Abstract
Fibrillarin, one of the major proteins of the nucleolus, plays several essential roles in ribosome biogenesis including pre-rRNA processing and 2′-O-ribose methylation of rRNA and snRNAs. Recently, it has been shown that fibrillarin plays a role in virus infections and is associated with viral RNPs. Here, we demonstrate the ability of recombinant fibrillarin 2 from Arabidopsis thaliana (AtFib2) to interact with RNAs of different lengths and types including rRNA, snoRNA, snRNA, siRNA and viral RNAs in vitro. Our data also indicate that AtFib2 possesses two RNA-binding sites in the central (138–179 amino acids) and C-terminal (225–281 amino acids) parts of the protein, respectively. The conserved GCVYAVEF octamer does not bind RNA directly as suggested earlier, but may assist with the proper folding of the central RNA-binding site.
INTRODUCTION
Fibrillarin is a 33–36 kDa nucleolar protein involved in multiple aspects of RNA biogenesis. It is a major protein in the fibrillar regions of the nucleolus where rRNA transcription and early pre-rRNA processing take place (1,2). Fibrillarin is also found in Cajal bodies (CBs), subnuclear dynamic particles that contain distinct components involved in RNA transcription and editing such as mRNA splicing and small nuclear/nucleolar ribonucleoprotein (sn/snoRNP) biogenesis (3,4). Fibrillarin is a core component of box C/D snoRNP particles, which are responsible for site-specific 2′-O-ribose methylation of rRNA and for pre-rRNA cleavage, and box C/D-containing small CB-specific RNPs (scaRNPs) that methylate snRNAs (5,6). Assembled sn/snoRNPs traffic through CBs before accumulating in both splicing speckles (sites of storage/assembly of splicing factors/complexes) and also in the dense fibrillar component of the nucleolus where early pre-rRNA processing events take place. In all cases, fibrillarin is found in multi-component complexes containing RNA (snoRNA, pre-rRNA) and proteins. In eukaryotes, the mature box C/D snoRNP complex competent for snRNA/pre-rRNA modification contains the RNA-binding 15.5-kDa protein and the two related proteins Nop56 and Nop58 in addition to fibrillarin and the snoRNA (5).
Fibrillarin is highly evolutionarily conserved with respect to sequence, structure and function (5,7–10). On the basis of the amino acid sequence alignment of fibrillarin proteins from different species, Aris and Blobel (7) suggested that fibrillarin contains several domains. Within the human fibrillarin protein (36 kDa—320 amino acids), three structure/functions regions have been predicted (7; Figure 1). The N-terminal region of fibrillarin is represented by a glycine- and arginine-rich (GAR) domain that is also found in fibrillarin from Saccharomyces cerevisiae, Xenopus laevis and Tetrahymena thermophila (7,9,11), but not in archaeal fibrillarin proteins (5,8). The GAR domain is methylated at arginine residues (12), possibly by the arginine methyltransferases found in complex with fibrillarin (13). The GAR domain is responsible for the interactions with various cellular (14,15) and viral proteins (16,17). The data concerning the role of the GAR domain in nucleolar localization are contradictory. Some findings demonstrate that the GAR domain is not required to target fibrillarin to the nucleolus or CBs, although it contains a nucleolar localization signal (3), but other results suggest that the GAR domain provides the nucleolar targeting of fibrillarin in plants and human cells (9,18).
Figure 1.
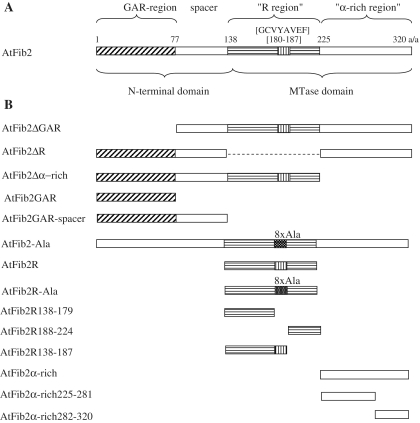
Schematic representation of AtFib2 protein (A) and its deletion mutants (B). The main fibrillarin regions predicted by Aris and Blobel (7) for human fibrillarin are as follows: N-terminal glycine–arginine-rich region (GAR), central region (referred to as R-region) that was suggested to provide RNA-binding activity via the conserved amino acid octamer GLVYAVEF, the spacer between these regions (spacer) and the C-terminal region containing predicted α-helices (referred to as α-rich-region). Domains identified by Wang et al. (8) for fibrillarin protein of Methanocaldococcus jannaschii are the N-terminal domain (mostly overlaps with GAR region) and the MTase domain comprising the central R-region and C-terminal α-rich region. The conserved GCVYAVEF octamer is indicated with vertical lines, and when replaced with eight alanines sequence (8× Ala)—with white dots on black. The positions of AtFib2 deletion mutants are indicated. Internal deletion in AtFib2ΔR mutant is shown by a dashed line.
Since the central part of the protein contains 90 amino acid residues with some similarity to the RNA recognition motif (RRM) present in various RNA-binding proteins, Aris and Blobel (7) have named this region a ‘putative RNA-binding domain’. The alignment with several RNA-binding proteins allowed them to identify an octameric region, which resembles a classical RNP1 motif within the RRM that directly participates in RNA-binding, (R/K)-G-(F/Y)-(G/A)-(F/Y)-(I/L/V)-X-(F/Y) (19). This is conserved among human, S. cerevisae and X. laevis fibrillarin proteins as a GLVYAVEF sequence (7).
The C-terminal region of the protein contains a short sequence (33 amino acid residues) predicted to form an α-helix structure. The C-terminal region of fibrillarin targets it to CBs (3,18) and interacts directly with Nop56 (20). Together, the central and C-terminal regions of the protein constitute a conserved Ado-Met-dependent methyltransferase (MTase)-like domain that contains S-adenosyl methionine (SAM, the methionine group donor) binding motifs and provides the MTase activity of fibrillarin (8,10,21,22).
In addition to the major functions in biogenesis of different RNAs and ribosomal subunits, the growing body of evidence demonstrates that fibrillarin is involved in infection by a range of different viruses. The nucleocapsid proteins of porcine arterivirus and avian coronavirus infectious bronchitis virus (IBV) interact with nucleolin and fibrillarin (23,17), and as a result the normal functions of these nucleolar proteins may be diverted for the benefit of the viruses. The infection of cells with IBV has been shown to disrupt nucleolar architecture (24) and cause arrest of the cell cycle in G2 and failure of cytokinesis (23,25,26). Plant viruses are also able to recruit fibrillarin to facilitate different stages of infection including virus movement, assembly of virus-specific RNP particles and possibly to combat host defence responses (see for review, 27). It has been shown that the ability of the plant groundnut rosette virus (GRV, the genus Umbravirus) to move long distances through the phloem, the specialized plant vascular system, strictly depends on the interaction of its movement protein (the ORF3 protein) with fibrillarin. This interaction is essential for nucleolar import of the ORF3 protein via CBs, re-localization of some fibrillarin from the nucleolus to cytoplasm, and assembly of cytoplasmic umbraviral RNP particles capable of long-distance spread and systemic infection (16,28,29). It has also been demonstrated that fibrillarin interacts with the multifunctional protein VPg of potato virus A (the genus Potyvirus) (30) and the 2b silencing suppressor protein of cucumber mosaic virus (the genus Cucumovirus) (31). These data indicate that interaction with fibrillarin may be a general property of various virus proteins from different taxonomic groups.
