Abstract
Daily mRNA oscillations of circadian clock genes largely depend on transcriptional regulation. However, several lines of evidence highlight the critical role of post-transcriptional regulation in the oscillations of circadian mRNA oscillations. Clearly, variations in the mRNA decay rate lead to changes in the cycling profiles. However, the mechanisms controlling the mRNA stability of clock genes are not fully understood. Here we demonstrate that the turnover rate of mouse Period3 (mPer3) mRNA is dramatically changed in a circadian phase-dependent manner. Furthermore, the circadian regulation of mPer3 mRNA stability requires the cooperative function of 5′- and 3′-untranslated regions (UTRs). Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) binds to both 5′- and 3′-UTR and triggers enhancement of translation and acceleration of mRNA decay. We propose the phase-dependent translation coupled mRNA decay mediated by hnRNP Q as a new regulatory mechanism of the rhythmically regulated decay of mPer3 mRNA.
INTRODUCTION
Circadian rhythms are daily physiological and behavioral oscillations observed in a variety of organisms. Circadian oscillations are driven by self-sustained time-keeping systems which are the intracellular clocks (1,2). These intracellular clocks consist of interacting positive and negative transcriptional and translational feedback loops of the clock genes (3–6). Daily oscillations in protein and/or mRNA levels are central features of the circadian genes (2,6). As for the underlying mechanism of mRNA cycling, a number of studies have shown that the oscillations of circadian genes are controlled at the transcriptional level (4,7–11). In Drosophila, nevertheless, it has been suggested that post-transcriptional regulations also contribute to the mRNA cycling (12–15). Furthermore, we previously demonstrated that 3′-untranslated region (UTR)-mediated mRNA decay played an essential role in mPer3 mRNA cycling, providing direct evidence for the post-transcriptional control of circadian mRNA oscillation (16).
As the quantity of mRNA is ultimately reflected to the amount of translated protein, the regulation of mRNA half-life is considered to be an important control point in gene expression. During the past decades, a large number of studies have identified cis-acting elements and trans-acting factors involved in the regulation of mRNA stability (17–21), and showed that the turnover rates of mRNAs could be regulated by internal or external stimuli, such as cytokines, hormones, hypoxia, or viral infection (17,22–24). Recently, it was reported that the circadian clock regulates the stability of CCR-LIKE(CCL) mRNA, one of the clock-controlled genes of Arabidopsis (25).
Variations in mRNA stability clearly lead to changes in the mRNA cycling profiles of circadian genes (16), however, little is known about the mechanisms controlling the half-lives of mRNA. To investigate the underlying mechanisms, we established several lines of stably transformed NIH3T3 cells that express luciferase (Luc) mRNA with the UTRs (5′-UTR or/and 3′-UTR) of mPer3 mRNA. Since many studies have shown that fibroblast cell lines, such as NIH3T3 and Rat-1, also contain an intrinsic circadian clock system, these cells have been used as appropriate experimental models to study the molecular mechanisms of the mammalian circadian clock (26–29).
Here, we present that the stability of mouse Period3 (mPer3) mRNA, one of the mammalian circadian genes, is dramatically changed in a circadian phase-dependent manner. Furthermore, hnRNP Q has a critical role for the robust mRNA oscillation of mPer3 and its translation. We suggest that the phase-dependent translation-coupled mRNA decay is involved in the regulation of the mRNA levels and oscillation pattern of mPer3. We demonstrate, for the first time in the field of circadian rhythm, that the cooperative function of the 5′- and 3′-UTRs is necessary and hnRNP Q plays a critical role in maintenance of the circadian oscillation of clock genes.
