Abstract
Whole comparative genomic hybridization (W-CGH) is a new technique that reveals cryptic differences in highly repetitive DNA sequences, when different genomes are compared using metaphase or interphase chromosomes. W-CGH provides a quick approach to identify differential expansion of these DNA sequences at the single-chromosome level in the whole genome. In this study, we have determined the frequency of constitutive chromatin polymorphisms in the centromeric regions of human chromosomes using a whole-genome in situ cross-hybridization method to compare the whole genome of five different unrelated individuals. Results showed that the pericentromeric constitutive heterochromatin of chromosome 6 exhibited a high incidence of polymorphisms in repetitive DNA families located in pericentromeric regions. The constitutive heterochromatin of chromosomes 5 and 9 was also identified as highly polymorphic. Although further studies are necessary to corroborate and assess the overall incidence of these polymorphisms in human populations, the use of W-CGH could be pertinent and of clinical relevance to assess rapidly, from a chromosomal viewpoint, genome similarities and differences in closely related genomes such as those of relatives, or in more specific situations such as bone marrow transplantation where chimerism is produced in the recipient.
Key words: comparative genomics, constitutive heterochromatin, human chromosomes, whole comparative genomic hybridization, W-CGH.
Introduction
Variations in the amount of constitutive heterochromatin that produce polymorphic and polytypic populations appear to be a general attribute of all eukaryote genomes.1 In the case of human beings, two unrelated individuals may share 99.9% of their genomic DNA sequence that is involved in producing specific proteins. Close to 0.1% difference is the result of genetic polymorphisms, which include single nucleotide polymorphisms and insertions/deletions.2 However, if large tandem arrays of repetitive DNA sequences, which are included in the constitutive heterochromatin, are included in the comparison, the genome differences involve millions of base pairs (bp) and can be observed under light microscopy.2 A variety of molecular and cytogenetic evidence supports the hypothesis that polymorphisms result from illegitimate recombination during DNA replication, leading to quantitative variability in constitutive heterochromatin and of most satellite DNA sequences.3,4
In the case of the human genome, polymorphic variants of nonacrocentric chromosomes usually occur at the para/pericentromeric heterochromatin of chromosomes 1, 9, 16 and the distal heterochromatin of the Y-chromosome.2 Highly repeated DNA sequences are C-band positive, and intraband discrimination among diverse DNA subfamilies occupying equivalent chromosome domains is possible using specific DNA probes via fluorescent in situ hybridization (FISH).5,6
It is now known that heterochromatin is not inert and is essential for cell and organism viability in multicellular eukaryotes.7 Some genes required for viability and fertility are thought to reside in heterochromatin, being elements required for normal chromosomal inheritance.8
The highly compact nature of the chromatin and the massive degree of repetition of DNA sequences means that identification of differences at the chromosomal level is not an easy task, because it requires the study of the complete genome of each individual, chromosome by chromosome, and the use of specific DNA probes for each DNA family.
Comparative genomic hybridization (CGH) is a suitable approach to identify differences in the contribution of genetic DNA copy number during the comparison of two distinct genomes; however, this technique does not provide information regarding differences in constitutive heterochromatin, because competitive subtraction by the addition of an unlabeled Cot-1 DNA fraction is used in the hybridization reaction.9,10 During the last few years, our research group has developed a method, whole comparative genomic hybridization (W-CGH), which allows the identification of chromosomal polymorphisms related to highly repetitive DNAs localized in constitutive heterochromatin. Using a common karyotype as a reference for DNA probe binding, W-CGH targets polymorphisms based on competition for hybridization between two different genomic DNAs. In this experimental system, subtraction of highly repetitive DNA sequences using unlabeled Cot-1 DNAs is not performed. W-CGH is ideal for the clear and rapid identification of relative levels of repetitive DNA sequences at all human centromeres.10 The relative size of the heterochromatic blocks is associated with the number of copies of each repetitive DNA sequence, a trait that is inherited in a Mendelian way.11 If the number of repeats in a sequence is consistently different between two individuals, then this chromosomal polymorphism in constitutive heterochromatin could be used as a good marker to discriminate or characterize cells and genomes of these individuals.
