Abstract
The Saccharomyces cerevisiae Uls1 belongs to the Swi2–Snf2 family of DNA-dependent ATPases and a new protein family of SUMO-targeted ubiquitin ligases. Here, we examine a physiological role of Uls1 and report for the first time its involvement in response to replication stress. We found that deletion of ULS1 in cells lacking RAD52 caused a synthetic growth defect accompanied by prolonged S phase and aberrant cell morphology. uls1Δ also progressed slower through S phase upon MMS treatment and took longer to resolve replication intermediates during recovery. This suggests an important function for Uls1 during replication stress. Consistently, cells lacking Uls1 and endonuclease Mus81 were more sensitive to HU, MMS and CPT than single mus81Δ. Interestingly, deletion of ULS1 attenuated replication stress-related defects in sgs1Δ, such as sensitivity to HU and MMS while increasing the level of PCNA ubiquitination and Rad53 phosphorylation. Importantly, Uls1 interactions with Mus81 and Sgs1 were dependent on its helicase domain. We propose that Uls1 directs a subset of DNA structures arising during replication into the Sgs1-dependent pathway facilitating S phase progression. Thus, in the absence of Uls1 other modes of replication fork processing and repair are employed.
INTRODUCTION
Replication forks often stall at specific sites in the genome, e.g. rDNA repeats or DNA lesions resulting from chemical or radiation damage. Homologous recombination repair (HRR) is a conserved process responsible for maintenance of genome stability. Products of genes belonging to the RAD52 epistasis group act in the repair of single-strand DNA (ssDNA) gaps or double-strand DNA breaks (DSBs) aiding in the restart of damaged or collapsed replication forks (1). However, the repair of such lesions must be tightly regulated because inappropriate, excessive or untimely recombination can lead to deleterious effects such as loss of heterozygosity or chromosome deletions and rearrangements (2). In Saccharomyces cerevisiae several proteins have been described as being implicated in the processing of stalled replication forks and control of recombination.
Three helicases were shown to control HRR: Srs2 and Sgs1, two well established helicases with anti-recombinogenic properties (3,4), and recently described Mph1 involved in the dissociation of D-loops formed by Rad51 recombinase (5). srs2Δ mutants display hyper-recombination phenotype (6) corroborated by later biochemical data showing that Srs2 protein disrupts Rad51 filament (7). It has been shown that Srs2 acts through interaction with the PCNA complex, a sliding clamp and processivity factor for replicative DNA polymerases, and directs the repair of stalled replication forks away from HRR and into post-replication repair (PRR)-dependent translesion synthesis (TLS) and template switching pathways (4,8). The sgs1Δ mutant is also characterized by mitotic hyper-recombination phenotype (9), sensitivity to genotoxins (10) and reduced replicative lifespan (11). These phenotypes can be rescued by overexpression of human BLM gene (12,13) underscoring conservation of function among members of RecQ helicase family. Srs2 and Sgs1, however, are not redundant, even though suppression of srs2Δ sgs1Δ lethality by deletion of RAD51 suggests partial functional overlap (14). Sgs1 overexpression can complement hyper-recombination and repair defects of srs2Δ mutant (15) but not vice versa, and it is suggested (16) that both helicases act preferentially at different stages or on different intermediates in HRR. Many functions performed by Sgs1 in DNA metabolism have recently been reviewed (17) and the complexity of phenotypes observed for sgs1Δ mutant is further elevated by the fact that Sgs1 can function alone as well as in complex with its interacting partners: DNA topoisomerase III, Top3 (18) and a stimulator of Top3 decatenation activity, Rmi1 (19,20). In DSB repair Sgs1 can be involved both upstream in the resection step of DNA ends (21,22) as well as downstream, with Top3–Rmi1, in the dissolution of double Holliday junctions (HJ) (23). Sgs1 colocalizes with DNA replication sites even in the absence of damage and is involved in the activation of intra-S checkpoint in response to hydroxyurea (HU) induced replication fork stalling. It stimulates checkpoint kinase, Rad53, activation independently of Top3, acting together with the S phase checkpoint mediator, Mrc1, in a pathway synergistic to a clamp loader subunit, Rad24 (10,24). Another important function of Sgs1 at replication forks is the regulation of recombination. Sgs1 slows the progression of replication forks and prevents deleterious HRR, especially in regions rich in natural pause sites, such as an rDNA array (25). In sgs1Δ mutant Rad51-dependent X molecules, containing HJ, accumulate at MMS damaged replication forks (26,27) since Sgs1–Top3–Rmi1 complex required for their resolution is compromised.
MUS81 and MMS4 have been isolated in a screen for genes required for viability in the absence of Sgs1 (28) and mutants in both were found to be defective in sporulation and sensitive to agents causing replication fork stalling and collapse. Together they encode a heterodimeric structure-specific endonuclease that cleaves branched DNA (29), preferably Y-shaped structures, D-loops and nicked HJ (30). This nuclease activity is enhanced by DNA-dependent ATPase, Rad54, which targets Mus81–Mms4 to substrates at perturbed replication forks (31). In summary, these biochemical data suggest that Mus81–Mms4 could cleave stalled or regressed forks leading to their collapse, but also process structures arising as a result of HRR action at arrested forks (29,31,32), consistent with a role both upstream and downstream in the restart of damaged replication forks. The synthetic lethality of sgs1Δ mus81Δ double mutant can be suppressed by deletion of HR genes suggesting that both proteins may act in non-redundant but overlapping pathways for the removal of toxic recombination intermediates. However, the generation times of such triple mutants are significantly higher than those of respective rad mutants, implying that Mus81 and Sgs1 also have roles that are independent of recombination (33).
Both Sgs1 and Mus81–Mms4 are required for the suppression of gross chromosomal rearrangements (GCR) (3,34). Recently, it has been shown that deletion of genes for SLX5 and SLX8 originally isolated by Mullen et al. (28), encoding a SUMO-targeted ubiquitin ligase (STUbL) complex (35,36), also resulted in even more substantial increase in GCR rate (37), implicating both proteins in the preservation of genomic stability. In agreement with this notion, it has been reported that Slx5 co-localizes with DNA damage-induced Rad52 foci and is recruited to DSB induced by HO endonuclease (38). The Slx5–Slx8 complex is also involved in the control of DSB repair at nuclear pores (39).