Thus, fibrillarin plays an important role in various processes in normal cells and during virus infections. In most cases, it functions as a part of different cellular and viral RNP complexes. Analysis of RNPs, immunoprecipitated with anti-fibrillarin antibodies, has shown fibrillarin to be associated with complexes containing all known box C/D snoRNAs in yeast and a plethora of snoRNAs in other organisms (32–40). Structural investigations of human fibrillarin (10) and the archaeal rRNA-modifying complex (8) did not determine whether and how fibrillarin directly interacted with RNA moieties. Indeed, it is generally accepted that the specificity of targeting of these complexes to the sites of modification of rRNA may not be determined by fibrillarin but rather by the human 15.5 kDa protein and/or Nop56/Nop58 (or their functional analogues in other organisms) and by the snoRNA components (5,41,42). However, since fibrillarin is the catalytic component of the C/D snoRNP, it is predicted to form critical contacts with the guide RNA (snoRNA) and target RNA (pre-rRNA) in the assembled C/D snoRNP complex. UV cross-linking of eukaryotic snoRNPs, where fibrillarin cross-linked to both D and D′ boxes, supported this suggestion (41). The data concerning the RNA-binding properties of fibrillarin in vitro and regions of the protein responsible for this activity are limited and contradictory. While some reports show that fibrillarin binds directly and specifically to snoRNAs in the absence of additional factors (43,44), other reports suggest that this interaction requires other cofactors such as yeast Snu13p (15.5 kDa in human) (42,45). Furthermore, the archaeal fibrillarin proteins are unable to bind box C/D guide snoRNAs in vitro (46–48).
Here, we have addressed the RNA-binding activity of fibrillarin in vitro. We have analysed plant fibrillarin since it has been shown to be involved in formation of RNP complexes with various RNAs—snoRNAs and rRNAs (33)—as well as with plant viral RNAs (16,28). In Arabidopsis thaliana there are two genes, AtFib1 and AtFib2, encoding nearly identical proteins conserved with eukaryotic fibrillarin proteins (49). We have characterized the interactions between recombinant fibrillarin 2 (AtFib2) and several RNA substrates that AtFib2 might be associated with in its various functions and have identified its RNA-binding regions.
MATERIALS AND METHODS
Cloning and mutagenesis
Several mutant variants of AtFib2 (16,28) were constructed as shown in Figure 1. DNA fragments coding for the mutant proteins shown in Figure 1 and wild-type AtFib2 (16,28) were cloned into the expression plasmid PGEX-KG (50), which was modified by the addition of a sequence encoding 6 histidines using XhoI and HindIII sites immediately before the stop codon. Substitution mutations were introduced using overlap PCR. The resulting proteins therefore contained a GST (glutathione S-transferase) tag fused to the N-terminus of AtFib2 and 6 histidines at its C-terminus.
Expression and purification of the proteins
The BL21 expression strain of Escherichia coli was transformed with the various plasmids encoding AtFib2 or its mutants. Protein expression induced by 1 mM IPTG was carried out for 4 h at room temperature. Proteins were purified on Ni-NTA Qiagen agarose by the denaturing method according to the manufacturer's protocol (Qiagen). For RNA-binding assays, the recombinant proteins were renatured, and urea was removed by dialysis of 0.1 ml protein sample against Milli-Q purified water for 1.5 h at room temperature with five changes of water. This method produced homogenous and non-aggregating recombinant AtFib2 protein monomers as shown previously by atomic force microscopy and ultracentrifugation in sucrose gradients (29). SDS–PAGE confirmed the monomeric nature of the recombinant AtFib2 protein (fused to GST and 6× His) (Supplementary Figure S1). Although E. coli-produced fibrillarin lacks post-translational modifications [for example, methylation (12)], this protein was able to form regular ring-like protein complexes with a virus protein, ORF3 (29) or functional (infectious) RNP complexes with virus RNA and ORF3 (16). These were similar in appearance to those detected in infected plants (16,29). Thus, post-translational modifications of AtFib2 are not required for at least some of its in vitro activities.
Electrophoretic mobility shift assay
The RNA-binding activities of AtFib2 and its mutant proteins were analysed by electrophoretic mobility shift assay (EMSA). RNA and protein were mixed at various molar ratios in RNA binding buffer (20 mM Tris–HCl, pH 7.5; 1 mM DTT; 3 mM MgCl2; 50 mM NaCl), incubated for 15 min on ice and loaded onto Tris–acetate agarose gels stained with ethidium bromide. Gels were photographed and the amount of free non-retarded RNA was quantified with Gel-Pro Analyzer (Version 3.1.00.00, Media Cybernetics). To determine the apparent Kd values, the data points were fitted using the Hill equation by SigmaPlot 11 [Systat Software Inc. (SSI)], where the RNA fraction bound = 1/(1 + (Kdn/[Protein concentration]n) and n represents the Hill constant. Three independent experiments were performed and the average Kd values and standard errors were determined.
For competition assays, U3 snoRNA was transcribed by T7 RNA polymerase (Fermentas T7 transcription kit K0411) and labelled with [a-32P]UTP. The labelled U3 snoRNA was preincubated with AtFib2 in binding buffer for 10 min, and then competitors were added and incubation was continued for 10 min. After electrophoresis, gels containing radiolabelled RNA were dried and exposed to a Phosphoimager (Fujifilm Fla-300), and the intensity of RNA bands were quantified (with Aida/2D Densitometry).
RNA substrates
Arabidopsis thaliana U3 snoRNA and U1 snRNA were transcribed by T7 RNA polymerase (Amersham Life Science) from PCR products containing a T7 promoter and the corresponding cDNA sequence (the constructs were kindly provided by S.H. Kim). pGEM7-derived RNA, and positive or negative RNA fragments (nucleotides 1–300) derived from green fluorescent protein (GFP) gene were synthesized using T7 promoter-driven transcription from pGEM7 plasmid constructs. Poly(A) was obtained from Reanal (Budapest, Hungary) and tRNA was supplied by Boehringer (Ingelheim, Germany). Viral RNA of tobacco mosaic virus (TMV) and potato virus X (PVX) were extracted from virus samples as described (51). GRV RNA was transcribed from the GRV cDNA clone, grpol, corresponding to the 5′-terminal 2556 nt of the GRV genome (52) by T7 polymerase using a mMESSAGE mMACHINE T7 kit (Ambion, Inc.). Positive or negative single-stranded (ss) short-interfering RNAs (21 nt) [siRNA171 (53)] were synthesized as RNA oligonucleotides by Syntol (Russia). Double-stranded (ds) RNAs were prepared by boiling equimolar quantities of complementary ssRNA substrates for 5 min and cooling for 3 h. 18S and 26S rRNAs were purified as follows. TAE 2% agarose gels containing ethidium bromide were prepared with two parallel rows of wells with a distance of 2–3 cm between the wells. Total RNA was extracted from Nicotiana benthamiana leaves as described (54) and loaded into the first row of wells along with the molecular weight markers. During the electrophoresis, the gel was inspected several times under the UV lamp and when the required band of RNA ran into the second row of wells, it was pipetted out and then precipitated with LiCl, resolved and its homogeneity was checked by the agarose electrophoresis.