The aim of the present study was to determine the frequency of polymorphisms linked to high levels of variation in the amount of repetitive DNA sequences in human chromosomes using W-CGH.
Materials and Methods
Study population
Five healthy individuals (four females and one male) aged 28–49 years, with a normal karyotype and who were referred to the Hospital Juan Canalejo, Coruña, Spain, were included in this study. Written informed consent was obtained from all subjects and approval was given by the Ethics Committee of the Hospital Juan Canalejo, Coruña, Spain and Centro de Investigación Biomédica del Noreste (CIBIN), Instituto Mexicano del Seguro Social (IMSS), where all the samples were processed.
Whole comparative genomic hybridization
Heparinized peripheral blood lymphocytes were cultured for 72 h in RPMI 1640 (GIBCO, Grand Island, NY, USA) medium supplemented with serum and antibiotics. Cells were arrested at metaphase via a 3 h treatment with 10 mg/mL colchicine. Chromosome slides were prepared by exposing the cell suspension to 0.075 M KCl for 20 min and were then fixed in methanol:acetic acid (3:1). Slides were denatured in 70% formamide/2× saline sodium citrate (SSC), pH 7 at 70°C for 2 min and dehydrated in increasing ethanol concentrations. DNA samples were extracted from blood samples collected in ethylenediaminetetraacetic acid (EDTA) using a DNA isolation kit for mammalian blood (Roche Diagnostics Corporation, Indianapolis, IN, USA). One microgram from each DNA sample was independently labeled with biotin-14-2′-deoxyuridine 5′-triphosphate (dUTP) and with digoxigenin-11-dUTP using a commercial nick-translation kit (Roche Diagnostics). After DNA labeling, probe-size fragments were tested in a 1% agarose gel to ensure that they fell within the range of 600–2,000 bp (this step was critical for the competitive hybridization of the probes). The probes were denatured at 70°C for 10 min and hybridized overnight to a denatured sample at 37°C. Slides were washed in 50% formamide/2× SSC (pH 7) for 5 min and twice in 2× SSC (pH 7) for 3 min. A nonspecific antibody-blocking solution was applied for 5 min at 37°C. The whole-genome probe mix was detected via 30 min incubation with fluorescein isothiocyanate (FITC)-labeled avidin (1:400) and a rhodamine-conjugated antidigoxigenin antibody (Roche Diagnostics). Finally, the slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1 µg/mL) in Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA).10
Combinations of labeled whole DNA genomes from different individuals were cohybridized on metaphase chromosomes from a single predefined individual (Table 1). DNA of the same individual labeled with biotin 14-dUTP and digoxigenin-11-dUTP was used as a control to test that all centromeric regions showed yellow fluorescence. Calibration of the intensity of red (R) and green (G) channels for the digital images was also performed using this internal control.
Table 1. Combinations of five different DNA samples labeled with digoxigenin and biotin and cohybridized on the chromosomes of a predefined individual. This results in nine different experiments.
| DNA labelling with biotine | DNA labelling with digoxigenin | |||||
|---|---|---|---|---|---|---|
| IND 1 | IND 2 | IND 3 | IND 4 | IND 5 | ||
| IND 1 | REF | EXP 1 | EXP 2 | EXP 3 | EXP 4 | |
| IND 2 | X | REF | EXP 5 | EXP 6 | EXP 7 | |
| IND 3 | X | X | REF | EXP 8 | EXP 9 | |
EXP, experiment; REF, reference; IND, individual.