Uls1 (Dis1–Ris1–Tid4), the second putative STUbL in S. cerevisiae (35), which belongs to the Swi2–Snf2 family of DNA-dependent ATPases, has been shown to antagonize silencing during mating-type switching (40). Although mutation of ULS1 causes accumulation of high molecular weight SUMO conjugates and double uls1Δ slx5Δ mutant shows synthetic growth defect (35), there is no biochemical evidence for a role of Uls1 in SUMO-dependent ubiquitination and/or preservation of genomic stability. Recently it has been shown that Uls1 acts together with Rad54 and Rdh54 to remove Rad51 recombinase from chromatin and that its translocase activity is required for this process (41). Interestingly, two protein paralogs from Schizosaccharomyces pombe, Rrp1 and Rrp2, showing 34 and 36% similarity, respectively, to the C-terminal portion of the S. cerevisiae Uls1, have been found to function in the Sfr1–Swi5 mediator complex-dependent branch of HR, described in S. pombe but conserved in mice and humans (42–44), and play a particularly important role in the rescue of stalled and/or collapsed replication forks in the absence of Rhp57 (Rad57 homolog of S. cerevisiae) (45). Based on these results, we sought to determine if Uls1 also shares a role in these processes.
In this study, we found that although the uls1Δ mutant does not exhibit sensitivity to genotoxic agents, its progression through S phase and resolution of chromosome replication intermediates is compromised when challenged with MMS. Uls1 is thus crucial for coping with replication stress which is especially evident in cells devoid of HR mediator Rad52 or endonuclease Mus81. In addition, we showed that deletion of ULS1 results in suppression of sgs1Δ phenotypes suggesting that Uls1 acts upstream in the Sgs1-dependent pathway to maintain genomic stability.
MATERIALS AND METHODS
Yeast strains, growth conditions and plasmids
Yeast strains used in this study are in the W303 background with a wild-type copy of RAD5 (Supplementary Table S1). Gene deletions were generated by PCR-based gene replacement method (46). Yeast transformations were done by the lithium acetate procedure (47). Yeast strains were grown in standard rich (YPD) medium or in selective synthetic minimal (SD) medium at 28°C (48). Doubling time calculations were carried out as previously described (49). For DNA damage sensitivity tests, cells were grown to mid-log phase and 10-fold serial dilutions were spotted onto YPD plates containing various concentrations of HU (Calbiochem), methyl methane sulfonate (MMS, Sigma-Aldrich) or camptothecin (CPT, Sigma-Aldrich). Plates were incubated at 28°C for 2–3 days and photographed. DNA damage sensitivity assays were repeated a minimum of three times. Cloning of the ULS1 gene on a centromeric plasmid (pGURA3_ULS1) was performed by the gap-repair procedure using W303-1A as a host strain and the split-marker vectors, pGRU and pGRA, as described elsewhere (50). Site-directed mutagenesis of ULS1 was conducted with QuickChange® kit (Stratagene) and confirmed by DNA sequencing.
Cell-cycle analysis, pulsed-field gel electrophoresis and microscopy
Cell-cycle synchronization and flow cytometry analysis of DNA content were performed as previously described (51). The fraction of cells remaining arrested in G1 was determined by an α-factor–nocodazole trap assay (51). The pulsed-field gel electrophoresis (PFGE) analysis of yeast chromosomes was performed as previously described (52). To analyze morphology of yeast strains, cells were fixed in 70% ethanol, treated with RNAse, followed by staining with Sytox green (Invitrogen) to visualize nuclei and observed with an Axio Imager M1 upright wide-field fluorescence microscope (Carl Zeiss, Germany) equipped with a 100× oil immersion objective (Zeiss Plan-Neofluar 100×/1.30), a GFP filter set and Nomarski interference contrast. Images were collected using a Zeiss AxioCam MRc digital color camera and processed with Zeiss AxioVision 4.5 software. Microscopy experiments were repeated three times with a minimum of 600 cells counted for each strain.
Protein analysis
Total protein was extracted by the trichloroacetic acid method as described previously (53). Protein extracts were resolved on 10% SDS–PAGE, blotted onto nitrocellulose membranes (Bio-Rad) and probed with the goat polyclonal anti-Rad53 (Santa Cruz Biotechnology, sc-6749) or the rabbit polyclonal anti-PCNA (kindly provided by Bruce W. Stillman, Cold Spring Harbor Laboratory) antibodies. Blotted membranes were stained for total protein with Ponceau S (Sigma-Aldrich) before immunodetection.
Recombination assays
The loss of the ADE2–CAN1 marker genes as a result of recombination between rDNA repeats was measured by scoring for canavanine (Sigma-Aldrich) resistance according to Burgess et al. (54). The recombination frequency between the δ repeats at the SUP4-o locus was determined by detecting the loss of URA3 marker gene on plates containing 5-flouroorotic acid (5-FOA, Zymo Research), as described elsewhere (55). Recombination experiments were repeated six times and results were subjected to t-test analysis.
