Abstract
Syncytin-1 and -2, human fusogenic glycoproteins encoded by the env genes of the endogenous retroviral loci ERVWE1 and ERVFRDE1, respectively, contribute to the differentiation of multinucleated syncytiotrophoblast in chorionic villi. In non-trophoblastic cells, however, the expression of syncytins has to be suppressed to avoid potential pathogenic effects. We studied the epigenetic suppression of ERVWE1 and ERVFRDE1 5′-long terminal repeats by DNA methylation and chromatin modifications. Immunoprecipitation of the provirus-associated chromatin revealed the H3K9 trimethylation at transcriptionally inactivated syncytins in HeLa cells. qRT-PCR analysis of non-spliced ERVWE1 and ERVFRDE1 mRNAs and respective env mRNAs detected efficient splicing of endogenously expressed RNAs in trophoblastic but not in non-placental cells. Pointing to the pathogenic potential of aberrantly expressed syncytin-1, we have found deregulation of transcription and splicing of the ERVWE1 in biopsies of testicular seminomas. Finally, ectopic expression experiments suggest the importance of proper chromatin context for the ERVWE1 splicing. Our results thus demonstrate that cell-specific retroviral splicing represents an additional epigenetic level controling the expression of endogenous retroviruses.
INTRODUCTION
Human endogenous retroviruses (HERVs) form a substantial part of the human genome. Despite the huge number of HERV copies, only a few intact open reading frames (ORFs) survived, avoiding the gradual accumulation of compromising mutations and/or deletions. Out of 18 intact env genes identified among various HERV families (1,2), four were shown to encode bona fide fusogenic retroviral envelope glycoproteins (1). The ERVWE1 provirus encodes a full-length envelope product dubbed syncytin-1, whose transmembrane (TM) subunit induces cell-to-cell fusion during differentiation of placental syncytiotrophoblast (3,4). Binding of the surface (SU) syncytin-1 subunit to the human sodium-dependent neutral amino acid transporters hASCT1 and hASCT2 is the first step of a complex cell fusion process (5). Syncytin-2 encoded by the provirus ERVFRDE1 is specifically expressed in the placenta and also displays fusogenic activity (6–8) in dependence on the carbohydrate transducer MFSD2 (9). Abnormalities of placental development and eclamptic disorders in pregnancy are often accompanied by altered syncytin-1 expression (10,11), whereas syncytin-2 acts rather as an immunosuppressor (12) and has been associated with abnormal trophoblast differentiation in placenta affected by trisomy 21 (13).
A high transcriptional activity of HERV elements has been detected in various healthy human tissues analyzed by a retrovirus-specific microarray (14) and quantitative RT–PCR (1). Expression of syncytins, however, is restricted to the placental trophoblast and only a weak expression was observed in the testes (3,4,6,15). Aberrant expression of syncytins outside of the placenta could trigger pathogenic changes, particularly chronic inflammation and inadvertent cell-to-cell fusions. Indeed, syncytin-1 has been detected in astrocytes and activated glial cells in the brain of multiple sclerosis patients (16,17). Syncytin-1 immunoreactivity has also been found in breast carcinomas and in higher-grade colorectal carcinomas (18,19). Cell-to-cell fusion in tumors may contribute to aneuploidy and promote gradual development of malignancies (20). Accordingly, syncytin-1 was shown to be involved in the fusion of breast cancer and endothelial cells (18). The hASCT1 and two receptors are widely expressed in many tissues, particularly in the liver and brain (21), and do not limit the fusogenic effect of syncytin-1. The MFSD2 receptor, in contrast, is expressed in the placenta and to a lesser extent in the testis and intestine (9).
Expression of syncytins must, therefore, be tightly controlled in order to avoid pathogenic fusions in non-placental tissues. The trophoblast-specific expression of syncytin-1 is controlled in part by the cyclic AMP-responsive ERVWE1 5′-LTR and in part by an upstream regulatory element (URE) of ∼400 bp containing the binding sites for AP2, Sp1 and Glial Cell Missing a (GCMa), the principal transcriptional factor of syncytin-1 and inductor of syncytiotrophoblast differentiation (22,23). Despite the tissue specificity of the URE, weak basal expression from ERVWE1 LTR can be observed after transient transfection into non-placental cells (23) and GCMa is not essential for this promoter activity (22). Therefore, the stringent suppression of syncytin-1 expression requires involvement of other mechanisms. We have shown that CpG methylation of ERVWE1 5′-LTR occurs in non-placental tissues and cell lines but not in trophoblastic cells from the placenta or choriocarcinoma BeWo cell line (24). These results were recently confirmed and also extended to ERVFRDE1 (25). Furthermore, CpG methylation of the ERVWE1 5′-LTR in non-placental cells is strongly resistant to demethylation by a DNA methyltransferase inhibitor and the reporter gene expression driven by ERVWE1 5′-LTR is efficiently suppressed by CpG methylation in transient expression assay (24). The whole picture of ERVWE1 suppression is, however, more complicated. At least in the testes, ERVWE1 transcripts have been detected (4) without any pathology. Because HERVs are known to keep the retroviral type of RNA splicing where only the spliced env mRNA serves for translation of the envelope glycoprotein, it is possible that ERVWE1 and ERVFRDE1 splicing adds another regulatory level to syncytins expression. The presence of two HERV-W-specific mRNA forms was previously detected in human placenta by northern blotting (4). Further analysis demonstrated the usage of two 5′-splice signals and three 3′-splice signals in the provirus ERVWE1 and splicing-out the 2.1 kb intron like insert (26). This splicing strategy of ERVWE1 has been conserved among all apes (27). The splicing of ERVFRDE1 utilizes the predicted splice site signals in the 5′-LTR and upstream of the env gene as described by sequencing of the 5′-RACE amplicon (8).
In the present study, we assessed the levels of genomic non-spliced and subgenomic spliced env mRNAs of both ERVWE1 and ERVFRDE1 in placental and non-placental cells by qRT–PCR. We demonstrate here that inefficient splicing together with epigenetic silencing by CpG methylation and inhibitory histone modifications suppress the expression of syncytins in non-placental cells. Based on these results, we suggest that the expression of endogenous retroviral glycoproteins in the mammalian genome can be efficiently controlled by tissue-specific pre-mRNA splicing.