Fluorescence microscopy and digital image analysis
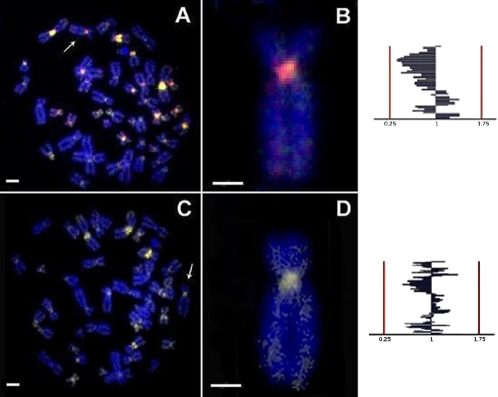
All slides were analyzed using a digital image analysis (DIA) platform based on a Leica DMLB fluorescence microscope (Leica Microsystems, Barcelona, Spain) and independent low-pass band filters for visualization of green, red, and blue fluorescence emissions. Images were acquired with a Leica DF-35 16 bit black and white charge-coupled device as 16-bit tagged image file format (TIFF) images. DIA for a semiquantitative assessment of the contribution of R and G fluorescence for each chromosome was performed in 50 different metaphases for each experimental approach. DIA was performed using the Q-Win image-analysis software (Leica Microsystems). Semiquantitative evaluation of the contribution of G and R fluorescence in metaphase chromosomes and interphase nuclei revealed the presence of color variations ranging between G and R on a scale of 0–255 gray levels. In general, pericentromeric regions showed yellow (G+R) fluorescence (Figure 1). We expected that G channel predominance would be indicative of a greater contribution of specific DNA sequences from G-labeled genomes, and vice versa. The comparison of two genomes should presumably show that the more abundant DNA sequence (e.g., R-labeled DNA) displaced the other DNA sequence (e.g., G-labeled DNA) by competition at a homologous chromosome region during hybridization. Fifty selected metaphases from each genome compared were digitized and analyzed. After background subtraction and G and R channel compensation, G and R channel values were plotted and compared for all chromosomes. Possible differences registered in the chromosomal arms were not detected using this experimental procedure because of blue quenching fluorescence produced after DAPI counterstaining.
Figure 1.
A) W-CGH in a human metaphase spread showing differential green or red dominance; B) chromosome 5 selected from the metaphase spread depicted in A shows red dominance; C) W-CGH in a human metaphase spread, where the pericentromeric heterochromatin blocks showed similar green and red fluorescence (yellow); D) chromosome 5 selected from the metaphase shown in C. The G/R ratio is indicated to the right of each chromosome. A and C scale bars: 10 µm; B and D scale bars: 5 µm.
Results
The comparison of two genomes after W-CGH shows that the most abundant DNA sequence in the whole DNA probe competes for hybridization successfully, displacing the other DNA sequence at an equivalent chromosome region. This results in a nonhomogeneous yellow color at the centromere region, with deviation to red or green depending on the contribution from each genome (compare selected chromosomes in Figure 1). Table 2 shows descriptive statistics (mean, standard deviation, maximum, minimum, and range) of the G/R variation of polymorphic and nonpolymorphic chromosomes in the nine different experiments described in Table 1. In our experimental conditions, this variable presented an internal error ranging from 0.23–0.90 in polymorphic, and 0.00–0.86 in nonpolymorhic chromosomes. Duplicate samples were tested in this study. There were no significant differences when the same processed samples were analyzed by two different observers, and in all the cases, identification of highly polymorphic centromeres coincided.
Table 2. Descriptive statistics for G/R variation in the nine different experiments.
