‘Dividing cells pass through a regular sequence of cell growth and division, known as the cell cycle’, according to a college textbook of biology published in 1983 [1], 5 years before the underlying principles of control were first laid bare during 1988, the annus mirabilis of cell cycle research [2,3]. One of the key architects of that revolution, Paul Nurse, was elected as President of the Royal Society in 2010, and this volume is intended in part as a tribute and in part as a reflection of what we now know, and what remains still to be found out about cell proliferation. Lest we forget that cells have fates other than their own reproduction, Pat O'Farrell [4] reminds us that many cells in our bodies survive for long periods in a quiescent state. He considers quiescence from the perspective of the developmental biologist, and sees growth factors as surrogates for nutritional signals. He surveys the complexity and the richness of growth control in higher eukaryotes, rightly pointing out what an important topic for future research this remains. This problem is also attacked from the perspective of the single starving cell by Yanagidai, who have been recently studying the effects of nitrogen or glucose deprivation on fission yeast [5]. It turns out that the results are daunting. Wild-type yeast undergoes two rapid divisions to generate quasi-spherical cells if they are suddenly deprived of nitrogen, and undergoes startling changes in intracellular morphology and metabolism that remain difficult to comprehend. Interestingly, defects in these adaptions are accompanied by cell death and hundreds of different genes, in many distant pathways, are required to respond to starvation or to ‘wake up’ when better times come along.
Long-term survival, we must remind ourselves, requires not only cell division but also sex. Dan Mazia, who was the guru of cell division of the 1950s and 1960s, put it thus: ‘More often than not, questions beginning with ‘Why’ are inane and of no service in scientific discourse. In biology, they sometimes make sense. If we ask why cells must divide, the answer can be given in terms of what happens if they do not. The answer is that they die, no matter what criterion of death we apply’ [6, pp. 82–83]. This applies to organisms as well as cells, of course, and it is still somewhat mysterious that we humans can all trace our ancestry back several billion years, yet we are all mortal. The continuity of the germ cells is something that successfully evolved, but is hard to explain and rarely examined. Yet, formation of gametes is amenable to genetic analysis, and van Werven & Amon [7] present an accessible and wide-ranging survey of the process in budding yeast, fission yeast and higher eukaryotes that makes clear what a delicately regulated process it is. Certain features seem to be common: the existence of a ‘master regulator’ whose activity depends critically on a very particular combination of nutritional or developmental signals, as well as downstream regulatory protein kinase cascades, in particular the TOR pathway and the cyclic AMP-dependent protein kinase. For some reason, functioning mitochondria are apparently also required for successful gamete formation in yeast and mice. Presumably, that has only been true since oxygen entered Earth's atmosphere two and a half billion years ago.
In ‘normal’ cell cycles, there is a gap between the end of mitosis and the start of DNA replication, and control of the G1 to S transition is an important point of no return in the cell cycle. Cross et al. [8] discuss the evolution of control networks at this stage of the cell cycle, comparing yeast with plants and animals. They find that many individual regulators have either undergone huge sequence divergence from the last common ancestor or have evolved from different origins. Despite this, the topology and dynamic properties of networks have striking similarities. Diffley [9] looks at the control of initiation of DNA replication and again notes the variety as well as the redundancy of mechanisms that ensure the genome is replicated once and only once each time a cell divides. Arguing from simple assumptions, he points out that suppression of re-replication must be close to 100 per cent efficiency and that a combination of mechanisms, each with a small but finite failure rate, is necessary to reduce the overall failure rate to acceptable levels.
High fidelity is also a major consideration for the control of key cell cycle transitions. The penalty for failure is high, the difference between success and failure is tiny, and mechanisms for assuring accuracy are numerous and robust. Having evolved over billions of years they may also be rather complicated and difficult to understand, as is the case with the so-called S-phase checkpoints, discussed in detail by Labib & De Piccoli [10]. As soon as people realized the importance of DNA and DNA replication for cells, in the early 1950s, they tested the effects of ionizing radiation and discovered that normal cells quickly stopped synthesizing DNA after X-ray damage (apart from very rare mutant individuals, who were extremely sensitive to X-irradiation and turned out—many years later—to carry mutations in the ATM protein kinase). These irradiated cells did not enter mitosis. After intense study, largely by geneticists, because biochemical analysis for such complex systems is for the most part too difficult, we are now aware of many if not most components of the S-phase checkpoint, but it is still difficult to appreciate how the system really works. Replication forks and collapsed replication forks are complicated structures and the details of how damage is sensed, signalled and repaired are complicated and only gradually being worked out in mechanistic detail. The virtue of Labib's account lies in its historical approach and his attention to describing the experiments that underlie our present understanding. Langerak & Russell [11] also discuss the effects of DNA damage on cell cycle progression and vice versa, concentrating on the mechanisms that repair double-strand breaks in DNA. These are largely twofold, non-homologous end joining (NHEJ), which tends to occur when DNA is broken during the G1 phase of the cell cycle, and homologous recombination (HR), a largely error-free repair process that uses sister chromatids to reconstruct lost DNA sequences. The latter requires production of long stretches of single-stranded DNA that search for neighbouring homologous DNA sequences and subsequently invade them. The abundance and the activity of a large cast of cofactors are regulated in such a way as to promote NHEJ during G1, when sister DNAs are absent, and HR during S and G2, when they are present.