MATERIALS AND METHODS
Plasmids
A two-step PCR was performed to generate the promoter/5′-UTR/luciferase (Luc)/3′-UTR/neomycin (Neo) vector. The fragment containing the mPer3 promoter region (4) and the 5′-UTR (accession no. NM_011067, version 1) was amplified from mouse (C57BL/6) genomic DNA with the forward primer (5′-CGGGGTACCCGCGCGTTATGTAAGGTACTCGGGGGCCTT-3′) and the reverse primer (5′-TTTGGCGTCTTCCATCCCGCCTGGCAGCCCTCAGCC-3′). The other fragment containing the N-terminus of the Luc-coding sequence was amplified from the pGL3 control vector (Promega) with the forward primer (5′-GGGCTGCCAGGCGGGATGGAAGACGCCAAAAACATAAAG-3′) and the reverse primer (5′-ATTTGTATTCAGCCCATATCG-3′). The second PCR fragment was digested with KpnI/NarI, cloned into the corresponding sites of the pGL3/3′-UTR vector (16), and designated as the promoter/5′-UTR/Luc/3′-UTR vector. The Neo-resistance gene preceded by the thymidine kinase promoter was amplified from the pMC1neo poly(A) vector (Stratagene) with the forward primer (5′-GCTCTAGAGCAGTGTGGTTTTGCAAGAGGAA-3′) and the reverse primer (5′-CAGGTCGACGGATCCGAACAAACG-3′). Following XbaI/SalI digestion, the fragment was cloned into the corresponding sites of the promoter/5′-UTR/Luc/3′-UTR vector.
To generate the promoter/5′-UTR/Luc/Neo vector, the SV40 poly(A) signal was amplified from the pGL3 control vector (Promega) with the forward primer (5′-GCGAATTCCGGCCGCTTCGAGCAGACATGAT-3′) and the reverse primer (5′-GCTCTAGATACCACATTTGTAGAGGTTTTAC-3′), and digested with EcoRI/XbaI. The mPer3 3′-UTR from the promoter/5′-UTR/Luc/3′-UTR vector was removed by digestion with EcoRI/XbaI and replaced with the SV40 poly(A) signal, to form the promoter/5′-UTR/Luc/SV40 poly(A) vector. Following XbaI/SalI digestion, the Neo-resistance gene was cloned into the restriction sites.
To generate Per3 1–357/NAT, the mPer3 5′-UTR was amplified with the forward primer (5′-CCCAAGCTTCCCGCACGGCCGGGCGCTGCT-3′) and the reverse primer (5′-CGCGGATCCCCCGCCTGGCAGCCCTCAGCC -3′) from mouse suprachiasmatic nuclei cDNAs. To generate serial deletion constructs, mPer3 5′-UTR fragments were amplified with forward primers 5′-CCCAAGCTTGCTGACCGCGCTCCCTGAGAGC-3′ for Per3 120–357/NAT, 5′-CCCAAGCTTCTCAGATGAGCGTGGTCGGCG-3′ for Per3 240–357/NAT, and the reverse primer 5′- CGCGGATCCCCCGCCTGGCAGCCCTCAGCC -3′ for both of deletion constructs. The amplified fragment was digested with HindIII/BamHI, and cloned into the corresponding sites upstream of the arylalkylamine N-acetyltransferase (AANAT) coding sequence of the pcNAT control vector. To generate SL-5UTR-NAT, stem-loop oligonucleotide (5′-AGCTTGCCTAGGCCGGAGCGCCCAGATCTGGGCGCTCCGGCCTAGGCA-3′) was annealed to form a duplex. After poly nucleotide kinase treatment, it was was attached by ligase to Per3 1–357/NAT construct which was cut with HindIII.
To generate 5′-UTR-firefly Luc (FLUC), the pCY2 vector was modified (30). Renilla Luc (RLUC) and the intergenic sequences were removed by cutting with HindIII/XbaI, and the 5′-UTR of mPer3 was ligated into the restriction enzyme sites. For in vitro binding followed by UV crosslinking, full-length and deletion fragments of the mPer3 5′-UTR were amplified as described above. The PCR products were digested with EcoRI and XbaI, and then inserted into the pSK′ vector (31), yielding pSK′ Per3 1–357, pSK′ Per3 120–357, pSK′ Per3 240–357.