RESULTS
Uls1 shows synthetic growth interaction with Rad52
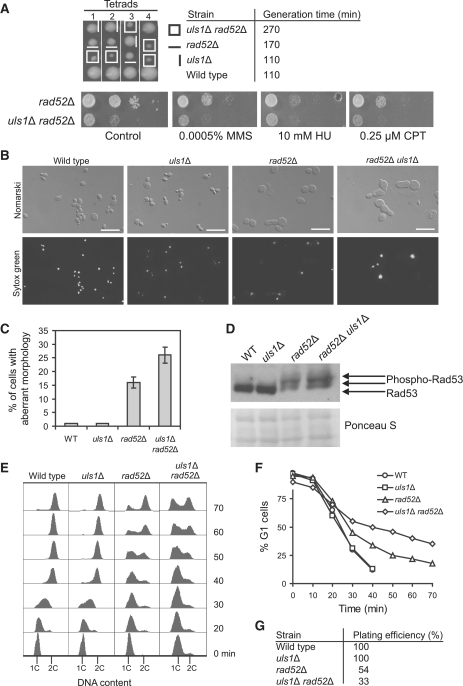
We have recently reported that two S. pombe protein paralogs, Rrp1 and Rrp2, have a role in the rescue of stalled and/or collapsed replication forks in the Sfr1–Swi5-dependent branch of HR, which acts in parallel to the second HR mediator complex Rhp55–Rhp57 (Rad55–Rad57 in S. cerevisiae) (45). Rrp1 and Rrp2 show significant similarity to the C-terminal portion of the S. cerevisiae Uls1 (45), a member of Swi2–Snf2 DNA-dependent ATPase family (40) and putative STUbL (35) (Supplementary Figure S1). In order to determine if Uls1 also has a role in replication-associated DNA damage response, we deleted the ULS1 gene in the W303 background and a growth of the resulting mutant was studied in the presence of DNA damaging agents. Similarly to single Δrrp1 and Δrrp2 mutants, the uls1Δ mutant had wild-type sensitivities to HU, MMS and CPT, agents that stall or collapse replication forks (Supplementary Figure S2). However, our S. pombe study demonstrated that both rrp1+ and rrp2+ genes exhibit genetic interactions with the HR mediator gene rhp57+; the double Δrrp1 Δrhp57 and Δrrp2 Δrhp57 mutants were more sensitive to HU, MMS and CPT than the single Δrhp57 mutant (45). On the other hand, both genes were epistatic to the recombinase gene, rhp51+ (RAD51 in S. cerevisiae). Thus, next we analyzed the sensitivity of double uls1Δ rad51Δ and uls1Δ rad57Δ mutants to DNA damaging agents. Double mutants were not more sensitive to HU, MMS and CPT than single rad51Δ and rad57Δ mutants, suggesting either a role of Uls1 in the Rad51-dependent HRR pathway or a function unrelated to DNA repair by HR (Supplementary Figure S2). We also examined the phenotype of uls1Δ in a rad52Δ background lacking the main HR mediator involved in multiple pathways of DSB repair, including Rad51-independent modes of DNA repair, as well as in a rad59Δ background devoid of single-strand annealing repair pathway (1). Deletion of ULS1 in the rad59Δ mutant did not change its level of sensitivity to DNA damaging agents (Supplementary Figure S2). Interestingly, we found that the double uls1Δ rad52Δ mutant showed a severe slow-growth phenotype with generation time of 270 min, significantly >170 min for rad52Δ and 110 min for uls1Δ and wild-type (Figure 1A). In contrast, deletion of ULS1 in rad52Δ did not affect the strain's sensitivity to DNA damaging agents.
Figure 1.
Synthetic growth defect of uls1Δ rad52Δ cells. (A) The uls1Δ rad52Δ double mutant exhibits severe slow growth phenotype as determined by tetrad analysis, measurements of doubling time and serial dilutions of indicated strains plated on YPD plates in the absence or presence of DNA-damaging agents. (B and C) The rad52Δ and uls1Δ rad52Δ strains accumulate in G2–M phase with aberrant morphology (enlarged cells with elongated, tubular buds). Asynchronously growing log-phase cells of indicated strains were fixed and stained with Sytox Green to visualize nuclei followed by microscopic observation to score the number of cells in each cell-cycle phase and characterize cell morphology. (D) Level of Rad53 phosphorylation as a marker of DNA damage checkpoint activation in the indicated strains. Rad53 was analyzed by western blotting with anti-Rad53 antibodies. Slow-migrating forms of Rad53 in rad52Δ and uls1Δ rad52Δ represent hyperphosphorylated Rad53. (E) Flow cytometry analysis reveals severely prolonged S phase in uls1Δ rad52Δ cells. Cells were synchronized in G1 by α-factor and released into fresh YPD media at 28°C to monitor S phase progression by flow cytometry. The positions of G1 (1C) and G2–M (2C) peaks are indicated below. (F) Percentage of cells remaining in G1 after release from α-factor synchronization was determined by an α-factor–nocodazole trap assay. (G) To determine number of cells able to form colonies, G1-synchronized cells used for analysis by an α-factor–nocodazole trap assay (Figure 1F) were diluted and approximately 100 cells were plated on YPD medium.
To further examine the synthetic growth interaction between Rad52 and Uls1, we compared cellular morphology in asynchronously growing wild-type, rad52Δ, uls1Δ, and uls1Δ rad52Δ strains. Wild-type and uls1Δ cells showed distribution of G1 (unbudded), S (small-budded) and G2–M (large-budded) phase cells typical for log-phase cultures. rad52Δ and uls1Δ rad52Δ mutants, however, accumulated in G2–M phase of cell cycle as enlarged cells with buds similar in size to the mother cell and a single DNA mass, indicative of prolonged G2–M cell-cycle arrest and impaired chromosome segregation (Figure 1B). In addition, abnormalities in cell morphology typical for rad52Δ were greatly exacerbated in uls1Δ rad52Δ as 25% of these cells formed elongated, tubular buds, virtually absent in wild-type and uls1Δ mutant (Figure 1B and C). Moreover, deletion of ULS1 in rad52Δ further increased phosphorylation of checkpoint effector kinase Rad53 (Figure 1D), suggesting accumulation of spontaneous DNA damage in these strains.
It has been shown that slowing down replication in S. cerevisiae by sublethal levels of HU or MMS induces pseudofilamentous growth dependent on checkpoint proteins Mec1, Rad53 and Swe1 (56). Thus, severe slow growth phenotype, increase in number of G2–M cells with elongated buds and hyperactivation of Rad53 in uls1Δ rad52Δ suggest that cells of this double mutant experience severe perturbations in replication progression. To test this hypothesis, we synchronized wild-type, uls1Δ, rad52Δ and uls1Δ rad52Δ cells in G1 with α-factor and released them into fresh YPD media to monitor cell-cycle progression by flow cytometry (Figure 1E). Both wild-type and uls1Δ completed S phase and reached 2C DNA content by 40 min, albeit uls1Δ with a slight delay. In contrast, rad52Δ cells replicated more slowly and accumulated in G2–M at 60–70 min after release, while the double uls1Δ rad52Δ mutant had not completed DNA synthesis even by that time. Moreover, deletion of ULS1 in the rad52Δ mutant increased the number of cells failing to enter S phase (Figure 1F), which is in a good agreement with reduced plating efficiency (Figure 1G) and growth (Figure 1A) observed for the double uls1Δ rad52Δ mutant when compared to the single rad52Δ mutant. This suggests that cells lacking Rad52 and Uls1 accumulate DNA damage leading to perturbation of cell-cycle progression and loss of viability. In sum, our results indicate that Uls1 function becomes especially important when cells suffer from replication stress due to the absence of Rad52.