A key issue in cell cycle studies has been the nature of the triggers for the onset of DNA replication and mitosis. Much to everyone's surprise, both turn out to be triggered by similar molecules, namely S- and M-phase-specific cyclin-dependent kinases (CDKs), and almost every article in this issue refers to these key cell cycle regulators. An important question about these enzymes, apart from their regulation, is their substrate specificity. In particular, and this was confusing in the early days of the modern era of cell cycle studies, how can it be that the same kinase initiates both S and M phase? Why, for example, do cells undergo DNA replication and not attempt to enter mitosis at the first appearance of CDK activity? Moreover, why do not cells re-replicate their chromosomes when a second rise in CDK activity triggers mitosis? Two explanations seemed possible at first: one was that different cyclins imbued Cdks with different properties, so S-phase cyclins promote S phase and M-phase cyclins catalyse mitosis. But an alternative, originally suggested by Paul Nurse and co-workers [12], is that it takes only a little Cdk activity to initiate S phase, but more to enter M phase. The available evidence suggests that the level of activity is indeed part of the story, as Uhlmann et al. [13] discuss in their article. However, this is only part of the story. Whether a cell enters to undergo DNA replication or mitosis in response to a rise in Cdk activity is as much determined by the presence or absence of substrates or structures for these kinases to work on. Thus, the reason why Cdks do not trigger S during G2 is that the pre-replication complexes required to initiate DNA replication are absent from this stage of the cell cycle. Likewise, G1 cells that have not yet replicated their DNA do not possess a pair of sister chromatids nor even the cohesion that will hold these together, and cannot undergo anything resembling a physiological mitosis until these have been produced.
Uhlmann et al. [13] take some trouble to examine whether ‘checkpoints’ impose order on the cell cycle, and conclude that, on the whole, they do not. We note, however, that the concept of checkpoint, while highly popular and therefore much abused in the literature, is often inappropriate in the context of the cell cycle as well as being rather fuzzy on close inspection. In an earlier generation, before yeast genetics was applied to cell cycle control, people like Dan Mazia used to talk about ‘Points of no return’ rather than ‘Checkpoints’. The idea of the checkpoint is that you may not proceed to the next process or event until the one in which you are presently engaged is complete: a quality control check. But there is something else as well—once you have finished a task and been allowed to pass on, you cannot go back. This applies equally to the G1–S, the G2–M and the metaphase to anaphase transitions. Uhlmann et al. [13] argue that even if the level of Cdks activity has an important role in determining entry into S or M, regulation of phosphatase activity plays an equally important part. The various thresholds for cell cycle transitions are set by ratios of kinase activity to phosphatase activity and not by kinase activity per se. This theme continues in the contribution from Domingo-Sasanes et al. [14], who restrict their discussion to the control of mitosis, but focus on the recently discovered role of greatwall kinase as a controller of protein phosphatases that both regulate and antagonize Cdks at the G2–M transition.
This brings us to the end of the cell cycle, or the beginning of the next, the metaphase to anaphase transition. Musacchio [15] entitles his piece ‘Spindle assembly checkpoint: the third decade’, inviting the query, what's taking you so long? The answer is that this is a very complicated piece of machinery involving both hardware (the kinetochore itself, and its connection with spindle microtubules) and software—the error correction mechanisms and surveillance mechanisms that constitute the spindle assembly checkpoint (SAC). Interestingly, this regulatory system, unlike other the so-called checkpoints, has little or no role in repairing the damage sensed and appears solely concerned with regulating cell cycle progression. At least three or four protein kinases (and presumably their counterpart phosphatases) are involved as well as specific regulatory proteins such as Mad2. Working out how the SAC functions will require greater understanding of kintetochore structure as well as further structural work on its target, the anaphase-promoting complex/cyclsome (APC/C). Nevertheless, it is clear that the structural approach has already been extremely illuminating. At the moment, it looks as if the mechanical basis for the tension sensor may be nothing more complicated than a substrate being pulled beyond the reach of a tethered kinase. We may hope that some of the other seemingly complex features of the mitotic checkpoint will proved to have a similarly simple basis, once we understand them.