| Experiment | X±SD | Maximum | Minimum | Range |
|---|---|---|---|---|
| 1 | 0.73±0.08 | 0.87 | 0.64 | 0.23 |
| 1′ | 1.03±0.014 | 1.05 | 1.02 | 0.03 |
| 2 | 1.03±0.41 | 1.98 | 0.67 | 1.31 |
| 2′ | 1.01±0.05 | 1.08 | 0.96 | 0.12 |
| 3 | 0.77±0.12 | 0.86 | 0.54 | 0.32 |
| 3′ | 0.98±0. 02 | 1.01 | 0.95 | 0.58 |
| 4 | 0.68±0.08 | 0.77 | 0.52 | 0.25 |
| 4′ | 1.03±0.04 | 1.09 | 1.00 | 0.86 |
| 5 | 0.88±0.21 | 1.32 | 0.61 | 0.71 |
| 5′ | 1.03±0.03 | 1.06 | 1.00 | 0.05 |
| 6 | 0.97±0.25 | 1.33 | 0.71 | 0.62 |
| 6′ | 1.01±0.00 | 1.01 | 1.01 | 0.00 |
| 7 | 1.17±0.18 | 1.47 | 0.72 | 0.75 |
| 7′ | 1.01±0.02 | 1.02 | 1.00 | 0.02 |
| 8 | 1.05±0.29 | 1.53 | 0.63 | 0.90 |
| 8′ | 0.99±0.02 | 1.01 | 0.98 | 0.03 |
| 9 | 1.07±0.26 | 1.41 | 0.73 | 0.68 |
| 9′ | 1.11±0.26 | 1.13 | 1.09 | 0.03 |
| Y-chromosome | 0.50±0.44 | 1.16 | 0.19 | 0.96 |
1 to 9, polymorphic chromosomes; 1′ to 9′, nonpolymorphic chromosomes.
To illustrate the similarities and differences in the results obtained for different chromosomes in all the compared genomes, both homologous chromosomes were studied in the 50 digital images. The ratio of G/R was obtained using the gray-level values detected along each chromosome and a G/R ratio >1.10 and <0.90 was considered as the cutoff value for defining differences in the contribution of each probe to the final hybridization. Using this criterion, we have summarized in Table 3 the average G/R values observed in the autosomes and on the sex chromosomes for all experiments. Chromosome (Chr)-6 exhibited the greatest level of polymorphism (5/9), followed by Chr-5 (4/9), Chr-9 (3/9), and Chr-1, Chr-14, and Chr-15 (2/9).
Table 3. Values of D/B in 22 autosomes, chromosome-X, and chromosome-Y in nine different experiments.
| Chromosome | EXP 1 | EXP 2 | EXP 3 | EXP 4 | EXP 5 | EXP 6 | EXP 7 | EXP 8 | EXP 9 | TOTAL |
|---|---|---|---|---|---|---|---|---|---|---|
| W-CGH | ♀D/♂B | ♀D/♀B | ♀D/♀B | ♀D/♀B | ♀D/♂B | ♀D/♂B | ♀D/♂B | ♀D/♀B | ♀D/♀B | |
| 1 | 1 | 1 | 0.73/0.77 | 1 | 1 | 0.81/0.83 | 1 | 1 | 1 | 2 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 3 | 0.76/0.79 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 4 | 1 | 1 | 1 | 1 | 1 | 1.16/1.22 | 1 | 1 | 1 | 1 |
| 5 | 1 | 1 | 1 | 0.67/0.73 | 0.78/0.86 | 1 | 1 | 1.33/1.40 | 1.27/1.27 | 4 |
| 6 | 1 | 1 | 0.73/0.77 | 1.15/1.11 | 1.23/1.32 | 1 | 1.18/1.03 | 1 | 1.26/1.26 | 5 |
| 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 9 | 1 | 0.81/0.80 | 1 | 1 | 1 | 1.12/1.13 | 0.74/0.71 | 1 | 1 | 3 |
| 10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 14 | 1 | 0.74/0.83 | 0.75/0.79 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 15 | 0.71/0.71 | 0.67/0.68 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 16 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 17 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 18 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 19 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1.27/1.37 | 1 |
| 20 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 21 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.72/0.75 | 1 |
| X | ND | 1 | 1 | 1 | ND | ND | ND | 1 | 1 | 0 |
| Y | 0.33 | ND | ND | ND | 0.22 | 0.32 | 0.28 | ND | ND | 4 |
Chromosomes with D/B values <0.90 and >1.10 are considered polymorphic; total, number of chromosomes polymorphic in the experiments.