Uls1 is required for S phase progression in the presence of DNA damage
To test the involvement of Uls1 in replication progression through damaged DNA template, we examined replication kinetics in wild-type and uls1Δ cells in the presence of MMS. Both strains were synchronized in G1, released into YPD or YPD with 0.03% MMS and analyzed by flow cytometry at 30-min intervals for DNA content (Figure 2A). Under control conditions the uls1Δ mutant progressed slightly slower through S phase than wild-type. When cells were treated with MMS, both wild-type and uls1Δ replicated slowly but the uls1Δ mutant reproducibly completed DNA synthesis with at least 30-min delay. A defect in DNA synthesis observed in the uls1Δ mutant was accompanied by slightly increased level of phosphorylated Rad53 in MMS-exposed cells indicating impaired processing of stalled replication forks (Figure 2B). These results suggest that Uls1 facilitates DNA replication, especially when DNA template is damaged.
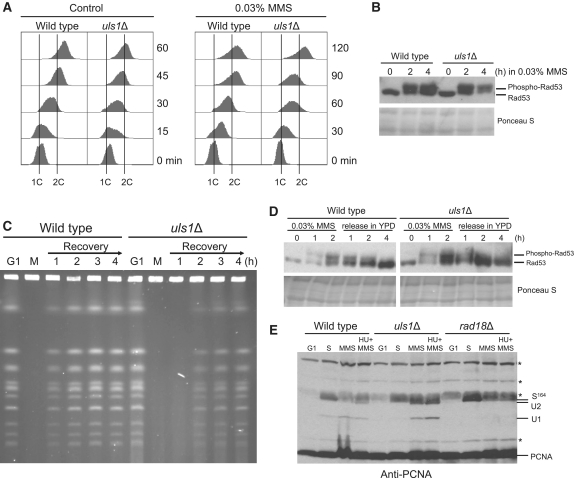
Figure 2.
DNA synthesis defect in the uls1Δ mutant. (A) uls1Δ cells exhibit S phase delay in the presence of MMS. G1-synchronized cells of wild-type and uls1Δ were released in the presence or absence of MMS and DNA content was measured by flow cytometry at the indicated time-points. (B) Rad53 phosphorylation is increased in uls1Δ cells in response to MMS. Protein samples were prepared from G1-synchronized cells before and after release in the presence of MMS at indicated time-points and analyzed by western blotting with anti-Rad53 antibodies. (C) Completion of DNA replication is delayed in uls1Δ after MMS treatment. DNA samples from cells arrested in G1 by α factor (G1), released and treated with 0.03% MMS (M) for 60 min and then recovering in fresh YPD media were subjected to PFGE analysis. (D) DNA damage checkpoint activation does not persist in uls1Δ following MMS treatment. Protein extracts were prepared from the wild-type and uls1Δ cultures treated with 0.03% MMS for 2 h, then washed and allowed to recover in fresh media. (E) PCNA mono- and polyubiquitination is increased in the absence of Uls1. Cells were synchronized in G1, allowed to enter S phase and after 30-min incubation in YPD media exposed to 0.03% MMS or 0.03% MMS plus 0.1 M HU for 2 h. The rad18Δ mutant lacking PCNA ubiquitination was analyzed as a control. Non-specific bands are depicted by asterisks.
To determine whether S phase delay observed in uls1Δ is caused by prolonged persistence of unresolved replication intermediates, wild-type and uls1Δ cells were arrested in G1 by α-factor, released in 0.03% MMS for 1 h and then allowed to recover in MMS-free media to monitor the fate of chromosomes by PFGE (Figure 2C). As expected, the intact chromosomal DNA isolated from G1 cells was separated as individual, well-defined bands, while exposure to MMS resulted in retention of the chromosomal DNA in the loading wells because of incomplete replication and presence of branched structures (57,58). During recovery period wild-type cells quickly resumed replication indicated by re-entry of intact chromosomes into the gel after 1 h from MMS release. Consistently with the flow cytometry data, chromosomes from the uls1Δ mutant were retained in the wells much longer suggesting a delay in resolving stalled replication forks and/or recombination intermediates generated in response to MMS (Figure 2C). After release from MMS no persistent activation of Rad53 was observed in uls1Δ mutant (Figure 2D) implying that in the absence of Uls1 MMS-induced replication intermediates do not lead to the generation of replication-associated DNA damage, which is in a good agreement with the lack of uls1Δ sensitivity to genotoxic agents (Supplementary Figure S2).
In S. cerevisiae, replication of damaged DNA requires the RAD6 pathway, which involves monoubiquitination of proliferating cell nuclear antigen (PCNA) on Lys164 by the Rad6–Rad18 E2–E3 ubiquitin-conjugating complex, which activates error-prone TLS, and/or polyubiquitination by the heterodimeric E2 Ubc13–Mms2 enzyme in concert with the RING finger E3 ubiquitin ligase Rad5, which triggers error-free lesion bypass pathway (59). Because of the putative role of Uls1 as a STUbL (35) belonging to the Swi2–Snf2 family of DNA-dependent ATPases with high similarity to Rad5 (40), we decided to examine post-translation modification status of PCNA in the uls1Δ mutant exposed to MMS in S phase (Figure 2E). We found that deletion of ULS1 causes elevated levels of mono- and polyubiquitinated PCNA when compared to wild-type cells. First, this suggests that Uls1 does not contribute to PCNA ubiquitination and does not channel DNA damage bypass to PRR pathway. Second, increased PCNA ubiquitination may reflect activation of a rescue pathway to cope with elevated replication stress caused by ULS1 deletion and/or may constitute one of alternative DNA damage bypass pathways triggered specifically in the absence of Uls1. As ULS1 is epistatic to RAD18, RAD5 and REV3 (Supplementary Figure S3), we favor the hypothesis that Uls1 has an upstream role in channeling replication fork processing and repair into the RAD6-independent pathway.