MATERIALS AND METHODS
Tissue samples
All samples of healthy testicular tissues and testicular tumors were obtained from the Department of Urology of the Third Faculty of Medicine, Charles University in Prague. Biopsies taken from patients with hormone-sensitive prostate cancer treated by bilateral orchiectomy represented healthy testicular tissue (T1-h, T2-h) with prevalence of spermiogenous epithelium from which the tunica albuginea was discarded. Histological inspection showed normal testicular tissue with decreased spermiogenesis because of the advanced age of these patients. Six testicular tumors were included to this study; histologically they are seminomas (T3-s, T4-s, T5-s and T7s), one of them with a big component of granulomatous multinuclear cells (T3-s), mixed germinal tumor with embryonal carcinoma and teratoma components (T-6t) and testicular lymphoma (T8-t). Histologically normal testicular tissues surrounding the tumors were obtained by macrodissection (T5-h, T6-h, T7-h and T8-h). Human ethics approval was granted by the Committee for Ethics, Handling Recombinant DNA and Clinical Research at the Institute of Molecular Genetics, and informed consents were obtained from the patients.
Cell culture procedures
Choriocarcinoma BeWo and JEG-3 cells were maintained in F-12 and MEM-D media mixed 1:1 (Sigma) supplemented with 1% NaHCO3, 10% FBS, 300 µg/ml glutamine, a mix of penicillin and streptomycin (100 µg/ml each), and 2.5 mg/ml amphotericin B at 37°C in humified atmosphere of 5% CO2. HeLa cells were grown under the same conditions with the FBS supplement of 5%. Activation of BeWo and JEG-3 cells was done with 40 μM forskolin (Sigma) for 48 h. Transfections of 2 × 105 JEG-3 and HeLa cells seeded on the six-well plates were done with FuGENE HD lipofection reagent (Roche) according to the manufacturer’s recommendation. Stable transfection of JEG-3 and HeLa cells was done with 0.2 µg of pcDNA3 plasmid DNA mixed with a 10-fold excess of cloned ERVWE1 DNA (see below). The pcDNA3 plasmid DNA alone was used for mock-transfection. Two days after transfection, the cells were selected for the resistance to G418 and expanded single cell clones were assayed in splice-specific RT–PCR after 3 weeks.
Cloning of the full-length ERVWE1 provirus
The 10 900 bp fragment of genomic placental DNA containing the ERVWE1 provirus together with the URE at the 5′-end was amplified using the Phusion High-Fidelity DNA Polymerase (Finnzymes) according to the manufacturer’s instructions. Primers and PCR conditions are given in Supplementary Data. The PCR fragment was ligated into the pGEM T-easy vector (Promega), verified by restriction digestion and partial sequencing.
Bisulfite analysis of cytosine methylation
Isolation of DNA samples, sodium bisulfite treatment, PCR and sequencing of PCR clones were performed essentially as described previously (24). See Supplementary Data for the primers for semi-nested PCR of ERVFRDE1 from bisulfite-treated DNAs. We analyzed 6–13 independent PCR clones with at least 95% conversion of cytosine outside of CpG dinucleotides.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP assay kit (Upstate Biochemicals, Temecula, CA). BeWo and HeLa cell lines, either non-transfected or stably transfected with cloned ERVWE1 proviral DNA, were cross-linked in the presence of 1% formaldehyde for 10 min at room temperature. Chromatin was sheared by pulse sonication at 40% amplitude for 6 min to obtain DNA fragments of ∼200–500 bp in a Digital Sonifier S-450D (Branson, Danbury, CT). The antibodies used for ChIP were as follows: anti-acetyl-histone H3 (Lys 9), anti-trimethyl-histone H3 (Lys 9), anti-trimethyl-histone H3 (Lys 36) (ab4441, ab8898 and ab9050, respectively, all from Abcam, Cambridge, UK) and anti-histone H3 (07-690 Millipore). For the control, we immunoprecipitated the cell extracts using normal rabbit IgG (Millipore-Upstate) instead of hyperimmune antibody. Immunoprecipitated chromatin was subjected to RT–PCR quantification using primers specific for the transcription start site in the 5′-LTR of the proviruses ERVWE1 and ERVFRDE1, for the central part of the ERVWE1 intron, for the 5′-end of the ERVWE1 exon, and for the GAPDH gene (see Supplementary Data for the sequence of primers). The amount of immunoprecipitated material was normalized to the input DNA. The enrichment values relative to the input were low but in accordance with values shown for the respective antibodies by the manufacturer.
Splice-specific quantitative RT–PCR
Total RNA was isolated by using the Tri® Reagent (Sigma) according to manufacturer’s instructions. One μg of total RNA was DNase-treated for 15 min and reverse transcribed in a total volume of 50 μl, with M-MLV Reverse Transcriptase (Gibco) and random hexamers for priming. One μl of the resulting cDNA was then used for the quantitative PCR based on the MESA GREEN qPCR MasterMix Plus for SYBR Assay Kit (Eurogentec) and a Chromo4 system for RT–PCR detection (Bio-Rad). Quantifications were performed for the ERVWE1 non-spliced and spliced transcripts, the ERVFRDE1 non-spliced and spliced transcripts, and for the RNA polymerase IIa (POLR2A) as a housekeeping reference gene. The volume of the reaction mixture was 20 μl with 400 nM final concentration of each primer (see the Supplementary Data for the sequence of primers). External standards were constructed by PCR using BeWo cDNA and transcript-specific primer sets. Resulting PCR fragments of ERVWE1 and ERVFRDE1 were cloned into the pGEM-TEasy (Promega) and verified by sequencing. Ten-fold serial dilutions of external standard plasmids containing the genomic or spliced cDNA of either ERVWE1 or ERVFRDE1, ranging from 10 to 106 copies per reaction, were used for construction of the calibration curves. Ten-fold serial dilutions of plasmid containing the fragment of the human POLR2A gene, ranging from 10 to 106 copies per reaction, were used for the construction of POLR2A calibration curve. We normalized the numbers of RNA copies of both proviruses placed in the reaction according to the estimated number of POLR2A transcripts. Cycling conditions for ERVWE1 and POLR2A were 5 min at 95°C, 45 cycles of 15 s at 95°C, 20 s at 55°C, 30 s at 60°C. Cycling conditions for ERVFRDE1 and POLR2A were 5 min at 95°C, 45 cycles of 15 s at 95°C, 60 s at 60°C. The negative controls included water as a template. All quantitative RT–PCRs were performed in triplicate. The specificity of the PCR products amplified was confirmed by melting curve analysis and by sequencing the PCR products. To assess the amount of contaminating exogenous DNA, either genomic or plasmid, we included reactions run in the absence of the reverse transcriptase as negative controls. The background values of these negative controls were subtracted from the results of repective reactions with reverse transcriptase.