Discussion
The results obtained in this study reinforce the validity of the W-CGH technique as a rapid method to assess genome differences between two compared genomes. The main values of this technique, in addition to its convenience of speed, are the identification of multiple differences in a single experiment and the possibility of comparing genomes of species where we lack information about genome characteristics. In fact, as demonstrated by Pita et al., there is the possibility of using this technique on interphase cells to recognize those differences that could be identified in parallel on the condensed chromosomes.12 This option has the outstandingly attractive possibility of testing for possible genome differences without the necessity of producing metaphase chromosomes. This technique has provided evidence for the presence of polymorphisms in insects where knowledge about the presence of these polymorphisms was low and mainly constrained to variations in C-banded regions.13 In mammalian species, unexplored polymorphisms were identified in swine, sheep, stallion, and donkey,14–16 and the technique also provided evidence for the presence of highly conserved repetitive DNA sequences on chromosome 9 when human and gorilla chromosomes were compared.17 Accordingly, W-CGH is a reliable tool for the comparison of the levels of polymorphism of repetitive DNA sequences in the human genome, giving a one-shot experimental hybridization for comparing two different genomes.
In human chromosomes, according to our results, these differences are particularly conspicuous at the pericentromeric regions of Chr-6, Chr-5, Chr-9, Chr-1, Chr-14, and Chr-15. Some of the chromosome domains identified in this experiment as highly polymorphic have also been reported as such using other techniques,18 and in particular, the constitutive heterochromatin on chromosomes 1, 9, and Y is known to be highly polymorphic largely because of the expansion differences existing in classical satellite DNA families.10 Human centromeric DNA families often exhibit highly variable array sizes on different chromosomes.19 Many exhibit variations of >500 kb, which render them amenable to visualization by W-CGH. Some chromosomes present markedly greater differences in the DNA families integrated within the constitutive heterochromatin. For example, α-satellite DNA presents size variations that range from 440 to 1550 kb in chromosome 118 and from 2250 to 4300 kb in chromosome 12.20 Differences displayed by other non-α DNA families may be even greater. For instance, β-satellite DNA harbors 30,000–60,000 copies of a 68 bp monomer located in the pericentromeric heterochromatin of chromosome 9 and in the acrocentric chromosomes 13, 14, 15, 21, and 22.21 Chromosomes harboring these DNA families are expected to exhibit greater differences during W-CGH analysis. Obviously, differences <500 bp are unlikely to be detected by W-CGH.
In addition to the rapid assessment of genome variability for identification of polymorphisms involving highly repetitive DNA sequences, the identification of those chromosomes showing the largest differences in tandem-repeat DNA sequences opens new possibilities for discriminating between two individuals in a different experimental context; for example, in sex-mismatched bone marrow transplantation where the recipient is always chimeric. In these cases, the W-CGH procedure could be used prior to the transplant to identify differences between donor and recipient. Once the polymorphic chromosomes have been identified, they can be targeted secondarily using specific DNA probes applied using a conventional FISH experimental environment. The use of single DNA probes for identification of a single chromosome would reveal differences in the size of the hybridized probe, allowing identification of the origin of the cell. Investigation of chimerism using this chromosome strategy has been performed elsewhere,12 using direct FISH experiments and also in situ digestion using restriction endonucleases, which uncovered certain polymorphisms at the chromosome level in Chr-9 and Chr-3.22,23 However, a prior W-CGH would increase the accuracy of discrimination between those chromosomes that could be putatively used for genome discrimination between donor and recipient.