RESULTS
RNA-binding activity of recombinant AtFib2
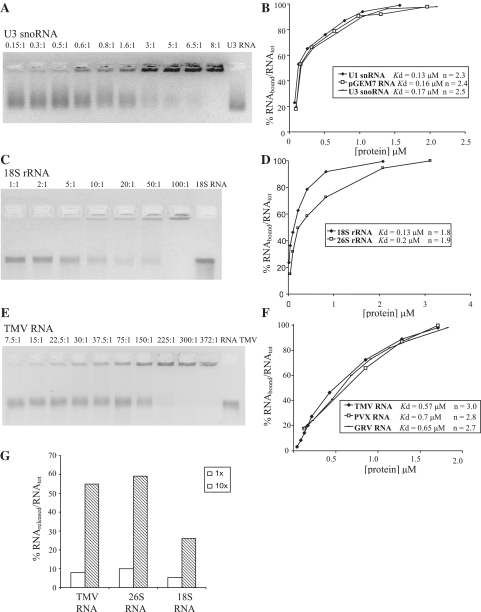
To analyse the RNA-binding properties of AtFib2, we tested its ability to interact with RNAs that fibrillarin is associated with during rRNA biogenesis: U3 snoRNA (one of the fibrillarin-associated snoRNAs (240 nt in length; 32,33) (Figure 2A and B) and 18S and 26S rRNAs, whose precursors are in vivo targets for the MTase activity of fibrillarin (6,32) (Figure 2C and D). Interaction with ‘non-specific’ RNAs such as genomic RNAs of several plant viruses was also tested (Figure 2E and F). RNA-binding data are given as EMSA images (protein:RNA molar ratios are indicated) and as binding curves obtained from the EMSA results (the percentage of RNA involved in complex with the protein is plotted versus the protein concentration in the sample). In the presence of a small amount of fibrillarin, the RNA molecules were found mostly in unbound form, while increasing the amount of fibrillarin in the binding reaction resulted in rapid transition of the RNAs to completely bound form (retained in the wells of 2% native agarose gel after electrophoresis); this was true for all RNAs tested. The absence of partially shifted bands, resulting from limited binding of RNA by fibrillarin between the completely bound and the free RNA in the gel (Figure 2A, C and E), suggests that most of the RNAs are either coated with fibrillarin or are not bound to fibrillarin at all; a phenomenon that is clearly protein concentration dependent. Moreover, immunoblotting analysis of the gels using antibody against fibrillarin further confirmed that the fraction of free RNA did not contain detectable fibrillarin molecules (data not shown). The Hill coefficients in a range of 1.8–3.0 for RNA-binding by fibrillarin also imply that RNA-binding by fibrillarin occurs in a co-operative manner (Figure 2B, D and F).
Figure 2.
AtFib2 binding to snoRNA, rRNA and viral RNA. EMSA of AtFib2 binding to snoRNA (A), rRNA (C) and viral RNA (E). Increasing amounts of AtFib2 (protein:RNA ratios indicated above each lane) were incubated in buffer for RNA binding (see ‘Materials and Methods’ section) with 0.25 µg of RNA (the used RNA is indicated above each gel) and loaded onto 2% non-denaturing agarose gel with ethidium bromide. The lane marked ‘RNA’ is the probe with no protein added. In all experiments, RNA was either free or retarded at the top of the gel. (B, D and F) Curves obtained from EMSA results, representing AtFib2 binding to various substrates. Kd and Hill coefficient (n) values are indicated for each substrate. (B) U3 snoRNA, U1 snRNA and 160 nt artifiсial RNA. (D) 18S and 26S ribosomal RNAs from N. benthamiana (F) Genomic RNAs of plant viruses (TMV, tobacco mosaic virus; PVX, potato virus X; GRV, groundnut rosette virus). The percentage of RNA involved in complex with the protein is plotted versus the protein concentration in the sample. (G) Competition binding assay of the AtFib2 to U3 snoRNA. The protein was incubated with [P32]-labelled U3 snoRNA (protein:RNA molar ratio is 8:1 that provides 100% binding of RNA) in the presence of equal amount (×1) or 10-fold (10×) excess of the following unlabeled competitors: TMV RNA, 26S rRNA or 18S rRNA. After incubation, samples were analyzed by electrophoresis on 2% non-denaturing agarose gel, and the amount of U3 snoRNA released from complex was determined (see ‘Materials and Methods’ section). The diagram shows the percentage of free U3 snoRNA depending on the type and the amount of competitor. The excess of the competitors is indicated by white (1×) or stroked (10×) columns.
The apparent Kd for U3 snoRNA binding by AtFib2 was estimated to be ∼0.17 μM. The full inclusion of U3 snoRNA into the complex with AtFib2 was observed at the protein:RNA molar ratio of 8:1 (Figure 2A and B; Table 1). We also tested the binding of AtFib2 to U1 snRNA (160 nt in length) and to a 160 nt RNA transcribed from the T7 promoter of pGEM7 plasmid (the latter was used as a non-specific substrate) (Figure 2B). AtFib2 was able to bind both these RNA substrates in a way similar to U3 snoRNA—the apparent Kd values were 0.13 μM for U1 snRNA and 0.16 μM for PGEM7 RNA (Figure 2B). The protein:RNA molar ratios that resulted in total RNA inclusion into the complex with AtFib2 protein was 6:1 for U1 snRNA and 10:1 for pGEM-7 RNA, and 50% of RNA binding was achieved at the ratio of 0.6:1–0.5:1 for all RNA substrates of this size.
Table 1.
RNA-binding characteristics of AtFib2 and its mutants
| Protein | U3 snoRNA |
TMV RNA |
||||||
|---|---|---|---|---|---|---|---|---|
| 100% ratioa | 50% ratiob | Apparent, Kd (µM) | n (Hill constant) | 100% ratioa | 50% ratiob | Apparent, Kd (µM) | n (Hill constant) | |
| AtFib2 | 8:1 | 0.6:1 | 0.17 ± 0.01 | 2.5 ± 0.18 | 300:1 | 100:1 | 0.57 ± 0.04 | 3.0 ± 0.3 |
| AtFib2-Ala | 8:1 | 0.65:1 | 0.18 ± 0.02 | 2.4 ± 0.2 | 300:1 | 100:1 | 0.57 ± 0.05 | 3.0 ± 0.2 |
| AtFib2ΔGAR | 8:1 | 0.6:1 | 0.17 ± 0.02 | 2.3 ± 0.3 | 300:1 | 100:1 | 0.57 ± 0.03 | 2.8 ± 0.3 |
| AtFib2ΔR | 10:1 | 0.6:1 | 0.17 ± 0.01 | 2.6 ± 0.34 | 360:1 | 120:1 | 0.7 ± 0.05 | 3.1 ± 0.3 |
| AtFib2Δα-rich | 11:1 | 0.6:1 | 0.17 ± 0.01 | 2.5 ± 0.24 | 360:1 | 120:1 | 0.7 ± 0.06 | 3.0 ± 0.3 |
| AtFib2GAR | No | No | No | No | No | No | No | No |
| AtFib2GAR-spacer | No | No | No | No | No | No | No | No |
| AtFib2R | 33:1 | 2.5:1 | 0.7 ± 0.08 | 2.0 ± 0.2 | 700:1 | 350:1 | 2.0 ± 0.03 | 2.3 ± 0.3 |
| AtFib2R-Ala | 41:1 | 12:1 | 3.4 ± 0.25 | 1.2 ± 0.2 | 870:1 | 650:1 | 3.7 ± 0.41 | 1.3 ± 0.3 |
| AtFib2R138–179 | 58:1 | 5:1 | 1.4 ± 0.12 | 1.8 ± 0.3 | 900:1 | 700:1 | 4.0 ± 0.22 | 1.5 ± 0.3 |
| AtFib2R138–187 | 64:1 | 16:1 | 4.5 ± 0.33 | 1.1 ± 0.24 | 900:1 | 700:1 | 4.0 ± 0.41 | 1.0 ± 0.3 |
| AtFib2R188–224 | No | No | No | No | No | No | No | No |
| AtFib2α-rich | 32:1 | 3:1 | 0.8 ± 0.06 | 2.0 ± 0.3 | 700:1 | 350:1 | 2.0 ± 0.18 | 2.5 ± 0.2 |
| AtFib2α-rich258–281 | 36:1 | 17:1 | 3.9 ± 0.4 | 1.0 ± 0.15 | 780:1 | 400:1 | 2.3 ± 0.2 | 1.5 ± 0.2 |
| AtFib2α-rich282–320 | No | No | No | No | No | No | No | No |
aProtein: RNA molar ratio of 100% RNA bound.