RESULTS
The histone modifications of active and silenced ERVWE1 and ERVFRDE1 loci
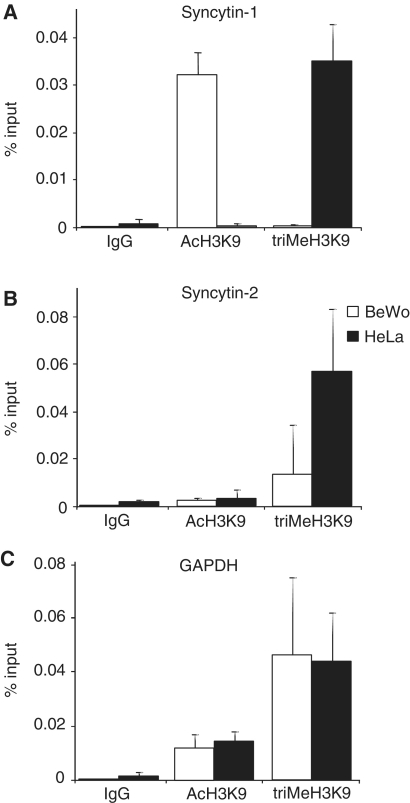
Post-translational histone modifications, particularly acetylation or trimethylation of histone H3 lysine 9 (H3K9), are signatures of active transcription or gene repression (28). To investigate this epigenetic level of transcriptional regulation, we compared H3K9 modifications associated with ERVWE1 and ERVFRDE1 5′-LTRs by ChIP in choriocarcinoma BeWo cells and cervical carcinoma HeLa cells, which show transcriptional activity or gene repression of both promoters, respectively. H3 acetylation was highly enriched in the 5′-LTR region of ERVWE1 in BeWo cells, but was negligible in HeLa cells. In contrast, trimethylation of H3K9 was extremely low in BeWo cells but highly enriched in HeLa cells (Figure 1A). The levels of H3 acetylation surrounding the 5′-LTR of ERVFRDE1 were low in both HeLa and BeWo cells. Trimethylation of H3K9 was high in HeLa cells and low in BeWo cells (Figure 1B). These results suggest that in BeWo cells, unlike ERVFRDE1, ERVWE1 is highly expressed, whereas both proviruses are efficiently repressed in HeLa cells. The level of H3K9 trimethylation correlates perfectly with the CpG methylation in the 5′-LTR region of both proviruses (24,25). The constitutively expressed GAPDH gene did not display any significant differences in H3K9 modifications between BeWo and HeLa cells (Figure 1C).
Figure. 1.
Acetylation and trimethylation of H3K9 in LTRs of syncytin-1 and -2 in BeWo and HeLa cells. ChIP assays were performed with antibodies against indicated histone modifications and the precipitated chromatin was subjected to qPCR with primers specific for (A) 5′-LTR of ERVWE1, (B) 5′-LTR of ERVFRDE1 and (C) coding region of human GAPDH, a constitutively expressed control gene. The results were normalized to IgG non-specific controls and shown as an average percentage of the input DNA. Data are presented as a mean ± SD from three triplicates.
Quantitative analysis of ERVWE1 and ERVFRDE1 proviral expression and splicing
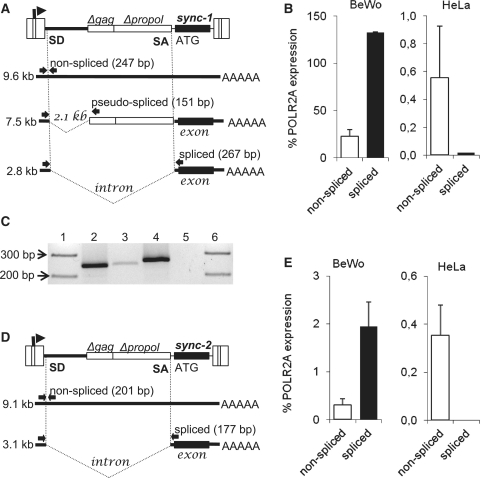
Next, we examined whether the expression levels of both syncytin loci correlate with the epigenetic status of their promoters. Splicing of the full-length genomic transcript into subgenomic env mRNA is necessary for the translation into a cleavable envelope glycoprotein. We, therefore, used the splice-specific qRT–PCR to assess the efficiency of splicing in BeWo cells, the best model of trophoblast differentiation, and in HeLa cells. The localization of splice signals in ERVWE1, the position of primers and the size of amplified products are shown in Figure 2A. In BeWo cells, we detected the presence of the full-length non-spliced transcript and retention of the 2.1 kb itron-like insert. The ratio of spliced and non-spliced transcripts was 6:1, and the expression of the spliced syncytin-1 transcript reached at least 130% of that of the housekeeping standard POLR2A. HeLa cells produced a very low level of the non-spliced ERVWE1 transcript and spliced transcripts did not raise above the background level (Figure 2B). This corresponds to the lack of syncytin-1 expression as previously detected by northern blotting and non-quantitative PCR (22,24). Minor DNA contamination was tightly controlled by including the RT-parallels (Figure 2C). We also detected the presence of pseudo-spliced mRNA without the 2.1 kb intron-like insertion in BeWo cells.
Figure. 2.
Expression and splicing of ERVWE1 and ERVFRDE1 proviruses in trophoblastic and non-placental cells. Splicing strategies of (A) ERVWE1 and (D) ERVFRDE1 are shown together with the positions of primers for splicing-specific PCR (arrows) and the sizes of diagnostic PCR products. The levels of non-spliced (open columns) and spliced (black columns) transcripts of (B) ERVWE1 and (E) ERVFRDE1 in BeWo and HeLa cells were estimated by qRT-PCR and are shown as the average percentage of the POLR2A expression ±SD from three triplicates. The low level of genomic DNA contaminating the RT–PCR product of non-spliced ERVWE1 mRNA in BeWo cells is shown (C). Lane 1, marker of molecular size; lane 2, non-spliced ERVWE1 mRNA RT+; lane 3, non-spliced mRNA RT−; lane 4, spliced mRNA RT+; lane 5, spliced mRNA RT−; lane 6, marker of molecular size.