One of the main constraints of this methodology is the possible existence of cross-hybridization between similar satellite DNA families present on different chromosomes. Stringency is critical, especially in human chromosomes, for the proper identification of single hybridization loci.24,25 For conventional FISH and when using a single DNA probe, the unbound probe or probe that has loosely bound to imperfectly matched sequences can be washed away to provide appropriate stringency, and in general, highly stringent conditions are advisable to achieve a high locus specificity.26 The conditions for probe binding are also critical to achieve a high specificity of the hybridized signal.27 The conditions for probe hybridization used in our protocol could be considered mild, because we used long probe incubation times and a relatively mild temperature (37°C) during hybridization. The use of 50% formamide/2× SSC for 5 min could also be considered mild conditions for removal of undesired DNA probe. However, we found that the results were repeatable using these conditions, and identification of the chromosomes involved in the polymorphisms was identical in different experiments. High-stringency probe hybridization and washing for probe removal after hybridization gave less stable results, and the amount of probe remaining on each chromosome varied greatly from one experiment to another.
Conclusions
The use of W-CGH is a quick experimental approach that allows identification, just by eye or using digital image analysis exploring the hybridized fluorescent signals on metaphase chromosomes, for a putative differential expansion of highly repetitive DNA sequences existing between two genomes. In the case of humans, we found that Chr-6, followed by Chr-5, Chr-9, Chr-1, Chr-14, and Chr-15, exhibited the larger level of polymorphism.
References
- 1.Jhon B. The biology of heterochromatin. In: Verma RS, editor. Heterocromatin, Molecular and Structural Aspects. Cambridge University Press; Cambridge, UK: 1988. pp. 1–147. [Google Scholar]
- 2.Hagymási K, Tulassay Z. The Human Genome Project, genetic viability and genetic epidemiology. Orv Hetil. 2005;146:2575–80. [PubMed] [Google Scholar]
- 3.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 4.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–20. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 5.Craig-Holmes AP, Moore FB, Shaw MW. Polymorphism of human C-band heterochromatin. I. Frequency of variants. Am J Hum Genet. 1973;25:181–92. [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Schwinger E, Mennicke K. Primed in situ labeling: sensitivity and specificity for detection of alpha-satellite DNA in the centromere regions of chromosomes 13 and 21. Cytogenet Cell Genet. 2001;95:28–33. doi: 10.1159/000057013. [DOI] [PubMed] [Google Scholar]
- 7.Vourćh C, Biamonti G. Transcription of salellite DNAs in mammals. Prog Mol Sub-cell Biol. 2011;51:95–118. doi: 10.1007/978-3-642-16502-3_5. [DOI] [PubMed] [Google Scholar]
- 8.Madon PF, Athalye AS, Parikh FR. Polymorphic variants on chromosome probably play a significant role in infertility. Reprod Biomed Online. 2005;11:726–32. doi: 10.1016/s1472-6483(10)61691-4. [DOI] [PubMed] [Google Scholar]
- 9.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–10. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- 10.Pita M, Fernández JL, Gosálvez J. Whole-Comparative Genomic Hybridization (W-CGH): 1- The quick overview of repetitive DNA sequences on a genome. Chromosome Res. 2003;11:673–9. doi: 10.1023/a:1025937726614. [DOI] [PubMed] [Google Scholar]
- 11.Verma RS. Heteromorphisms in heterochromatin. In: Verma RS, editor. Heterochromatin, molecular and structural aspects. Cambridge University Press; Cambridge, UK: 1988. pp. 276–92. [Google Scholar]
- 12.Pita M, Fernández JL, Gosálvez J. Whole-comparative genomic hybridization and “cell code” estimation: an application for assessment of cellular chimerism. Eur J Med Res. 2007;12:206–11. [PubMed] [Google Scholar]