bProtein: RNA molar ratio of 50% RNA bound.
No indicates the protein showed no RNA-binding, and therefore, no kinetic parameters were calculated.
Values are averages for three individual experiments. Standard deviations are given for apparent Kd and Hill coefficient.
AtFib2 interacted with 18S and 26S rRNAs from N. benthamiana in a similar manner (Figure 2C and D). The apparent Kd values were 0.13 μM for 18S rRNA and 0.2 μM for 26 S rRNA. The full RNA binding was observed at the protein:RNA molar ratio of 80:1 for 18S rRNA and 150:1 for 26S rRNA. The 50% of RNA binding was achieved at the protein:RNA molar ratio of 6:1 for 18S rRNA and of 10:1 for 26S rRNA.
The fact that AtFib2 was shown to be a component of plant viral movement RNP (16,28) prompted us to test the interactions between fibrillarin and viral RNAs. As shown in Figure 2E and F, fibrillarin bound genomic RNAs of various plant viruses (TMV, PVX or GRV) with similar efficiency. The apparent Kd for TMV RNA was ∼0.57 μM. Full retardation protein:RNA molar ratio was ∼300:1 (Figure 2E and F). These data suggest that AtFib2 has slightly lower affinity for viral RNAs than for rRNAs. However, the viral RNA-binding characteristics of fibrillarin were similar to those of viral movement proteins that form RNP complexes with these viral RNAs (55–57).
AtFib2 RNA binding was additionally analysed in competition assays. Figure 2G shows that a radiolabelled U3 snoRNA can be released from the complex with AtFib2 by TMV RNA, 18S rRNA and 26S rRNA, although 18S rRNA was a less efficient competitor.
Within the snoRNP complex, SAM (the methyl group donor) is attached to fibrillarin (8,10,22). Therefore, we have tested the influence of this ligand on the AtFib2 binding to U3 snoRNA, pGEM7 RNA and TMV RNA. The addition of SAM [15-fold molar excess compared with AtFib2 concentration as in (48)] had no effect on AtFib2 binding to any of these RNA substrates (data not shown).
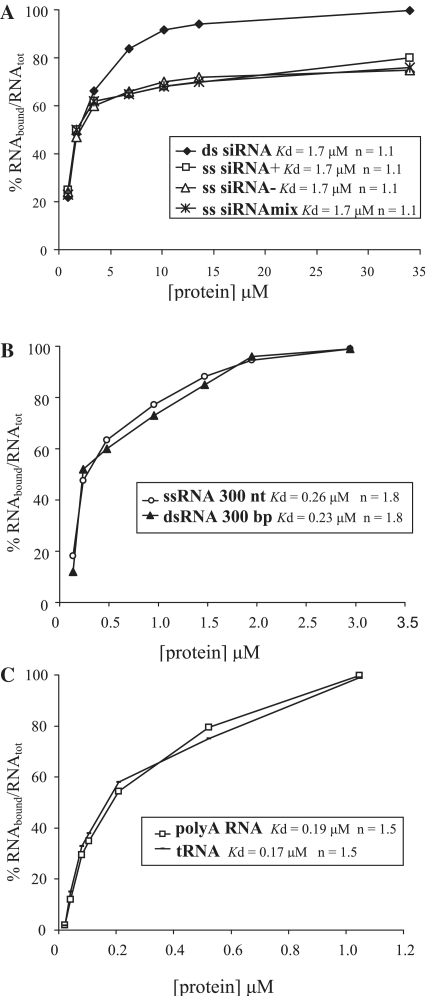
Several well-studied silencing suppressors (p19, p21 and HC-Pro) inhibit the intermediate step of RNA silencing via binding to siRNAs (58). Some recent indications that fibrillarin may be involved in the process of suppression of gene silencing induced by viruses (30) prompted us to probe its ability to interact with siRNAs. Fibrillarin was able to bind both single-stranded (separate or mixed but non-annealed positive and negative) and double-stranded 21 nt siRNAs (Figure 3A) with the similar value of the apparent Kd of ∼1.7 μM. However, while at the ratio of 1 molecule of AtFib2 per 1 molecule of siRNA 50% of both ss or double-stranded RNAs was bound by fibrillarin and the full retardation ratio for double-stranded siRNA was 20:1, complete binding of ss siRNA was not achieved at all: ∼20% of ss molecules remained unbound (Figure 3A). To determine whether these differences in binding are due to generic differences in the structure of ss or double-stranded RNAs, we tested the binding of AtFib2 to 300-nt long dsRNA or ssRNA fragments (derived from the GFP gene). Remarkably, both these RNAs demonstrated similar affinity to fibrillarin (Kd of ∼0.26 μM and of 0.23 μM, correspondingly, Figure 3B), suggesting no clear preferences of fibrillarin for ss or double-stranded RNAs. Furthermore, the binding behaviour of AtFib2 for a highly structured RNA substrate (tRNA) and to a 80 nt poly(A) sequence, which lacks secondary structure under conditions tested (pH 8.0), was also essentially similar (Kd were 0.19 μM for poly(A) and 0.17 μM for tRNA) (Figure 3C). Thus, it remains unclear why ss siRNAs show lower maximum binding than double-stranded siRNAs, although it cannot be ruled out that a portion of ss siRNA molecules (∼20%) may have a conformation unfavourable for binding by fibrillarin.
Figure 3.
AtFib2 binding to dsRNA and ssRNA: (A) small 21 nt dsRNA, positive ssRNA, negative ssRNA and mixture of non-annealed positive and negative RNAs, (B) 300-nt dsRNA and ssRNA corresponding to the GFP coding sequence (nucleotides 1–300) and (C) highly structured RNA (tRNA) and non-structured RNA [80 nt long poly(A)]. Curves are obtained from EMSA results.