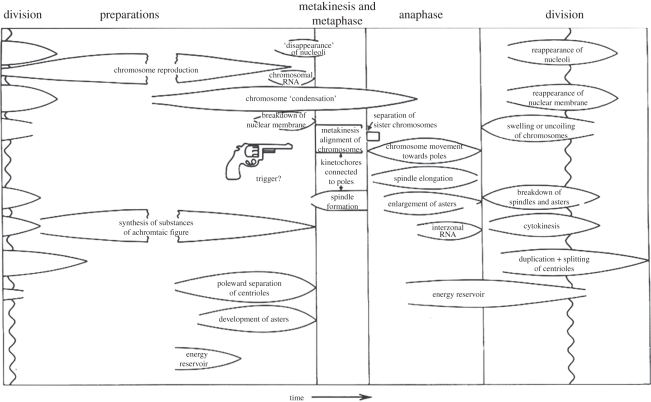
David Barford's [16] magisterial review of the APC/C depends even more so on structural determination, but his recent impressive advances required him to work out ways of making large quantities of this enormous, complicated multi-subunit complex. As he describes, we can begin to see how the thing works, although mysteries still remain, particularly in its control. This is connected with the previous paper, of course, because somehow the SAC can reliably amplify a signal from a single kinetochore to inhibit millions of APC/Cs, and somehow (as anyone who has ever watched cells enter anaphase can testify) the inhibition is lifted when all the chromosomes are properly aligned on the metaphase plate, such that it looks as though someone fired a starting pistol (figure 1) to signal chromosome separation.
Figure 1.
Mazia's diagram of mitosis with the starting pistol (‘Trigger?’). Adapted from Mazia [6].
Pines & Hagan [17] revisit many of the points raised in the preceding articles from a wide-ranging perspective, but they go on to stress the importance of intensely local conditions controlling physically distant processes. A particular concern of theirs is the spindle pole in fission yeast and its role as the place where a commitment to mitosis is made first, and the centrosome in animal cells, which they argue plays a central role in the control of mitotic entry. They make a plea for more quantitative studies in cell biology, and urge the development of reporters that can monitor the local activity of protein kinases and phosphatases in living cells.
Hyman's article [18] takes up the same theme from a ‘systems biology’ perspective. Actually, one of his main concerns is people's understanding of exactly what is systems biology. Tony makes the important point that both time and space cover vast ranges of scale in biology. Molecular movements in proteins occur on the microsecond timescale, yet it takes years for a human to reach sexual maturity. Or look at Barford's beautiful pictures of the APC/C elsewhere in this issue and remind yourself that a human on the same scale would be roughly twice as big as the Earth. Hyman makes the same plea as Pines and Hagan: more emphasis on quantitative data is necessary to begin to make sense of biological models. He also provides a useful definition for systems biology: ‘It is the approach of collecting quantitative biological information at one level of complexity, and using it to build models that describe the next level of complexity’. Thus, he claims that ‘systems biology is an approach, not a field’. The more we learn about the complexity of the regulatory network controlling cell proliferation the more useful are the systems biology approaches in the cell cycle field.
The final contribution to this collection by Kronja & Orr-Weaver [19] covers one of the less familiar areas of cell cycle control, namely the control of expression of mRNA at the level of translation. It turns out that there is quite a lot to say, not only in the authors' favourite model system, the Drosophila egg, zygote and early embryo, but also in yeast, frogs and even human cells. It has been known for some time that translation of normal capped mRNAs declines during mitosis, whereas internal ribosome entry sites (IRES) are preferentially used, and some of the important examples of this switch have recently come to light; their underproduction causes faults in cytokinesis. The majority of well-established and better worked-out examples do come from eggs and early embryos, however, which is probably not surprising considering that transcriptional control of gene expression is largely absent in these (typically) enormous cells. The regulatory networks are quite complicated, as can be seen from figure 2 of this review. One suspects that there is rather a lot to be learned still in this area.
Altogether, this collection of articles provides a kind of partial snapshot of the current state of understanding in some of the most active areas of enquiry in the broad field of the cell cycle. For the most part, the general principles are reasonably well defined, but the precise details of molecular mechanism in many cases prove harder to pin down than perhaps one might have expected 25 years ago. Moreover, there remain considerable tracts of uncharted territory, that is, areas where even the basic principles are difficult to comprehend. First and foremost among these is the problem of the regulation and coordination of cell growth, and the relationship between growth control and division control, which was arguably the starting point when Paul Nurse decided to work on fission yeast with the late Murdoch Mitchison. It is perhaps appropriate to end with an acknowledgement of Murdoch's contribution to this field. Apart from definitively defining the field in the title of his 1971 monograph ‘The Biology of the Cell Cycle’ [20] (the term was not in common currency before this, surprisingly enough), Murdoch served as a stimulating, quizzical, generous mentor to Paul Nurse as well as two of the editors of this issue, Kim Nasmyth and Bela Novak. Murdoch took a keen interest in the cell cycle field until the very end, and his passing marks the end of an exciting and heroic era.