Uls1 shows genetic interactions with Mus81 and Sgs1
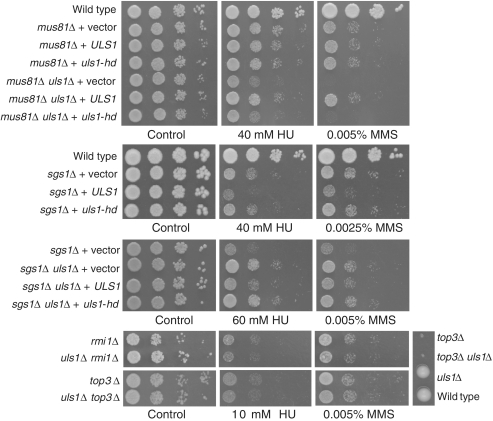
Many proteins are involved in replication fork progression, stabilization and restart, sometimes additionally, positively or negatively regulating HR events during replication. It has been shown that this function is fulfilled, among others, by anti-recombinogenic helicases such as Sgs1 (3) and Srs2 (4). Together with Mus81–Mms4 and Yen1 nucleases (58,60) as well as a third helicase, Mph1 (61), they play a crucial role in stalled replication fork processing. Having established that cells lacking Uls1 display replication defect in the presence of MMS, we decided to identify Uls1-mediated DNA repair pathway by examining the genetic interactions among Mph1, Mus81, Sgs1, Srs2, Yen1 and Uls1 in the presence of HU, MMS and CPT (Figure 3 and Supplementary Figure S4). We found that lack of ULS1 had no effect on the phenotype of mph1Δ, srs2Δ, yen1Δ mutants (Supplementary Figure S4). In contrast, deletion of ULS1 in the mus81Δ background conferred pronounced additional sensitivity to all DNA-damaging agents tested. This synergistic effect was comparable to that described recently for the mus81Δ yen1Δ mutant (58,60), which together with the epistatic relationship between uls1Δ and yen1Δ (Supplementary Figure S3), could point to the role of Uls1 in a Yen1-related pathway. However, we also observed partial rescue of HU and MMS sensitivity of sgs1Δ by concomitant deletion of ULS1, which has not been observed in sgs1Δ yen1Δ (60). Interestingly, this suppression was specific to Sgs1 function not shared with Rmi1–Top3 complex as it was not observed in the double uls1Δ top3Δ and uls1Δ rmi1Δ mutants (Figure 3).
Figure 3.
Deletion of ULS1 suppresses DNA damage sensitivity of sgs1Δ but not rmi1Δ–top3Δ and shows synergistic effect with mus81Δ for decreased resistance to DNA damaging agents. Mutants were transformed with the centromeric plasmid bearing the wild-type ULS1 gene (pGURA3_ULS1) or the mutated version uls1-D1108A,E1109A (pGURA3_uls1-hd) encoding the catalytic inactive Uls1 (uls1-hd). Growth test in the presence of HU, MMS and CPT was performed as in Figure 1.
Next we checked whether the observed increase of sensitivity in the uls1Δ mus81Δ double mutant and the suppression of DNA damage sensitivity in the uls1Δ sgs1Δ double mutant were indeed the result of deleting ULS1. We cloned the ULS1 gene from genomic DNA by a plasmid rescue as described in Materials and Methods, and used it to transform single mus81Δ and double uls1Δ mus81Δ mutant strains. We showed that acquisition of wild-type copy of ULS1 restored the sensitivity to HU, MMS and CPT of the double mutant to the level of the single mus81Δ (Figure 3). Similarly, we found that introduction of wild-type copy of ULS1 into the uls1Δ sgs1Δ double mutant increased its sensitivity to HU and MMS to the level of single sgs1Δ, confirming the role of ULS1 deletion in the suppression of sgs1Δ sensitivity to replication inhibitors (Figure 3). Interestingly, the presence of extra copies of ULS1 in the single sgs1Δ mutant further increased its sensitivity to HU and MMS (Figure 3). Both observations suggest that Uls1 may act upstream in the Sgs1-dependent DNA repair pathway during replication stress.
To determine whether the lack of ATP-dependent DNA helicase activity of Uls1 is responsible for the phenotypes observed in the uls1Δ mus81Δ and uls1Δ sgs1Δ double mutants, we generated two point mutations in the conserved ATPase motif located in the N-terminal part of Uls1 helicase domain (uls1-D1108A,E1109A or uls1-hd) to obtain helicase–ATPase deficient protein (62) (Supplementary Figure S1). Expression of uls1-hd mutant neither complemented the increased sensitivity of uls1Δ mus81Δ to DNA damaging agents nor reversed suppression of DNA damage sensitivity in uls1Δ sgs1Δ (Figure 3). Moreover, expression of uls1-hd was not toxic in sgs1Δ in the presence of HU and MMS. This strongly suggests that DNA translocase activity of Uls1 is required for regulation of replication-associated DNA repair.