The splicing strategy of the ERVFRDE1 provirus and positions of primers for splice-specific RT–PCR are shown in Figure 2D. BeWo cells expressed the spliced transcript of ERVFRDE1 at much lower levels than ERVWE1 and reached only 2% of POLR2A expression level, while the ratio of spliced over non-spliced transcript was similarly high at 7:1. Again, the transcription of ERVFRDE1 in HeLa cells was low: the non-spliced transcript expression reached 0.35% of POLR2A expression, which was comparable with BeWo cells, and the spliced transcript was not detected (Figure 2E). In conclusion, the transcription of ERVWE1 and ERVFRDE1 reflects the epigenetic status of their LTRs, and the cell-specific RNA splicing acts as an additional regulatory mechanism in the control of syncytin-1 and -2 expression.
The expression of syncytin-1 and cell-to-cell fusion efficiency were shown to be augmented by forskolin, adenylate cyclase activator. In our splice-specific qRT–PCR assay, stimulation of BeWo cells with forskolin increased ERVWE1 transcription with the ratio of spliced over non-spliced transcripts 5:1. There was only weak, statistically insignificant forskolin stimulation in JEG-3 cells (Supplementary Figure S1A). Even more striking was the shift of ERVFRDE1 transcription and splicing in BeWo cells, which increased up to 25% of POLR2A, and the ratio of spliced over non-spliced transcript was 40:1 (Supplementary Figure S1B). No effect of the forskolin treatment was observed in HeLa cells (data not shown).
Transcription and splicing of ERVWE1 and ERVFRDE1 in the testes and germ cell tumors
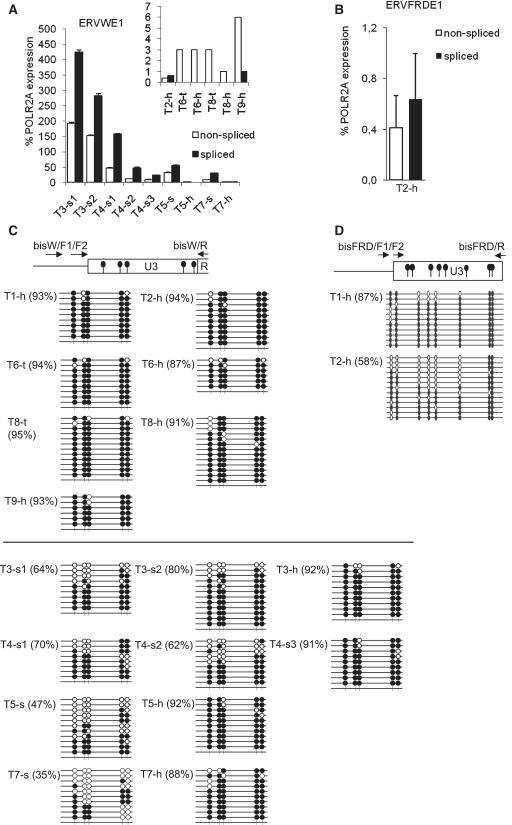
A weak signal of syncytin-1 mRNA was previously detected in the testes and testicular cancer (4,29). Therefore, the testes might be an organ where the transcriptional suppression of syncytin loci by CpG methylation and chromatin modifications is leaky. In this tissue, the additional expression control due to differential splicing, reported above, would be of biological importance. In order to explore this possibility, we performed splice-specific qRT–PCR of ERVWE1 and ERVFRDE1 sequences in total RNA samples prepared from healthy testes. As shown in Figure 3A, the level of non-spliced ERVWE1 transcripts reached only between 1% and 6% of the POLR2A transcripts with a hardly detectable presence of the spliced transcript. The transcription of ERVFRDE1 in the testes was even weaker and reached ∼1% of POLR2A transcripts with roughly equimolar amounts of non-spliced and spliced transcripts (Figure 3B). In addition to two biopsies of healthy testes, we analyzed the expression of ERVWE1 in several biopsies from six different testicular germ cell tumors (Figure 3A). In several samples of four seminomas, the expression levels of both non-spliced and spliced transcripts were highly increased in contrast to normal tissues and comparable or even higher by several fold to POLR2A expression. The ERVWE1 expression was variable in biopsies taken from different parts of the same tumor. The splicing was efficient in all tumor samples with a ratio of spliced over non-spliced transcripts varying between 1.9 and 4.0. In two samples of non-seminoma testicular cancer, we did not observe any significant increase of either ERVWE1 expression or splicing.
Figure. 3.
Transcription, splicing and CpG methylation of ERVWE1 and ERVFRDE1 in the healthy testes and testicular germ cell tumors. The levels of non-spliced (open columns) and spliced (black columns) transcripts of (A) ERVWE1 and (B) ERVFRDE1 in two samples of healthy testes, T2-h and T9-h, in two samples of granulomatous seminoma 3, T3-s1 and T3-s2, three samples of seminoma 4, T4-s1, T4-s2 and T4-s3, in tumor samples and adjacent healthy tissues of seminoma 5 and 7, T5-s, T5-h, T7-s and T7-h, in tumor sample and adjacent healthy tissue of mixed germ cell tumor 6, T6-t and T6-h and in tumor sample and adjacent healthy tissue of testicular lymphoma 8, T8-t and T8-h were estimated by qRT–PCR and are shown as the average percentage of the POLR2A expression ± SD from three triplicates. The samples of healthy testes and non-seminoma tumors are shown in the inset with different scale of the y-axis. The CpG methylation status of the 5′-LTR U3 region of (C) ERVWE1 and (D) ERVFRDE1 in three samples of healthy testes, T1-h, T2-h and T9-h, and in aforementioned samples of seminomas and non-seminoma tumors was examined by the bisulfite sequencing technique. Methylated CpG sites are indicated by solid circles, unmethylated CpG sites are indicated by open circles, numbers in parentheses depict the percentage of methylated CpG dinucleotides.