References
- 1.Curtis H. G. 1983. Biology, 4th edn. New York, NY: Worth; ISBN-13: 9780879011864 [Google Scholar]
- 2.Dunphy W. G., Brizuela L., Beach D., Newport J. 1988. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 54, 423–431 10.1016/0092-8674(88)90205-X (doi:10.1016/0092-8674(88)90205-X) [DOI] [PubMed] [Google Scholar]
- 3.Gautier J., Norbury C., Lohka M., Nurse P., Maller J. 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54, 433–439 10.1016/0092-8674(88)90206-1 (doi:10.1016/0092-8674(88)90206-1) [DOI] [PubMed] [Google Scholar]
- 4.O'Farrell P. H. 2011. Quiescence: early evolutionary origins and universality do not imply uniformity. Phil. Trans. R. Soc. B 366, 3498–3507 10.1098/rstb.2011.0079 (doi:10.1098/rstb.2011.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanagida M., Ikai N., Shimanuki M., Sajiki K. 2011. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Phil. Trans. R. Soc. B 366, 3508–3520 10.1098/rstb.2011.0124 (doi:10.1098/rstb.2011.0124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazia D. 1961. Mitosis and the physiology of cell division. In The cell, vol. III (eds Brachet J., Mirsky A. E.), pp. 77–413 New York, NY: Academic Press [Google Scholar]
- 7.van Werven F. J., Amon A. 2011. Regulation of entry into gametogenesis. Phil. Trans. R. Soc. B 366, 3521–3531 10.1098/rstb.2011.0081 (doi:10.1098/rstb.2011.0081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F. R., Buchler N. E., Skotheim J. M. 2011. Evolution of networks and sequences in eukaryotic cell cycle control. Phil. Trans. R. Soc. B 366, 3532–3544 10.1098/rstb.2011.0078 (doi:10.1098/rstb.2011.0078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diffley J. F. X. 2011. Quality control in the initiation of eukaryotic DNA replication. Phil. Trans. R. Soc. B 366, 3545–3553 10.1098/rstb.2011.0073 (doi:10.1098/rstb.2011.0073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labib K., De Piccoli G. 2011. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Phil. Trans. R. Soc. B 366, 3554–3561 10.1098/rstb.2011.0071 (doi:10.1098/rstb.2011.0071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langerak P., Russell P. 2011. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Phil. Trans. R. Soc. B 366, 3562–3571 10.1098/rstb.2011.0070 (doi:10.1098/rstb.2011.0070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern B., Nurse P. 1996. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12, 345–350 10.1016/S0168-9525(96)80016-3 (doi:10.1016/S0168-9525(96)80016-3) [DOI] [PubMed] [Google Scholar]
- 13.Uhlmann F., Bouchoux C., López-Avilés S. 2011. A quantitative model for cyclin-dependent kinase control of the cell cycle: revisited. Phil. Trans. R. Soc. B 366, 3572–3583 10.1098/rstb.2011.0082 (doi:10.1098/rstb.2011.0082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo-Sasanes M. R., Kapuy O., Hunt T., Novak B. 2011. Switches and latches: a biochemical tug-of-war between the kinases and phosphatases that control mitosis. Phil. Trans. R. Soc. B 366, 3584–3594 10.1098/rstb.2011.0087 (doi:10.1098/rstb.2011.0087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musacchio A. 2011. Spindle assembly checkpoint: the third decade. Phil. Trans. R. Soc. B 366, 3595–3604 10.1098/rstb.2011.0072 (doi:10.1098/rstb.2011.0072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barford D. 2011. Structural insights into anaphase-promoting complex function and mechanism. Phil. Trans. R. Soc. B 366, 3605–3624 10.1098/rstb.2011.0069 (doi:10.1098/rstb.2011.0069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pines J., Hagan I. 2011. The renaissance or the cuckoo clock. Phil. Trans. R. Soc. B 366, 3625–3634 10.1098/rstb.2011.0080 (doi:10.1098/rstb.2011.0080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyman A. A. 2011. Whither systems biology? Phil. Trans. R. Soc. B 366, 3635–3637 10.1098/rstb.2011.0074 (doi:10.1098/rstb.2011.0074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kronja I., Orr-Weaver T. L. 2011. Translational regulation of the cell cycle: when, where, how and why? Phil. Trans. R. Soc. B 366, 3638–3652 10.1098/rstb.2011.0084 (doi:10.1098/rstb.2011.0084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchison M. 1971. The biology of the cell cycle. Cambridge, UK: Cambridge University Press [Google Scholar]