Cells lacking both Uls1 and Mus81 exhibit prolonged G2–M delay
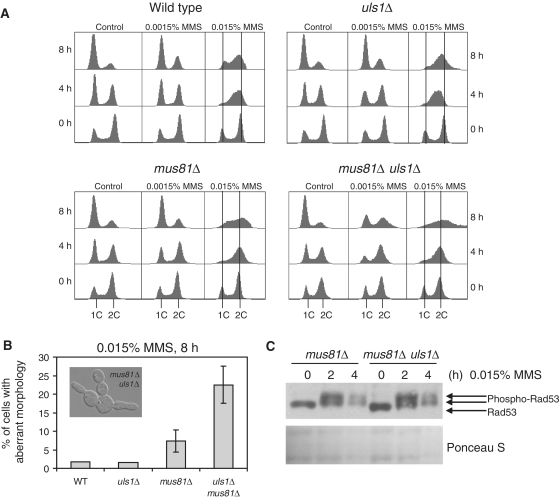
To investigate in greater detail the interactions of Uls1 with Mus81, we monitored the effect of MMS on the cell-cycle progression of wild-type and uls1Δ, mus81Δ and uls1Δ mus81Δ mutants (Figure 4A). Asynchronous cultures were used, since it has been shown that G1-arrested mus81Δ cells released into various concentrations of MMS could not be distinguished from wild-type (60). In the presence of low concentration of MMS (0.0015%), wild-type and single mutants showed no perturbation of cell-cycle progression, while the uls1Δ mus81Δ double mutant exhibited G2–M arrest (Figure 4A). When 0.015% MMS was added, wild-type cells first accumulated in G2–M phase but were able to adapt and resume cycling. The uls1Δ mutant progressed much slower than wild-type through DNA replication in the presence of high concentration of MMS, confirming our data obtained for cultures synchronized in G1 (Figure 2). In contrast, both mus81Δ and uls1Δ mus81Δ strains did not exhibit this slowing phenotype but accumulated with a 2 C DNA content during 4-h incubation in MMS. However, after 8 h in MMS mus81Δ restarted cell cycle, while uls1Δ mus81Δ showed a broadened peak of 2C DNA content, suggesting cell segregation defects and aberrant mitosis due to the persistence of unresolved recombination intermediates formed in S phase (Figure 4A) (63). Consistently, in the G2–M arrested population of uls1Δ mus81Δ we observed a 3-fold increase in the number of cells with morphology defects as compared to the mus81Δ single mutant (Figure 4B). Apart from enlarged cells with big buds and unsegregated DNA characteristic for mus81Δ and mus81Δ yen1Δ (60), many cells with multiple, elongated buds or filament-like projections were present (Figure 4B). The level of Rad53 phosphorylation induced by the incubation in 0.015% MMS was comparable in all four studied strains, indicating that the observed increase of sensitivity of uls1Δ mus81Δ did not result from a checkpoint defect (Figure 4C). We conclude that Uls1 is involved in an alternative Mus81-independent pathway for repair of replication-associated DNA damage.
Figure 4.
The mus81Δ uls1Δ double mutant arrests at G2–M with aberrant cell morphology during MMS treatment. (A) The mus81Δ uls1Δ double mutant accumulates with the G2–M DNA content after 8-h incubation in MMS. (B) The mus81Δ uls1Δ strain exhibits 3-fold increase of cells with morphology defects in the presence of MMS. (C) Western blotting of Rad53 reveals intact checkpoint activation in tested strains after MMS treatment. Asynchronous cells of indicated strains were exposed to MMS and analyzed by flow cytometry to measure DNA content, by microscopy to score the number of cells with altered morphology and used for protein extraction to detect Rad53.
Deletion of ULS1 suppresses several defects of sgs1Δ
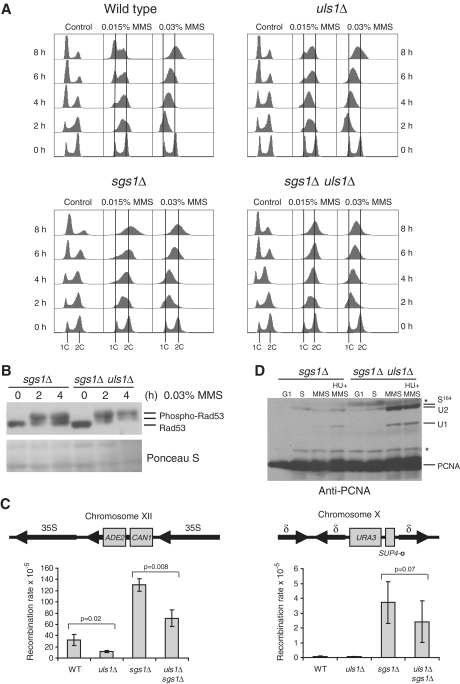
Next we investigated the mechanism of DNA damage sensitivity suppression conferred by ULS1 deletion in the sgs1Δ background suggesting the role of Uls1 in the Sgs1-dependent DNA damage bypass. Since uls1Δ cells exhibited a significant delay of S phase progression in the presence of MMS (Figure 2A) and in the absence of Rad52 (Figure 1E), we compared the effect of MMS on the cell-cycle progression of asynchronous cultures of sgs1Δ and double uls1Δ sgs1Δ mutants (Figure 5A). In the presence of 0.015% MMS, we did not see any slowing defect in the sgs1Δ mutant, only persisting G2–M arrest, resembling that of uls1Δ mus81Δ strain (compare Figure 4A). The uls1Δ sgs1Δ double mutant also accumulated with a 2C DNA content at 8 h in 0.015% MMS but in a much sharper peak, typical for G2–M checkpoint arrest suggesting decrease of unsolved recombination intermediates. Interestingly, in the presence of higher concentration of MMS (0.03%), S phase progression was significantly slower in the uls1Δ sgs1Δ double mutant compared to the single sgs1Δ (Figure 5A). This was accompanied by increased Rad53 phosphorylation induced by 0.03% MMS in uls1Δ sgs1Δ compared to sgs1Δ suggesting the involvement of checkpoint pathway in the observed phenomena (Figure 5B).
Figure 5.
The uls1Δ mutation attenuates sgs1Δ defects in DNA damage response. (A) Flow cytometry analysis of DNA content in asynchronous cells treated with MMS revealed accumulation of sgs1Δ uls1Δ in S phase. (B) Deletion of ULS1 upregulates checkpoint activation in sgs1Δ. Analysis of Rad53 phosphorylation in response to MMS exposure was performed as described in Figure 2. (C) Mutation of ULS1 decreases recombination rate in both wild-type and sgs1Δ. Recombination rates were calculated by measuring either the number of cells resistant to FOA and thus exhibiting the loss of URA3 marker from the SUP4-o locus or the number of cells resistant to canavanine and forming red colonies on YPD plates as a result of recombination between rDNA repeats leading to the simultaneous loss of ADE2 and CAN1 marker genes. All experiments were repeated six times with three independent clones each time. Standard deviations are indicated. (D) High increase of PCNA mono- and polyubiquitination in sgs1Δ after deletion of ULS1. Analysis of PCNA modifications was conducted as described in Figure 2.