Previous studies showed the correlation between transcriptional suppression and promoter hypermethylation of ERVWE1 in non-placental cells (24,25). We, therefore, assayed the methylation status of 5′-LTR regions of the ERVWE1 and ERVFRDE1 (Figure 3C and D) proviruses in healthy testes and testicular germ cell tumors using the sodium bisulfite technique. We designed the forward primers for semi-nested PCR complementary to the unique genomic sequences adjacent to the 5′-LTRs to ensure the selective amplification of ERVWE1 and ERVFRDE1 loci (Figure 3C and D). In two samples of healthy testicular tissues, the ERVWE1 5′-LTRs were found to be heavily methylated (Figure 3C) in all analyzed clones. The overall percentage of methylated CpGs was >90%, even higher than that in other non-placental tissues or cell lines reported in our previous study (24). None out of 20 sequenced clones was hypomethylated. The 5′-LTRs of ERVFRDE1 provirus displayed a slightly lower level of CpG methylation reaching 87 or 58% of methylated CpGs in two samples of healthy testes (Figure 3D). The observed increase of the ERVWE1 expression in seminomas was accompanied with a slight hypomethylation of its 5′-LTR varying between 38% and 91% of methylated CpGs (Figure 3C). There is, however, no correlation between the level of CpG demethylation and the ERVWE1 expression and we did not find completely unmethylated 5′-LTR copies in all seminoma samples expressing ERVWE1. No hypomethylation was observed in non-seminoma germ cell tumor samples, which do not express ERVWE1 (Figure 3C). We conclude that 5′-LTRs of ERVWE1 and ERVFRDE1 are methylated in the testes and that the weak expression of syncytin-1 in healthy testes can be observed despite this epigenetic status. The inefficient splicing of ERVWE1 transcripts prevents improper syncytin-1 expression in normal testes but not in seminomas, where both non-spliced and spliced forms of ERVWE1 mRNAs occur.
Splicing of ectopically expressed ERVWE1 mRNA
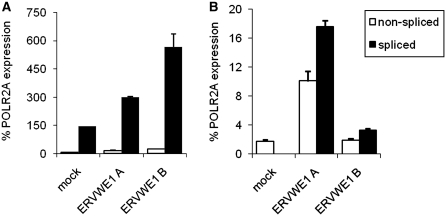
The inefficient splicing of ERVWE1 in the testes and HeLa cells suggests the regulatory potential of HERV RNA processing. In order to test it experimentally, we quantified the spliced and non-spliced transcripts overexpressed ectopically from a non-methylated full-length ERVWE1 proviral clone containing the URE at the 5′-end. The whole ERVWE1 provirus, amplified by PCR and cloned into a plasmid, was stably transfected into JEG-3 and HeLa cells and the resulting transcripts were quantified by splice-specific qRT–PCR. As shown in Figure 4A, the ectopic transcription of the exogenously introduced ERVWE1 provirus in two stably transfected clones of JEG-3 cells was two to four times higher than the transcription of the endogenous ERVWE1 in mock-transfected JEG-3 cells. The splicing of endogenous transcripts was efficient, comparable with BeWo cells (Figure 2B), and the level of non-spliced transcript did not increase significantly in transfected cells, suggesting that the exogenous copies integrated in different genomic locations are prone to transcription and splicing in the cells of trophoblastic origin. In a parallel experiment, two independent HeLa cell clones, ERVWE1A and ERVWE1B, with stable expression of ERVWE1 from ectopically integrated proviruses displayed less efficient splicing with only slight excess of spliced over non-spliced transcripts (Figure 4B). Although the stable ectopic expression of ERVWE1 in HeLa cells was much lower than the endogenous expression in JEG-3 cells and reached just 20 and 4% of the POLR2A, the efficiency of splicing from the exogenous copies was in striking contrast to the absence of splicing in mock-transfected HeLa cells. Altogether, these results indicate that the integration of the unmethylated ERVWE1 provirus into ectopic chromosomal positions and different chromatin context alleviates the blocks of transcription and splicing that are present at the endogenous ERVWE1 locus. Both transcription and splicing, however, lack the positive tissue-specific control and their efficiencies do not reach the values observed in trophoblastic JEG-3 cells.
Figure. 4.
mRNA splicing of ectopically expressed ERVWE1 in JEG-3 (A) and HeLa (B) cells. The pcDNA3 plasmid DNA alone was used for mock-transfected control. Expression of non-spliced (open bars) and spliced (solid bars) ERVWE1 mRNA from de novo integrated proviral DNA was quantified in two independent cell clones of both JEG-3 and HeLa cells by splice-specific qRT–PCR in two independently repeated experiments. Data are presented as the average percentage of the POLR2A expression ±SD from three parallels.