Collectively, these data suggest that fibrillarin binds RNA with no or little sequence or structure specificity [dsRNA and ssRNAs of different lengths, snoRNA, snRNA, rRNAs, virus RNAs, tRNA, poly(A), etc.]. However, certain RNAs show some differences in the affinity to fibrillarin. For instance, fibrillarin interacts with long viral RNAs less effectively, than with rRNAs of similar length, and the least effective binding was demonstrated for very short RNAs (21 nt) suggesting that some unidentified yet structure/sequence preference of RNA binding by fibrillarin cannot be ruled out.
Identification of the RNA-binding sites within the AtFib2 protein
The high homology between the human and Arabidopsis fibrillarin proteins (∼70% similarity) (9) allowed us to make the schematic representation of the AtFib2 molecule on the basis of a slightly modified scheme suggested by Aris and Blobel (7) for the human fibrillarin (Figure 1A). This scheme shows the three main regions of the protein (indicated above the protein scheme): the N-terminal GAR domain, the central region containing the putative RNA-binding domain (R-region) and the C-terminal region containing the predicted sequence rich in α-helices. On the basis of sequence similarity (7,8) and X-ray crystallography data (10), the central and C-terminal regions were combined into an MTase domain, while the GAR domain and the following spacer were shown as the N-terminal domain. Based on this putative structure of AtFib2, we generated a series of deletion or substitution mutants to address RNA-binding (Figure 1B).
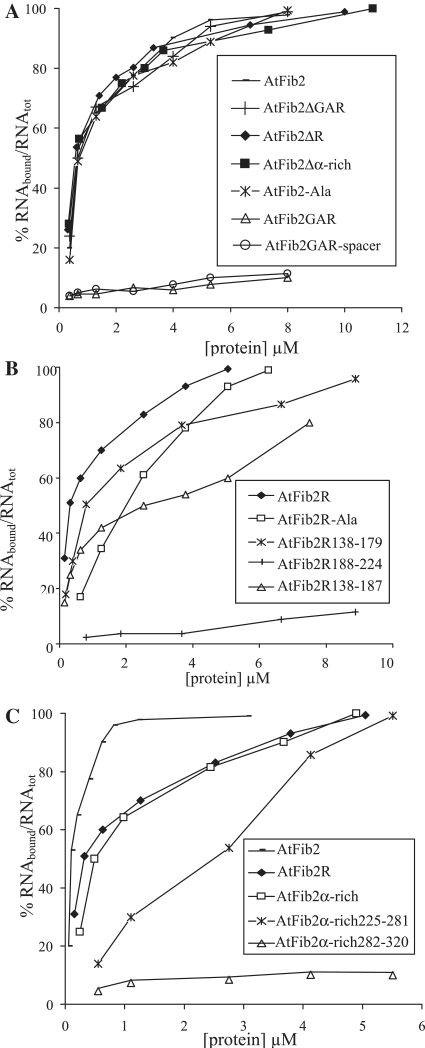
In the following experiments, the RNA-binding activity of At2Fib mutants was analysed using U3 snoRNA (240 nt) and genomic TMV RNA (6400 nt) as substrates. Figure 4 describes the dependence of U3 snoRNA binding by the protein variants on the concentration of the protein in the sample. Table 1 presents the protein:RNA molar ratios for full and 50% RNA binding for both U3 snoRNA and TMV RNA substrates and the values of apparent Kd.
Figure 4.
Analysis of RNA-binding activity of AtFib2 mutants (using U3 snoRNA). The percentage of RNA included in complex with the protein is plotted versus the protein concentration in the sample. Mutants (and wild-type protein) compared on one plot are indicated. (A) Comparison of U3 snoRNA binding of AtFib2 protein and its mutant variants: AtFib2-Ala, AtFib2ΔGAR, AtFib2ΔR, AtFib2Δα rich, AtFib2GAR and AtFib2GAR-spacer. (B) U3 snoRNA binding by isolated R region (AtFib2R mutant protein) and its variants with deletions and substitutions: AtFib2R-Ala, AtFib2R138–179, AtFib2R138–187 and AtFib2R188–225. (C) Comparison of U3 snoRNA binding by the isolated R region (AtFib2R mutant protein), α-rich region (AtFib2α-rich) or deletion variants of α-rich region: AtFib2α-rich225–281 and AtFib2α-rich282–320.
It was suggested previously (7) that the conserved octamer in the R-region could provide the RNA-binding function of fibrillarin. However, the substitution of the GCVYAVEF octamer with alanines had no effect on RNA binding to U3 snoRNA by the full-length AtFib2 protein (Figure 4A, mutant AtFib2-Ala, Table 1). Interestingly, the deletion of any of three previously predicted domains (7) also did not lead to reduction of RNA-binding activity (Figure 4A and Table 1). Indeed, the mutated fibrillarin proteins with a deleted GAR region (AtFib2ΔGAR), R-region (AtFib2ΔR) or α-rich region (AtFib2Δα-rich) demonstrated the same values of apparent Kd as the full-length protein (∼0.17 μM) for U3 snoRNA binding (Figure 4A). In the case of TMV RNA, the values of apparent Kd for AtFib2ΔR and AtFib2Δα-rich mutants were slightly higher (∼0.7 μM versus 0.57 μM for the full-length protein). These results suggested that there is not a simple relationship between the regions of fibrillarin and RNA binding and that the RNA-binding properties of AtFib2 may reside in more than one region of the protein.
To examine this possibility, we have constructed mutants comprising the separate regions of the protein (Figure 1B) and studied their RNA-binding activity. Figure 4 demonstrates the binding of U3 snoRNA by the AtFib2 mutants. The binding parameters for the interaction of the AtFib2 mutants with U3 snoRNA and with TMV RNA are given in Table 1. The GAR domain (AtFib2GAR) showed no RNA-binding activity by itself or together with the spacer that connects it to the R-region (AtFib2GAR-spacer) (Figure 4A). At the same time, both the R region (AtFib2R) and the α-rich region (AtFib2α-rich) bound RNA in a similar manner (Figure 4B and C; Table 1). The binding parameters of these mutants differed from that of the full-length protein, but were similar to each other—the apparent Kd values were 0.7–0.8 μM for U3 snoRNA and 2 μM for TMV RNA. The complete inclusion of RNA into the complex required higher protein:RNA molar ratios than for the full-length At2Fib (33:1 for U3 snoRNA and 700:1 for TMV RNA, Table 1). This effect could be partially due to the smaller size of the molecules of the mutant proteins compared to full-length fibrillarin, requiring more molecules to cover the given RNA. Interestingly, the substitution of the GCVYAVEF octamer with a sequence containing eight alanine residues reduced (but did not abolish) the RNA-binding of the isolated R region (AtFib2R-Ala) (Figure 4B)—the apparent Kd values were 3.4 μM and 3.7 μM versus 0.7 μM and 2 μM for U3 snoRNA and TMV RNA, respectively. The full retardation ratios were 40:1 for U3 snoRNA and 870:1 for TMV RNA (Figure 4B and Table 1).
Collectively, these data demonstrate the existence of two independent RNA-binding regions in the fibrillarin molecule: the first within the central region and the second within the C-terminal region, and that the activity of the central RNA-binding region is not directly provided by the conserved GCVYAVEF octamer, but that this does contribute to binding perhaps by assisting the proper folding of the RNA-binding domain resided in the R region of AtFib2.
