 98.Vlach J., Hennecke S., Amati B. 1997. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 16, 5334–5344 10.1093/emboj/16.17.5334 (doi:10.1093/emboj/16.17.5334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sánchez-Díaz A., González I., Arellano M., Moreno S. 1998. The Cdk inhibitors p25rum1 and p40SIC1 are functional homologues that play similar roles in the regulation of the cell cycle in fission and budding yeast. J. Cell. Sci. 111, 843–851 [DOI] [PubMed] [Google Scholar]
- 100.Taylor I. A., Treiber M. K., Olivi L., Smerdon S. J. 1997. The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1 A resolution. J. Mol. Biol. 272, 1–8 10.1006/jmbi.1997.1229 (doi:10.1006/jmbi.1997.1229) [DOI] [PubMed] [Google Scholar]
- 101.Xu R. M., Koch C., Liu Y., Horton J. R., Knapp D., Nasmyth K., Cheng X. 1997. Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis. Structure 5, 349–358 10.1016/S0969-2126(97)00192-5 (doi:10.1016/S0969-2126(97)00192-5) [DOI] [PubMed] [Google Scholar]
- 102.Zheng N., Fraenkel E., Pabo C. O., Pavletich N. P. 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666–674 10.1101/gad.13.6.666 (doi:10.1101/gad.13.6.666) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Regan L., Blot J., Fry A. M. 2007. Mitotic regulation by NIMA-related kinases. Cell Div. 2, 25. 10.1186/1747-1028-2-25 (doi:10.1186/1747-1028-2-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gladfelter A. S., Hungerbuehler A. K., Philippsen P. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172, 347–362 10.1083/jcb.200507003 (doi:10.1083/jcb.200507003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hungerbuehler A. K., Philippsen P., Gladfelter A. S. 2007. Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells. Eukaryot. Cell 6, 473–486 10.1128/EC.00273-06 (doi:10.1128/EC.00273-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fritz-Laylin L. K., et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642 10.1016/j.cell.2010.01.032 (doi:10.1016/j.cell.2010.01.032) [DOI] [PubMed] [Google Scholar]