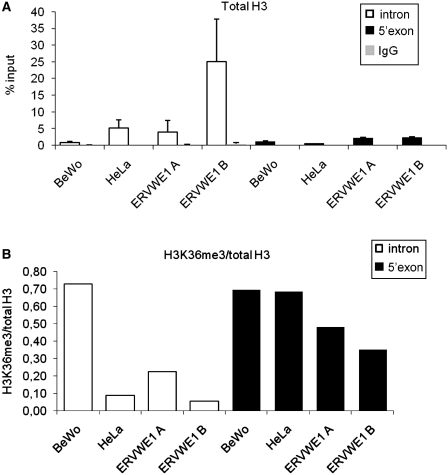
H3K36me3 at the ERVWE1 intron/exon boundary
Among the histone post-translational modifications, the trimethylation of lysine 36 of H3 (H3K36me3) has been reported to be enriched on exons and to mark the expressed exons (30–32). Differences in H3K36me3 levels were also shown at actively used alternative exons of the FGFR2 gene (33). We, therefore, compared the H3K36me3 at the central part of intron versus 5′-part of the ERVWE1 exon, i.e. the beginning of the syncytin-1 coding sequence, in BeWo and HeLa cells. Because the differences in H3K36me3 described in ChIP analyses of single genes or in ChIP-on-chip experiments are low and reflect the nucleosome density (34), we compared the occupancy of total H3 at the same sites and normalized the levels of H3K36me3. As shown in Figure 5A, the H3 occupancy of the ERVWE1 in BeWo cells was similarly low at both the intron and the exon. In HeLa cells, there was, however, significantly increased H3 occupancy at the intron and low at the 5′-end of the exon. The relative level of H3K36 in BeWo cells was similarly high along the intron/exon boundary (Figure 5B). In contrast, the relative level of H3K36me3 in HeLa cells increased 7-fold along this boundary, reaching in the exon approximately the same level as observed in BeWo cells (Figure 5B). We further tested the H3 occupancy and H3K36me3 level in two independent HeLa cell clones with stable insertions of ectopic ERVWE1. Here, we took into account the increased copy numbers of ERVWE1, up to five proviruses introduced into unknown genomic positions. The value of H3 occupancy at the intron in the first clone, ERVWE1A, was comparable with intact HeLa cells and provided the increased proviral copy number, we can expect the presence of proviruses with low, BeWo-like H3 occupancy (Figure 5A). This corresponds with basal expression and splicing of ERVWE1 in this clone (Figure 4B). The second clone, ERVWE1B, displayed a very high value of H3 enrichment and provided the increased proviral copy number, we expected that in this case most proviruses display increased H3 occupancy comparable with intact HeLa cells (Figure 5A). Accordingly, the ERVWE1 expression in this clone was low and only slightly increased over the intact HeLa cells (Figure 4B). The values of H3 occupancy at the 5′-end of the exon were low in both clones, only slightly increased in comparison with intact HeLa cells, which corresponds with the increased proviral copy number (Figure 5A). Finally, the relative level of H3K36me3 at the intron differed in these two clones, being substantially higher in the clone ERVWE1A and reaching ∼30% of H3K36me3 density of BeWo cells (Figure 5B). Altogether, we propose that efficient expression and splicing of the ERVWE1 provirus are accompanied by a high level of H3K36me3 at the intron and at the intron/exon boundary. In contrast, high H3 occupancy at the intron but not at the subsequent exon indicates transcriptionally silent provirus and inefficient splicing.
Figure. 5.
H3 occupancy (A) and H3K36me3 (B) in the intron and 5′-exon end of ERVWE1 in BeWo and HeLa cells. Using the anti-H3K36me3 and anti H3 antibodies we immunoprecipitated chromatin from BeWo and HeLa cells and analyzed the amount of immunoprecipitated intron fragment (open bars) and 5′-exon end (solid bars) by qPCR. HeLa cells were either intact or with stably integrated ectopic ERVWE1 (ERVWE1 A, B). (A) The anti-H3CT antibody was used to quantify the total H3 associated with intron and 5′-exon end of ERVWE1 as an average percentage of the input DNA. IgG non-specific controls are shown (gray bars) as an average percentage of the input DNA. Data are presented as a mean ± SD from three triplicates. (B) The enrichment for H3K36 was normalized to H3 occupancy and the ratio of H3K36me3/total H3 in intron (open bars) and 5′-exon end (solid bars) of ERVWE1 is shown.
DISCUSSION
The critical point in cell differentiation is stringent regulation of tissue-specific genes, which restricts gene expression only to the correct cell type and correct developmental stage. In this study, we compared the mechanisms of epigenetic control, i.e. CpG methylation, chromatin modifications, and splicing of two HERVs encoding syncytins, fusogenic retroviral glycoproteins involved in the development of placenta. It was shown previously (24,25) that CpG methylation-dependent transcriptional suppression of syncytins might be the general regulatory mechanism protecting non-placental tissues from inadvertent cell-to-cell fusion. We add now that suppressive chromatin modifications strengthen this control. CpG methylation of retroviral LTRs has already been coupled with transcriptional suppression of e.g. Rous sarcoma virus (RSV), murine leukemia virus, human immunodeficiency virus type 1 (HIV-1), human T-cell leukemia virus, and HERV-K (35–40). Heavy CpG methylation of ERVWE1 in non-placental cells is accompanied by deacetylation and trimethylation at H3K9, which results in a strong resistance against reactivation by 5-azacytidine and trichostatin A (24). Similar epigenetic marks and resistance were described in the latent reservoir of human immunodeficiency virus (37) and in long-term silenced retroviral vectors (41). Transcriptional suppression of ERVWE1 is, however, leaky in the testes and the tight control of splicing prevents formation of translatable syncytin-1 mRNA and expression of its protein product.
Promoter methylation of ERVWE1 and ERVFRDE1 does not correlate with their weak but detectable transcription in the testes (4,6). This might be caused by some testis-specific and male germ line-specific epigenetic mechanisms of transcriptional control leading to increased transcription noise (42). Transcription of HERV-Ws in the germ line, at least in the past, is reflected by the presence of numerous HERV-W-derived processed pseudogenes in the human genome (43,44). Furthermore, very low expression of ERVWE1 was also observed in HeLa cells and lymphoblastic MCF cells with heavily methylated 5′-LTR (Figure 2, Supplementary Figure S2). Taken together, epigenetic suppression of syncytins is not absolute and additional mechanisms, e.g. splicing and processing of ERVWE1 mRNA, must be involved in their control.
Splicing of genomic ERVWE1 mRNA was already shown to be inefficient in the testes as only the pseudo-spliced form could be seen on the Northern blot (4). Smallwood et al. (45) demonstrated the increasing level of pseudo-spliced ERVWE1 mRNA in term placenta and suggested that the relative amounts of spliced and pseudo-spliced mRNAs could regulate syncytin-1 expression during pregnancy. Finally, Gimenez et al. (29) found increased levels of spliced ERVWE1 mRNA in several testicular tumors. Based on these findings, we focused on splice-specific quantitative analysis of ERVWE1 and ERVFRDE1 expression in trophoblastic and non-placental cells. We describe here a very stringent control of splicing at both syncytin loci, which, in the case of ERVWE1, is very robust under extremely high ectopic expression of the cloned provirus. Importantly, we have found deregulation of splicing in biopsies of seminomas. In contrast to previous studies (4,26,29,46), we detected the full-length and non-spliced ERVWE1 transcript. We controlled the DNA contamination in our splice-specific qRT–PCR and sequenced the amplified product. These controls confirm that our detection of non-spliced transcript does not come from the genomic DNA or from other HERV-W members. We can only speculate why this transcript has not been detected so far, and further research will be needed to fully explain the splicing strategy of ERVWE1 and the system of choice of the spliced fragment length.