- 13.Pita M, Zabal-Aguirre M, Arroyo F, Gosálvez J, López-Fernández C, De La Torre J. Arcyptera fusca and Arcyptera tornosi repetitive DNA families: whole-comparative genomic hybridization (W-CGH) as a novel approach to the study of satellite DNA libraries. J Evol Biol. 2008;21:352–61. doi: 10.1111/j.1420-9101.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- 14.Cortés-Gutiérrez EI, Dávila-Rodríguez MI, López-Fernández C, Fernández JL, Gosálvez J. Alkali-labile sites in sperm cells from Sus and Ovis species. Int J Androl. 2008;31:354–63. doi: 10.1111/j.1365-2605.2007.00781.x. [DOI] [PubMed] [Google Scholar]
- 15.Dávila-Rodríguez MI, Cortés-Gutiérrez EI, López-Fernández C, Pita M, Mezzanotte R, Gosálvez J. Whole-comparative genomic hybridization in domestic sheep (Ovis aries) breeds. Cytogenet Genome Res. 2009;124:19–26. doi: 10.1159/000200084. [DOI] [PubMed] [Google Scholar]
- 16.Gosálvez J, Crespo F, Vega-Pla JL, López-Fernández C, Cortés-Gutiérrez EI, Dávila-Rodriguez MI, et al. Shared Y chromosome repetitive DNA sequences in stallion and donkey as visualized using whole-genomic comparative hybridization. Eur J Histochem. 2010;54:e2. doi: 10.4081/ejh.2010.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pita M, Gosálvez J, Gosálvez A, Nieddu M, López-Fernández C, Mezzanotte R. A highly conserved pericentromeric domain in human and garilla chromosome. Cytogenet Genome Res. 2009;126:253–58. doi: 10.1159/000251962. [DOI] [PubMed] [Google Scholar]
- 18.Waye JS, Durfy SJ, Pinkel D, Kenwrick S, Patterson M, Davies KE, et al. Chromosome-speciphic alpha satellite DNA from human chromosme 1: hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics. 1987;1:43–51. doi: 10.1016/0888-7543(87)90103-0. [DOI] [PubMed] [Google Scholar]
- 19.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 20.Baldini A, Rocchi M, Archidiacono N, Miller OJ, Miller DA. A human alpha satellite DNA subset specific for chromosome 12. Am J Hum Genet. 1990;46:784–8. [PMC free article] [PubMed] [Google Scholar]
- 21.Waye JS, Willard HF. Human β-satellite DNA: genomic organization and sequence definition of a class of highly repetitive DNA. Proc Natl Acad Sci USA. 1989;86:6250–4. doi: 10.1073/pnas.86.16.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández JL, Pereira S, Campos A, Gosálvez J, Goyanes V. An extra band within the human 9qh+ region that behaves like the surrounding constitutive heterochromatin. J Med Genet. 1994;31:632–4. doi: 10.1136/jmg.31.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buño I, Fernández JL, López-Fernández C, Díez-Martín JL, Gosálvez J. Sau3A in situ digestion of human chromosome 3 pericentromeric heterochromatin I. Differential digestion of alpha-satellite1 DNA sequences. Genome. 2001;44:120–7. doi: 10.1139/gen-44-1-120. [DOI] [PubMed] [Google Scholar]
- 24.Durm M, Haar F M, Hausmann M, Ludwig H, Cremer C. Optimization of fast-fluorescence in situ hybridization with repetitive alpha-satellite probes. Z Naturforsch C. 1996;51:253–61. doi: 10.1515/znc-1996-3-418. [DOI] [PubMed] [Google Scholar]
- 25.Trowell HE, Nagy A, Vissel B, Choo KH. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet. 1993;2:1639–49. doi: 10.1093/hmg/2.10.1639. [DOI] [PubMed] [Google Scholar]
- 26.Vorsanova SG, Yurov YB, Soloviev IV, Demidova IA, Malet P. Rapid identification of marker chromosomes by in situ hybridization under different stringency conditions. Anal Cell Pathol. 1994;7:251–8. [PubMed] [Google Scholar]
- 27.Guerin-Reverchon I, Chardonnet Y, Chignol MC, Thivolet J. Study of stringency conditions for human papillomavirus DNA detection on cell lines, frozen and paraffin-embedded tissue sections by in situ hybridization with biotinylated probes. Histochemistry. 1990;93:637–43. doi: 10.1007/BF00272207. [DOI] [PubMed] [Google Scholar]