To more accurately localize the sites of RNA-binding activity within fibrillarin, parts of the R and α-rich regions (Figure 1B) were expressed, purified and tested for RNA-binding activity. The N-terminal part of the R region upstream of the conserved octamer (AtFib2R138-179) showed reduced RNA-binding activity (the apparent Kd was 1.4 μM for U3 snoRNA and 4 μM for TMV RNA) (Figure 4B). The N-terminal part of R region including the conserved octamer (mutant protein AtFib2R138-187) showed even more reduced ability in binding of U3 snoRNA (apparent Kd is 4.5 μM), while the apparent Kd for TMV RNA did not change (Figure 4B and Table 1) suggesting that the octamer may be interfering with the ability of the N-terminal part of the R region to bind snoRNA. The C-terminal part of the R region (AtFib2R188-224) did not show RNA-binding activity at all (Figure 4B).
The N-terminal part of the α-rich region of AtFib2 (AtFib2α-rich-225-281) showed RNA-binding activity (Figure 4C and Table 1), though slightly lower (Kd was 3.9 μM for U3 snoRNA and 2.3 μM for TMV RNA) than that of the whole α-rich region of AtFib2. Interestingly, the binding of viral RNA (TMV RNA) was similar for both AtFib2α-rich and AtFib2α-rich225-281 (with Kd of 2 μM for AtFib2α-rich and of 2.3 μM for AtFib2α-rich225-281). The C-terminal part of the α-rich region (AtFib2α-rich-282-320) did not show RNA-binding activity with any RNA substrate (Figure 4C and Table 1).
Thus taken together, our data demonstrate that AtFib2 possesses two RNA-binding sites in the central (138–179 amino acids) and C-terminal (225–281 amino acids) parts of the protein, respectively, both of which exhibit low sequence specificity of RNA-binding activity in vitro. It is worth noting that like wild-type AtFib2, all the AtFib2 mutant proteins able to bind RNA exhibited Hill coefficients that were >1 with all RNA substrates, indicating that both AtFib2 RNA-binding sites provide co-operative RNA binding (Table 1).
DISCUSSION
Our results clearly demonstrate that AtFib2 interacts with RNAs of various sizes and nature in a co-operative manner. However, the possibility of specific RNA binding by fibrillarin could not be completely ruled out because fibrillarin displays certain differences in RNA-binding preferences that may be involved in various biological processes. Moreover, this work has been carried out with bacterially produced recombinant AtFib2 protein, which lacked posttranslational modifications. This protein was shown to be able to fulfil at least some of fibrillarin functions in vitro such as formation of regular ring-like protein complexes with the GRV ORF3 protein or functional (infectious) RNP particles with GRV RNA and ORF3 (16,29; see also ‘Materials and Methods’ section). However, it cannot be completely ruled out that the recombinant AtFib2 protein may have less specificity to RNA binding than intact AtFib2.
We have also found that there are two RNA-binding regions within the molecule of AtFib2 protein: one located in the central region comprising 138–179 amino acid residues and the other in the C-terminal α-helix region comprising 225–281 amino acid residues. The GCVYAVEF octamer does not take part in interactions with RNA directly, but may be required for the proper conformation of the central RNA-binding region. Both RNA-binding regions were able to independently interact with various RNAs with no or little specificity. Interestingly, in our experiments the deletion of either of the RNA-binding regions had no effect on the parameters of RNA-binding of the protein. This suggests that the two regions function independently, and that the deletion of one of them does not destroy or damage the conformation of the other. The GAR domain together with the following spacer was incapable of RNA-binding.
The most prominent process in which fibrillarin plays a crucial role is site-specific 2′-O-ribose methylation of rRNA and snRNAs. Low specificity of RNA binding by the plant fibrillarin is in good agreement with the data suggesting that the crucial role in targeting of the methylation snoRNP complex to the specific sites of pre-rRNAs is played by other core components (5,20,32,59). The 15.5 kDa protein initiates snoRNP assembly by specific recognition and binding of K-turn in the C/D site of snoRNAs (45). Then, Nop58 and fibrillarin are independently recruited to the snoRNA (41,45). In cross-linking experiments, Nop58 was in contact with the C box and showed a specific interaction (41). Nop56 is involved in the formation of snoRNP complex via interaction with fibrillarin and within the complex establishes direct and sequence-specific interactions with C′ box of snoRNA (20,41,45). The mature snoRNP complex is directed to the site of methylation by sequence-specific duplex formation between guide snoRNA and pre-rRNA. Fibrillarin is the only core component of snoRNP that showed no specificity in RNA-binding (41). Fibrillarin has been cross-linked to the D, D′ and C′ box. In contrast to the situation with the other core proteins, mutations within the C′ or D′ boxes did not influence the binding of fibrillarin (41).
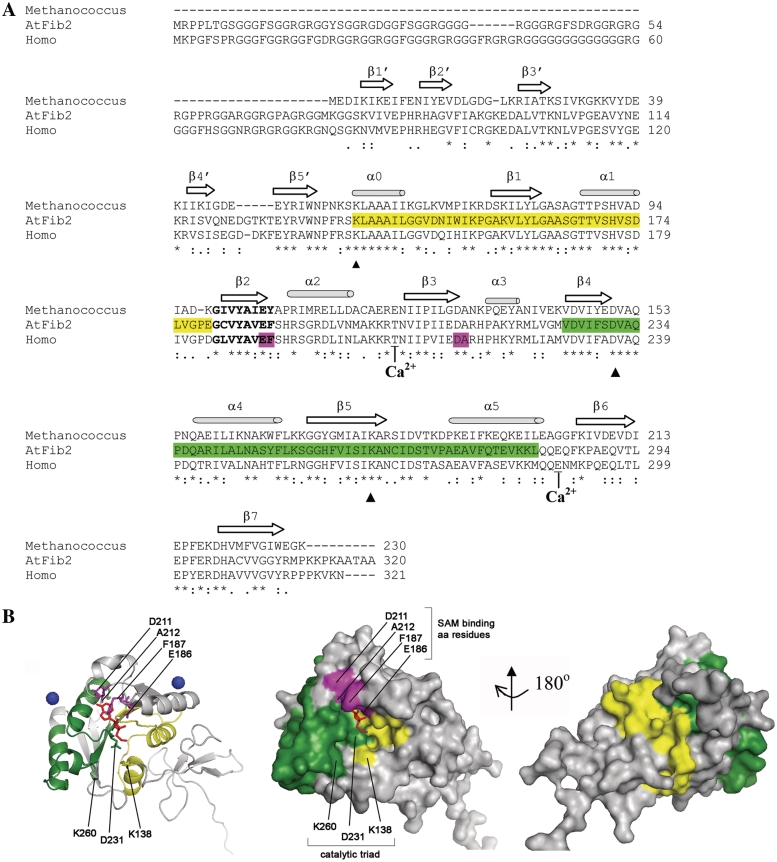
Remarkably, both of the RNA-binding regions identified in this work (amino acid 138–179 and amino acid 225–281) are located within the MTase domain of the fibrillarin comprising the central (R) and C-terminal regions of fibrillarin (Figures 1A and 5A). The fact that fibrillarin is highly conserved among different species (7–9) has allowed us to apply the structural data obtained for the fibrillarin proteins of other species to AtFib2 in order to evaluate the structure of RNA-binding sites and their location on the molecule of fibrillarin (Figure 5A). Fibrillarin proteins belong to the superfamily of the Rossmann-fold SAM-dependent MTases (8). The nine sequence motifs that are strongly conserved among the MTases, including conserved SAM-binding motif and the catalytic triad/tetrad [K-D-K-(H)], have also been revealed in fibrillarin proteins of various species (60,61). Despite the rather low total homology in the amino acid sequence, these Rossmann-fold MTases share strikingly similar structure. It comprises a seven-stranded β-sheet flanked by α-helices to form an α−β−α sandwich (62). The distinction of the R- and α-rich regions in MTase domain of fibrillarin (7) is consistent with the tertiary structure of Rossmann-fold MTases, and the presence of two RNA-binding sites in fibrillarin is in agreement with the suggested dimeric nature of the MTase domain (62). Min et al. (10) while studying the crystal structure of human fibrillarin discovered that it binds two Ca2+ ions. The divalent cations are often essential for polynucleotide binding. These data also might reflect the existence of two independent binding sites for RNA in fibrillarin.