In comparison with the results by Gimenez et al. (29), we observed only a slight demethylation of 5′-LTR of ERVWE1 in seminomas with released ERVWE1 expression and splicing. The comparison of both studies is, however, difficult because of the small number of tumor samples and various degrees of differentiation. The weak correlation between ERVWE1 expression and hypomethylation of its 5′-LTR could be explained by exceptional epigenetic features of male germ cells such as high level of CpG methylation, presence of rare histone variants, protamine occupancy, mimicking methylcytosine by hydroxymethylcytosine, etc. We analyzed whether the deregulation of transcription and splicing in testicular cancer was associated with the expression of GCMa, but we did not detect any GCMa mRNA using RT–PCR (data not shown). The deregulation of syncytin-1 splicing probably occurs not only in testicular germ cell tumors, particularly seminomas, but also in other various human cancers, e.g. in breast carcinomas (18) and endometrial carcinomas (46).
The regulatory potential of retroviral splicing generally results from the need in infectious retroviruses to balance between genomic mRNA and spliced subgenomic mRNA(s) and produce the right proportion of protein component assembling into new functional virions. HIV-1 translates its own trans-activator, Tat, from a spliced mRNA transcript and downregulation of HIV-1 splicing is observed during the provirus persistence in memory cells (47). Another example of specific splicing control is the suppression of env splicing of RSV in mammalian cells (48). It was suggested that mammalian cells lack chicken-specific trans factor(s) binding to cis regulatory sequences, which could explain the suppression of env splicing (48–50), and the non-permissiveness of these cells for RSV replication (51). In replication-defective HERVs, the balance of non-spliced and spliced transcripts is not important any more but the cis splice signals can be adopted for cell-specific control of splicing and expression of retroviral glycoproteins. Cell-specific splicing in endogenous retroviruses remains to be studied systematically but in the light of our recent findings, it might be a useful indicator of biological activity of HERVs with intact env genes, which do not fulfill the criteria for sensu stricto syncytins. Blaise et al. (52) identified a new four-member family of HERVs, HERV-P(b), with one full-length env gene, ZFERV-like env, fusogenic in HeLa cells. Its expression was observed in many healthy tissues without any significant specificity for the placenta. Out of six full-length env genes of the young HERV-K(HML-2) family, the env encoded by HERV-K108 was also shown fusogenic in human embryonic kidney cells as well as in mouse, hamster and cat cells (53). Similarly, the rest of 18 intact env ORFs, which did not score yet in cell fusion assays, might be proven as biologically relevant in the future. Description of their splicing in various tissues could identify particular cell types where these retroviral glycoproteins play any role in cell functioning.
The striking contrast of splicing efficiency in HeLa cells as an example of epithelial tumors and in germ cell tumors including choriocarcinoma cell lines (Figures 2 and 3) can be used to analyze the mechanism controling endogenous ERVWE1 pre-mRNA splicing. Cell-specific splicing factors can be involved but our results point to the role of chromatin context and, in particular, the modifications of histones associated with exon/intron boundary. At least partially efficient splicing in HeLa cells can be observed after the stable integration of the unmethylated cloned ERVWE1 provirus (Figure 4) into a new chromosomal position different from the endogenous locus at 7q21-22. The availability of trophoblast-specific transcription factors binding to the URE sequence could further increase the efficiency of splicing in stably transfected HeLa, and experiments with ectopic expression of GCMa are highly needed to explain the control of ERVWE1 splicing. The requirement of chromatin context results probably from the co-transcriptional occurence of pre-mRNA splicing and elongation rate dependent on the chromatin configuration. RNA polymerase II and transcription activators associate with splicing factors (31,54) and slow elongation through the intron/exon boundary or even stalled RNA polymerase II gives more time to recognize the splicing signals, which is particularly important for the suboptimal signals used for alternative exons inclusion (54–56). In our study, we have shown the high density of H3K36me3 along the intron–exon boundary of ERVWE1 in BeWo cells with high expression and efficient splicing. This is rather unusual as most studies demonstrated 3′-exon enrichment of H3K36me3. The 5′-intronic sequence in retroviruses, however, is of coding potential in non-spliced transcripts and behaves as an alternative exon. A more extensive study along the provirus ERVWE1 also comprising the 2.1 kb intron-like insertion could elucidate this point. Our approach using the HeLa cell clones with various expression of ectopic proviruses could also help to better understand the blocks of ERVWE1 transcription and splicing; we, however, need the exact analysis of copy numbers and genomic positions of proviruses in individual clones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Czech Science Foundation (grant No. 301/09/2031); Academy of Sciences of the Czech Republic (grant No. IAA500520709 and AV0Z50520514); INSERM, Institut Paoli-Calmettes. Funding for open access charge: Czech Science Foundation (grant No. 301/09/2031).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Věra Hoserová for the technical assistance and Jan Svoboda for critical reading of the article.