Increased mitotic recombination rate is one of major phenotypes of sgs1Δ mutant so we sought to determine if deletion of ULS1 had any effect on this trait. We measured the rate of recombination by monitoring loss of the ADE2 and CAN1 marker genes in the rDNA array and the URA3 gene located at the SUP4-o locus surrounded by several δ repeats (Figure 5C). As previously found (49), deletion of SGS1 conferred high level of recombination at both sites. Interestingly, frequency of recombination was 2-fold decreased in uls1Δ sgs1Δ and uls1Δ strains at the rRNA locus. The effect at the SUP4-o locus was not statistically significant due to high variability of data between experiments in our hands but we reproducibly observed decrease in recombination frequency in uls1Δ sgs1Δ cells compared to the sgs1Δ single mutant. Since hyperrecombination phenotype of sgs1Δ results from DNA replication-associated defects this strengthens our conclusion that Uls1 is involved in the Sgs1 pathway for dealing with replication stress. We should emphasize, however, that even though deletion of ULS1 can rescue many of sgs1Δ mutant phenotypes, however the suppression does not reach the wild-type level, so there are other aspects of Sgs1 activity which are clearly Uls1-independent.
Having established that in the uls1Δ mutant the level of ubiquitinated PCNA is significantly increased, we hypothesize that channeling of replication-associated DNA damage repair into PRR pathway in the absence of Uls1 may be responsible for the suppression of sgs1Δ sensitivity to HU and MMS. Accordingly, the analysis of PCNA modifications revealed that the sgs1Δ mutant exhibited low level of ubiquitinated PCNA in the presence of MMS, while the uls1Δ sgs1Δ double mutant was characterized by accumulation of mono- and polyubiquitinated forms of PCNA (Figure 5D) suggesting that indeed in these cells repair of replication-associated DNA damage is channeled into PRR. We conclude that this, together with increased checkpoint activation in the double mutant providing additional time to cope with DNA damage, contributes to the observed rescue of sgs1Δ DNA damage sensitivity by concomitant ULS1 deletion.
DISCUSSION
Uls1 belongs to the Swi2–Snf2 family of DNA-dependent ATPases whose other members like the RING-finger E3 ubiquitin ligase Rad5 (59), to which it shows a high degree of homology (40), or the chromatin remodeling complex Ino80 (64) are implicated in DNA repair and replication fork processing, so we sought to determine if Uls1 fulfills a similar role. In this study, we found that the strain deleted for ULS1 does not show any sensitivity to DNA damaging agents suggesting that Uls1 has no role in DNA damage repair. However, the mutant proceeds slower than wild-type through S phase and exhibits delayed completion of DNA replication after MMS treatment as seen on the PFGE gels. Interestingly, deletion of ULS1 leads to synthetic growth defect and dramatic elongation of S phase in rad52Δ background under standard conditions. uls1Δ rad52Δ mutant accumulated in G2–M phase of cell cycle with increased number of cells exhibiting aberrant morphology and flow cytometry profiles, indicative of the defects in S phase progression and chromosome segregation. Slow growth of rad52Δ mutant results from accumulation of ssDNA lesions and failure to repair spontaneous DSB arising infrequently in wild-type during replication (33). Together these results strongly point to the role of Uls1 protein in dealing with replication stress.
We next examined the effect of deleting ULS1 in the background of several genes involved in replication fork metabolism and found no synergism with mph1Δ, srs2Δ, rad5Δ and yen1Δ, but demonstrated interesting genetic interactions with mus81Δ and sgs1Δ, both dependent on Uls1 ATPase activity as the uls1-hd strain behaved like the uls1Δ mutant. The uls1Δ mus81Δ double mutant was characterized by highly elevated sensitivity to MMS, HU and CPT, as well as a more pronounced G2–M arrest with a 3-fold increase in the number of cells with morphology defects as compared to the mus81Δ single mutant, suggesting that Uls1 and Mus81 endonuclease act in complementary pathways. The RecQ helicase Sgs1 also works in parallel to Mus81 in DNA replication and repair, and interestingly we observed partial suppression of HU and MMS sensitivity of sgs1Δ mutant by simultaneous deletion of ULS1. Surprisingly, this suppression was specific to Sgs1 function not shared with Rmi1–Top3 complex, as it was not visible in the double uls1Δ rmi1Δ nor in the uls1Δ top3Δ mutant. Deletion of ULS1 in the sgs1Δ background not only rescues its sensitivity to HU and MMS but also considerably slows down S phase progression under MMS treatment as compared to the single sgs1Δ mutant, probably due to enhanced Rad53 phosphorylation observed in uls1Δ sgs1Δ. Recently it has been shown that in S. pombe, apart from canonical checkpoint genes, three additional genes, mus81+, rqh1+ (SGS1) and sfr1+, were necessary to slow replication in the presence of MMS (65) but this ability was not correlated with MMS sensitivity of tested strains, the latter resulting rather from DNA damage repair defects in G2. Failure to slow replication may instead contribute to increased genomic instability as is seen in human patients lacking S phase DNA damage checkpoint (66), a phenotype also observed in both mus81Δ and sgs1Δ mutants. We found that deletion of ULS1 partially complemented the hyperrecombination in sgs1Δ mutant at rRNA locus with only mild albeit consistent effect at SUP4-o indicating that slowing of S phase progression might have resulted in a decrease in genomic instability in the double mutant.