Figure 5.
Localization of the RNA-binding sites in the AtFib2 structure. (A) Sequence alignment of fibrillarin homologues from various species was performed using ClustalW2 multiple sequence alignment (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The sources of aligned sequences are as follows: Methanocaldococcus jannaschii (Methanocaldococcus) (accession number AAB98690.1), Arabidopsis thaliana (AtFib2) (accession number AAG10104.1) and Homo sapience (Homo) (accession number AAA52453.1). The similarity scores between AtFib2 and Metanocaldococcus and human fibrillarin homologues are 42 and 70%, respectively. Asterisks mark the identical residues in all sequences, colons mean the conserved substitutions, dots mean the semi-conserved substitutions. Elements of the secondary structure are labelled as in (8,62). The GCVYAVEF motif is shown in bold letters. The two RNA-binding regions determined here are marked with yellow (site within the R-region) or green (site within the ‘α-rich’ region) in the sequence alignment and structures below. The calcium-binding sites in the human fibrillarin (10) are indicated. The MTase catalytic triad K138/D231/K260 is indicated with filled triangle. The amino acid residues that were shown to interact with the SAM analogue in human fibrillarin (E186-F187 and D211-A212) are shown in purple. (B) The localization of the RNA-binding domains on the three-dimensional model of AtFib2 protein. A homology model of AtFib2 based on the structure of human fibrillarin [PDB 2IPX (10)] was built using MODELLER 9v7 with the graphic interface Easy Modeller 2.0. The structure does not include the GAR region as it is absent from the crystal structure of human fibrillarin (10). The localization of the RNA-binding regions is depicted in cartoon form and on a molecular surface in the same orientation (the surface models are given in two projections—looking from the side of SAM-binding pocket and turned 180o around the vertical axis). The RNA-binding regions are coloured as in (A). The amino acid residues that were shown to interact with SAM analogue in human fibrillarin (E186-F187 and D211-A212) are shown in purple and labelled. Amino acid residues of the MTase catalytic triad of AtFib2, K138/D231/K260 are also labelled. Ligands are indicated as follows: two Ca2+ ions are shown as blue spheres and the SAM analogue is shown as red sticks. Figures were prepared using PyMOL (www.pymol.org).
To get an insight into the architecture of AtFib2 RNA-binding sites, we built a three-dimensional model of AtFib2 based on the available structure of the human fibrillarin (10). The model does not include a GAR region, as it is absent from the crystal structure of human fibrillarin (10). Figure 5B shows this model of AtFib2 depicted in cartoon form and as a molecular surface. Interestingly, both of the AtFib2 RNA-binding sites identified in this work are spatially separated and primarily exposed on the surface of the molecule (Figure 5B). The amino acid residues that were shown to interact with SAM analogue in human fibrillarin (10), E186-F187 and D211-A212 are located outside of the RNA-binding sites but in close proximity to them (Figure 5B). Furthermore, amino acid residues of the MTase catalytic triad of AtFib2, K138/D231/K260 reside within the R- (K138) and α-rich- (D231 and K260) RNA-binding sites (Figure 5A and B). Thus, it seems conceivable that the catalytic triad, SAM-binding site and adjoining parts of the RNA-binding domains form the united pocket of fibrillarin. Although the predicted structural model of AtFib2 active sites requires experimental verification, it provides a rationale for the involvement of two RNA-binding sites in formation of the MTase catalytic centre in fibrillarin. It is likely that these two RNA-binding sites (in a concert with other proteins, 15.5 kDa protein, Nop56/Nop58) can interact with guide (snoRNAs and scaRNAs) and substrate (rRNA, snRNAs) RNA molecules, respectively, to place their duplex containing the methylation site closer to the active SAM-binding centre. However, given the apparently low specificity of the fibrillarin RNA-binding sites, recognition of specific sequences on the guide and substrate RNAs by fibrillarin could be presumably determined by other components of snoRNPs (scaRNPs). At the same time, the lack of stringent specificity in RNA binding may account for interactions of fibrillarin with a wide range of guide (snoRNAs, scaRNAs) and substrate (rRNA, snRNAs) RNA molecules.
In addition to its major role in modification of rRNA and snRNAs, fibrillarin is also involved in other functions. Recent data indicate that interaction with fibrillarin is a general property of various virus proteins that is not restricted to one or two virus taxonomic groups [see for review (27)]. However, such interactions may have quite diverse molecular implications for different viruses being required at various phases of virus infection cycle such as virus movement, replication or counter defence (27). This may suggest novel, unexpected natural functions for fibrillarin that are hijacked or affected by viruses at different stages of infection for needs of the viruses. The ability of this nucleolar protein to bind a broad spectrum of various RNAs may be a key factor determining these functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International Association for the promotion of co-operation with scientists from the New Independent States of the former Soviet Union (Fellowship no. 04083-2809 to D.V.R.); Russian Foundation for Basic Research (grant no. 10-04-00522-а to N.O.K. and D.V.R.); Russian Ministry of Education and Science (contract no. 02.740.11.5145 to M.T., N.O.K. and D.V.R.); Scottish Government (to M.T. and J.W.S.B.). Funding for open access charge: The James Hutton Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S.H. Kim for providing the cDNA of AtFib2 gene and cDNA constructs of the U1 and U3 RNAs. We thank E.N. Dobrov and O.V.Borisova for providing the preparations of TMV and PVX viruses.