REFERENCES
- 1.de Parseval N, Lazar V, Casella J-F, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope genes. J. Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blond J-L, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 5.Lavillette D, Marin M, Ruggieri A, Mallet F, Cosset FL, Kabat D. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 2002;76:6442–6452. doi: 10.1128/JVI.76.13.6442-6452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaise S, Ruggieri A, Dewannieux M, Cosset FL, Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J. Virol. 2004;78:1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargas A, Moreau J, Landry S, LeBellego F, Toufaily C, Rassart E, Lafond J, Barbeau B. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 2009;392:301–318. doi: 10.1016/j.jmb.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 9.Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. A placenta-specific receptor for the fusogenic endogenous retrovirus-derived, human syncytin-2. Proc. Natl Acad. Sci. USA. 2008;105:17532–17537. doi: 10.1073/pnas.0807413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knerr I, Beinder E, Rascher W. Syncytin, a novel human endogenous retroviral gene in human placenta: evidence for its dysregulation in preeclampsia and HELLP syndrome. Am. J. Obstet. Gynecol. 2002;186:210–213. doi: 10.1067/mob.2002.119636. [DOI] [PubMed] [Google Scholar]
- 11.Lee X, Keith JC, Jr, Stumm N, Moutsatsos I, McCoy JM, Crum CP, Genest D, Chin D, Ehrenfels C, Pijnenborg R, et al. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta. 2001;22:808–812. doi: 10.1053/plac.2001.0722. [DOI] [PubMed] [Google Scholar]
- 12.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. Placental synyctins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl Acad. Sci. USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malassine A, Frendo J-L, Blaise S, Handschuh K, Gerbaud P, Tsatsaris V, Heidmann T, Evain-Brion D. Human endogenous retrovirus-FRD envelope protein (syncytin 2) expression in normal and trisomy 21-affected placenta. Retrovirology. 2008;5:6. doi: 10.1186/1742-4690-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005;79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frendo JL, Olivier D, Cheynet V, Blond JL, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, Wallace JL, Deacon RM, Warren K, Power C. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nature Neurosci. 2004;7:1088–1095. doi: 10.1038/nn1319. [DOI] [PubMed] [Google Scholar]
- 17.Mameli G, Astone V, Arru G, Marconi S, Lovato L, Serra C, Sotgiu S, Bonetti B, Dolei A. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus but not Human herpesvirus 6. J. Gen. Virol. 2007;88:264–274. doi: 10.1099/vir.0.81890-0. [DOI] [PubMed] [Google Scholar]
- 18.Bjerregaard B, Holck S, Christensen JJ, Larsson LI. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006;63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen JM, Christensen IJ, Nielsen HJ, Hansen U, Bjerregaard B, Talts JF, Larsson L-I. Syncytin immunoreactivity in colorectal cancer: Potential prognostic impact. Cancer Lett. 2009;280:44–49. doi: 10.1016/j.canlet.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 21.Arriza JL, Kavanaugh MP, Fairman WA, Wu YN, Murdoch GH, North RA, Amara SG. Cloning and expression of a human neutral amino acid transporter with structural similarity to the glutamate transporter gene family. J. Biol. Chem. 1993;268:15329–15332. [PubMed] [Google Scholar]
- 22.Yu C, Shen K, Lin M, Chen P, Lin C, Chang GD, Chen H. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 2002;277:50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- 23.Prudhomme S, Oriol G, Mallet F. A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression. J. Virol. 2004;78:12157–12168. doi: 10.1128/JVI.78.22.12157-12168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matoušková M, Blažková J, Pajer P, Pavlíček A, Hejnar J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exptl. Cell. Res. 2006;312:1011–1020. doi: 10.1016/j.yexcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Gimenez J, Montgiraud C, Oriol G, Pichon J-P, Ruel K, Tsatsaris V, Gerbaud P, Frendo J-L, Evain-Brion D, Mallet F. Comparative methylation of ERVWE1/Syncytin-1 and other human endogenous retrovirus LTRs in placenta tissues. DNA Res. 2009;16:195–211. doi: 10.1093/dnares/dsp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blond J-L, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl Acad. Sci. USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Gimenez J, Montgiraud C, Pichon J-P, Bonnaud B, Arsac M, Ruel K, Bouton O, Mallet F. Custom human endogenous retrovirus dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010;38:2229–2246. doi: 10.1093/nar/gkp1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nature Struct. Mol. Biol. 2009;16:990–996. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 33.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicng by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nature Struct. Mol. Biol. 2010;17:1495–1500. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hejnar J, Plachý J, Geryk J, Machoň O, Trejbalová K, Guntaka RV, Svoboda J. Inhibition of the Rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: different sensitivity in permissive chicken cells versus mammalian cells. Virology. 1999;255:171–181. doi: 10.1006/viro.1998.9597. [DOI] [PubMed] [Google Scholar]
- 36.Lorincz MC, Schübeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blažková J, Trejbalová K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koiwa T, Hamano-Usami A, Ishida T, Okayama A, Yamaguchi K, Kamihira S, Watanabe T. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002;76:9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2) J. Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInerney JM, Nawrocki JR, Lowrey CH. Long-term silencing of retroviral vectors is resistant to reversal by trichostatin A and 5-azacytidine. Gene Ther. 2000;7:653–663. doi: 10.1038/sj.gt.3301155. [DOI] [PubMed] [Google Scholar]
- 42.McCarrey JR, Geyer CB, Yoshioka H. Epigenetic regulation of testis-specific gene expression. Ann. NY Acad. Sci. 2005;1061:226–242. doi: 10.1196/annals.1336.025. [DOI] [PubMed] [Google Scholar]
- 43.Marques A, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PloS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavlíček A, Pačes J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smallwood A, Papageorghiou A, Nicolaides K, Alley MK, Jim A, Nargund G, Ojha K, Campbell S, Banerjee S. Temporal regulation of the expression of syncytin (HERV-W), maternally imprinted PEG10, and SGCE in human placenta. Biol. Reprod. 2003;69:286–293. doi: 10.1095/biolreprod.102.013078. [DOI] [PubMed] [Google Scholar]
- 46.Strick R, Ackermann S, Langbein M, Swiatek J, Schubert SW, Hashemolhosseini S, Kosheck T, Fasching PA, Schild RL, Beckmann MW, et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J. Mol. Med. 2007;85:23–38. doi: 10.1007/s00109-006-0104-y. [DOI] [PubMed] [Google Scholar]
- 47.McLaren M, Marsh K, Cochrane A. Modulating HIV RNA processing and utilization. Front. Biosci. 2008;13:5693–5707. doi: 10.2741/3110. [DOI] [PubMed] [Google Scholar]
- 48.Berberich SL, Macias M, Zhang L, Turek LP, Stoltzfus CM. Comparison of Rous sarcoma virus RNA processing in chicken and mouse fibroblasts: evidence for double-spliced RNA in nonpermissive mouse cells. J. Virol. 1990;64:4313–4320. doi: 10.1128/jvi.64.9.4313-4320.1990. (Erratum in J. Virol. 64, 6360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 1998;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNally MT. RNA processing control in avian retroviruses. Front. Biosci. 2008;13:3869–3883. doi: 10.2741/2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svoboda J, Hejnar J, Geryk J, Eleder D, Vernerová Z. Retroviruses in foreign species and the problem of provirus silencing. Gene. 2000;261:181–188. doi: 10.1016/s0378-1119(00)00481-9. [DOI] [PubMed] [Google Scholar]
- 52.Blaise S, de Parseval N, Heidmann T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology. 2005;2:19. doi: 10.1186/1742-4690-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dewannieux M, Blaise S, Heidmann T. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 2005;79:15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl Acad. Sci. USA. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.