A model has been proposed by Fabre et al. (33) where, apart from its role in dissolution of recombination intermediates with Top3–Rmi1, Sgs1 would aid in replication fork progression through obstacles and regulate the restart of stalled forks by non-recombinogenic mechanisms to prevent initiation of HR. The function of Sgs1 with Top3 and Rmi1 in HR repair is separable from processing DNA replication intermediates and can be uncoupled by deleting a single aspartic acid residue (49). Also, even though the synthetic lethality of sgs1Δ mus81Δ double mutant can be suppressed by deletion of HR genes, suggesting that both proteins may act in non-redundant but overlapping pathways for the removal of toxic recombination intermediates, the generation times of such triple mutants are significantly higher than those of respective rad mutants. This would imply that Mus81 and/or Sgs1 also have roles that are independent of recombination (33). As described above uls1Δ rad52Δ mutant exhibits synthetic growth defect and the generation time of sgs1Δ rad52Δ double mutant is also increased relative to the single rad52Δ mutant (67–69), reinforcing the notion that Sgs1 and Uls1 might participate in the upstream events at replication forks. It has been shown that human BLM helicase by itself can mediate replication fork regression (70), dissociate D-loops (71) and promote disruption of inactive hRad51 filament (72) thus preventing the formation of toxic recombination intermediates and channeling replication associated structures away from HR. Given the conservation of function among RecQ helicases, Sgs1 may possess similar biochemical activities facilitated by Uls1. Alternatively, Sgs1 might limit early Rad51-dependent recombination at the fork and thus suppress genome rearrangements, but have no role in Rad51-independent, but Rad52-dependent processes of fork restart, as has recently been shown for S. pombe Sgs1 homolog, Rqh1 (73). This activity would involve disassembling D-loops or Rad51 filaments but not dissolution of HJ.
Uls1 can thus act upstream of Sgs1, for example providing chromatin environment facilitating its activity related to regulation of replication fork progression, not Top3–Rmi1-dependent dissolution of recombination intermediates. In its absence other factors might take over and divert replication fork restart into Sgs1-independent pathways, such as error-free PRR, Mus81 or Esc2–Mph1 related HR repair (63), thus alleviating HU and MMS sensitivity and hyperrecombination phenotype of the sgs1Δ mutant strain. Interestingly, deletion of ULS1 not only results in increased levels of PCNA polyubiquitination in a single mutant but also in the double uls1Δ sgs1Δ mutant, indicative of the channeling of replication associated lesions into PRR pathway, which could possibly contribute to the observed rescue of sgs1Δ phenotype described in this paper. It would be of great interest to determine in the future what are the roles of the chromatin remodeling and SUMO-dependent ubiquitin ligase activities in the phenotypes observed upon ULS1 deletion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Higher Education, Warsaw, Poland (grant N302158737); partially by statutory funds (1014/5/IBR/2011). Funding for open access charge: Ministry of Science and Higher Education, Warsaw, Poland (grant N302158737).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Serge Gangloff, Rodney Rothstein and Bruce W. Stillman for sharing yeast strains and reagents, Jerzy Piątkowski for the help with tetrad dissection, Jadwiga Jabłońska for the assistance with recombination assays, and Johanne M. Murray and Antony M. Carr for helpful discussions. RW thanks Grzegorz Zapotoczny for the initial work in this project.
REFERENCES
- 1.Krogh BS, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. Rev. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 3.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 4.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 5.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 8.Schiestl RH, Prakash S, Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 12.Yamagata K, Kato J, Shimamoto A, Goto M, Furuich Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo SJ, Tatebayashi K, Ohsugi I, Shimamoto A, Furuichi Y, Ikeda H. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–625. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 14.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 15.Mankouri HW, Craig TJ, Morgan A. SGS1 is a multicopy suppressor of srs2: functional overlap between DNA helicases. Nucleic Acids Res. 2002;30:1103–1113. doi: 10.1093/nar/30.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branzei D, Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 2007;21:3019–3026. doi: 10.1101/gad.1624707. [DOI] [PubMed] [Google Scholar]
- 17.Ashton TM, Hickson ID. Yeast as a model system to study RecQ helicase function. DNA Repair. 2010;9:303–314. doi: 10.1016/j.dnarep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc. Natl Acad. Sci. USA. 2011;108:4944–4949. doi: 10.1073/pnas.1014240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Matulova P, Marini V, Burgess RC, Sisakova A, Kwon Y, Rothstein R, Sung P, Krejci L. Cooperativity of Mus81/Mms4 with Rad54 in the resolution of recombination and replication intermediates. J. Biol. Chem. 2009;284:7733–7745. doi: 10.1074/jbc.M806192200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 36.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair. 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Cook CE, Hochstrasser M, Kerscher O. The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle. 2009;8:1080–1089. doi: 10.4161/cc.8.7.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Buchman AR. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell. 2010;39:862–872. doi: 10.1016/j.molcel.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akamatsu Y, Jasin M. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan J, Chen J. The role of the human SWI5-MEI5 complex in homologous recombination repair. J. Biol. Chem. 2011;286:9888–9893. doi: 10.1074/jbc.M110.207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dziadkowiec D, Petters E, Dyjankiewicz A, Karpiński P, Garcia V, Watson A, Carr AM. The role of novel genes rrp1+ and rrp2+ in the repair of DNA damage in Schizosaccharomyces pombe. DNA Repair. 2009;8:627–636. doi: 10.1016/j.dnarep.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 48.Kaiser C, Michaelis S, Mitchel A. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press. NY: Cold Spring Harbor; 1994. Methods in yeast genetics. [Google Scholar]
- 49.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast. 1996;12:1439–1457. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1439::AID-YEA37%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 51.Wysocki R, Javaheri A, Allard S, Sha F, Côté J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. NY: Cold Spring Harbor; 2001. Cold Spring Harbor Laboratory Press. [Google Scholar]
- 53.Delaunay A, Isnard AD, Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang YW, Kang CM. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol. Biol. Cell. 2003;14:5116–5124. doi: 10.1091/mbc.E03-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 58.Ho CK, Mazón G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair. 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair. 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Schürer KA, Rudolph C, Ulrich HD, Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mankouri HW, Ngo HP, Hickson ID. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:1683–1694. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Willis N, Rhind N. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol. Biol. Cell. 2009;20:819–833. doi: 10.1091/mbc.E08-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- 67.Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 69.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 71.van Brabant AJ, Ye T, Sanz M, German JL, III, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 72.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Fréon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.