REFERENCES
- 1.Warner JR. The nucleolus and ribosome formation. Curr. Opin. Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 2.Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 3.Snaar S, Wiesmeijer K, Jochemsen AG, Tanke HJ, Dirks RW. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 2000;151:653–662. doi: 10.1083/jcb.151.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spector DL. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 5.Reichow SL, Hamma T, Ferré-D′Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 7.Aris JP, Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmuneantisera. Proc. Natl Acad. Sci. USA. 1991;88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanocaldococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pih KT, Yi MJ, Liang YS, Shin BJ, Cho MJ, Hwang I, Son D. Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 2000;123:51–58. doi: 10.1104/pp.123.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min J, Wu H, Zeng H, Loppnau P, Weigelt J, Sundstrom M, Arrowsmith CH, Edwards AM, Bochkarev A, Plotnikov AN. Human fibrillarin. A Resource for Studying Biological Macromolecules. Protein Data Bank. 2006 http://www.pdb.org/pdb/home/home.do. PDB ID 2IPX. [Google Scholar]
- 11.David E, McNeil JB, Basile V, Pearlman RE. An unusual fibrillarin gene and protein: structure and functional implications. Mol. Biol. Cell. 1997;8:1051–1061. doi: 10.1091/mbc.8.6.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagida M, Hayano T, Yamauchi Y, Shinkawa T, Natsume T, Isobe T, Takahashi N. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins {α}3 and {beta}1 that is independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 2004;279:1607–1614. doi: 10.1074/jbc.M305604200. [DOI] [PubMed] [Google Scholar]
- 14.Nicol RL, Frey N, Olson EN. From the sarcomere to the nucleus: role of genetics and signaling in structural heart disease. Annu. Rev. Genomics Hum. Genet. 2000;1:179–223. doi: 10.1146/annurev.genom.1.1.179. [DOI] [PubMed] [Google Scholar]
- 15.Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 16.Kim SH, MacFarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JWS, Taliansky M. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl Acad. Sci. USA. 2007;104:11115–11120. doi: 10.1073/pnas.0704632104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo D, Wootton SK, Li G, Song C, Rowland RR. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levitskii SA, Mukhariamova KSh, Veiko VP, Zatsepina OV. Identification of signal sequences determining the specific nucleolar localization of fibrillarin in HeLa cells. Mol. Biol. 2004;38:405–413. [PubMed] [Google Scholar]
- 19.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Lechertier T, Grob A, Hernandez-Verdun D, Roussel P. Fibrillarin and Nop56 interact before beining co-assembled in box C/D snoRNPs. Exp. Cell Res. 2009;315:928–942. doi: 10.1016/j.yexcr.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Hardin JW, Reyes FE, Batey RT. Analysis of a Critical Interaction within the Archaeal Box C/D Small Ribonucleoprotein Complex. J. Biol. Chem. 2009;284:15317–15324. doi: 10.1074/jbc.M901368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:13808–13813. doi: 10.1073/pnas.0905128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Wurm T, Britton P, Brooks G, Hiscox JA. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dove BK, You JH, Reed ML, Emmett SR, Brooks G, Hiscox JA. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell Microbiol. 2006;8:1147–1157. doi: 10.1111/j.1462-5822.2006.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dove BK, Brooks G, Bicknell KA, Wurm T, Hiscox JA. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 2006;80:4147–4156. doi: 10.1128/JVI.80.8.4147-4156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taliansky ME, Brown JWS, Rajamäki ML, Valkonen JPT, Kalinina NO. Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv. Virus Res. 2010;77:119–158. doi: 10.1016/B978-0-12-385034-8.00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SH, MacFarlane S, Kalinina NO, Rakitina D, Ryabov E, Gillespie T, Haupt S, Brown JWS, Taliansky M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canetta E, Kim SH, Kalinina NO, Shaw J, Adya AK, Gillespie T, Brown JWS, Taliansky M. A plant virus movement protein forms ring-like complexes with the major nucleolar protein, fibrillarin, in vitro. J. Mol. Biol. 2008;376:932–937. doi: 10.1016/j.jmb.2007.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajamäki ML, Valkonen JP. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell. 2009;21:2485–2502. doi: 10.1105/tpc.108.064147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González I, Martínez L, Rakitina DV, Lewsey MG, Atienzo FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T. Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to its RNA silencing suppressor activity. Mol. Plant Microbe Interact. 2010;23:294–303. doi: 10.1094/MPMI-23-3-0294. [DOI] [PubMed] [Google Scholar]
- 32.Dunbar DA, Wormsley S, Lowe TM, Baserga SJ. Fibrillarin-associated box C/D small nucleolar RNAs in Trypanosoma brucei. Sequence conservation and implications for 2′-O-ribose methylation of rRNA. J. Biol. Chem. 2000;275:14767–14776. doi: 10.1074/jbc.m001180200. [DOI] [PubMed] [Google Scholar]
- 33.Saez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverria M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 2004;25:7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 35.Kiss-Laszlo Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 36.Sollner-Webb B, Tycowski KT, Steitz JA. Ribosomal RNA processing in eukaryotes. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA. Structure, Evolution, Processing, and Function in Protein Biosynthesis. Boca Raton, FL: CRC Press; 1996. pp. 469–490. [Google Scholar]
- 37.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 38.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EG. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyc K, Steitz JA. U3, US and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113––3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tycowski KT, Shu M-D, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 41.Cahill NM, Friend K, Speckmann W, Li ZH, Terns RM, Terns MP, Steitz JA. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fatica A, Galardi S, Altieri F, Bozzoni I. Fibrillarin binds directly and specifically to U16 box C/D snoRNA. RNA. 2000;6:88–95. doi: 10.1017/s1355838200991623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh S, Ghosh R, Das P, Chattopadhyay DJ. Expression and purification of recombinant Giardia fibrillarin and its interaction with small nuclear RNAs. Protein Expr. Purif. 2001;21:40–48. doi: 10.1006/prep.2000.1319. [DOI] [PubMed] [Google Scholar]
- 45.Watkins NJ, Ségault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 46.Hardin JW, Batey RT. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specifity. Nucleic Acids Res. 2006;34:5039–5051. doi: 10.1093/nar/gkl644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barneche F, Steinmetz F, Echeverria M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 2000;275:27212–27220. doi: 10.1074/jbc.M002996200. [DOI] [PubMed] [Google Scholar]
- 50.Guan K-L, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 51.Chapman SN. Tobamovirus isolation and RNA extraction. Methods Mol. Biol. 1998;81:123–129. doi: 10.1385/0-89603-385-6:123. [DOI] [PubMed] [Google Scholar]
- 52.Taliansky ME, Robinson DJ, Murant AF. Complete nucleotide sequence and organization of the RNA genome of groundnut rosette umbravirus. J. Gen. Virol. 1996;77:2335–2345. doi: 10.1099/0022-1317-77-9-2335. [DOI] [PubMed] [Google Scholar]
- 53.Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48:1050–1060. doi: 10.1093/pcp/pcm074. [DOI] [PubMed] [Google Scholar]
- 54.Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q, Palukaitis P. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology. 1996;216:71–79. doi: 10.1006/viro.1996.0035. [DOI] [PubMed] [Google Scholar]
- 56.Kalinina NO, Rakitina DV, Yelina NE, Zamyatnin AA, Stroganova TA, Klinov DV, Prokhorov VV, Ustinova SV, Chernov BK, Schiemann J, et al. RNA-binding properties of the 63 kDa protein by the triple gene block of Poa semilatent hordeivirus. J. Gen. Virol. 2001;82:2569–2578. doi: 10.1099/0022-1317-82-10-2569. [DOI] [PubMed] [Google Scholar]
- 57.Taliansky ME, Roberts IM, Kalinina N, Ryabov EV, Raj SK, Robinson DJ, Oparka KJ. An umbraviral protein, involved in longdistance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 2003;77:3031–3040. doi: 10.1128/JVI.77.5.3031-3040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lakatos L, Csorba T, Pantoaleo V, Champman EJ, Carrington JC, Liu Y-P, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;80:5747–5756. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lafontaine DL, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- 61.Purta E, O'Connor M, Bujnicki J, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 62.Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of AdoMet-dependent MTases. In: Cheng X, Blumenthal RM, editors. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific, SIN; 1999. pp. 1–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.