Cell culture and generation of stable cell lines
The NIH3T3 cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin in a humidified atmosphere containing 5% CO2 at 37°C.
To generate the NIH-mPer3 promoter-5′-UTR-Luc-3′-UTR and NIH-mPer3 promoter-5′-UTR-Luc stable cell lines, the promoter/5′-UTR/Luc/3′-UTR/Neo and promoter/5′-UTR/Luc/Neo vectors were introduced to NIH3T3 fibroblasts by using the calcium phosphate precipitation method, respectively. After 2 days, we started selection with 800 µg G418/ml (Invitrogen). Neomycin-resistant clones were isolated by the standard procedure. The resulting cell lines were maintained in DMEM supplemented with 10% bovine calf serum, 1% penicillin–streptomycin and 200 µg G418/ml.
Dexamethasone shock and mRNA decay kinetics
The dexamethasone shock was done as previously described (16). In brief, ∼1.5 × 105 cells/well were seeded in 12-well plates. When the cells reached confluence, the medium was exchanged with a medium containing 100 nM dexamethasone. After 2 h, this medium was replaced with complete medium. To examine the mRNA oscillation profiles, cells were harvested at the indicated times and stored at −70°C until total RNA was extracted.
To examine the mRNA decay kinetics in the rising phase, actinomycin D was added to a final concentration of 5 µg/ml after 18 h of dexamethasone shock. At the indicated time, the cells were harvested and kept at −70°C until total RNA was extracted. For mRNA decay kinetics in the declining phase, actinomycin D was added after 30 h of dexamethasone shock.
In vitro RNA synthesis and the luciferase assay
For mRNA transfection, 5′-UTR-FLUC was linearized by digestion with EcoRI. This plasmid contains a 20-nt long poly (A) sequence upstream of the EcoRI site. Reporter mRNA was in vitro transcribed with SP6 RNA polymerase (Roche) in the presence of the cap analog. The RLUC control mRNA reporter was linearized by digestion with XbaI. Firefly and Renilla Luc activities were determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
Transient transfection and the AANAT assay
For plasmid transfection, cells were seeded in 24-well plates at a density of 1.0 × 105 cells/well a day prior to transfection. Transfection was carried out using Metafectene (Biontex) according to the manufacturer's instructions. After incubation for 24 or 36 h, cells were harvested for further experiments. For mRNA transfection, NIH3T3 cells were seeded in 24-well plates at a density of 1.0 × 105 cells/well on the day prior to synchronization. We transfected 2 μg of capped reporter mRNA into NIH3T3 cells at the indicated time points using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After incubation for 6 h, the cells were harvested for further experiments.
AANAT activity was determined as previously described (32). In brief, the transfected cells were disrupted by five times of freezing and thawing cycle. After centrifugation, the supernatant was incubated in the presence of 2.5 mM tryptamine–HCl, 25 μM acetyl CoA and 1 μl [3H] acetyl CoA (3.6 Ci/mmol, 250 μCi/ml) at 37°C for 30 min. The reaction was stopped by dilution with 50 mM sodium phosphate buffer. The amount of radiolabeled acetyltryptamine was determined using a liquid scintillation counter.
Quantitative Real-time RT–PCR
Quantitative real-time RT–PCR was performed as previously described (16). In brief, total RNA was isolated by using the TRI Reagent (Molecular Research Center). RNA was reverse transcribed by using Moloney murine leukemia virus reverse transcriptase (Roche Applied Science) according to the manufacturer's instructions. For detection and quantification, the MyiQ™ Real-Time PCR Detection System (Bio-Rad) was used. The sequences of the forward and reverse primers were as follows: endogenous mPer3, 5′-TTGTCAGGTTGGCCTTCTCT-3′ and 5′-GGCATCCTAGCAGAGGTGAG-3′; Luc, 5′-GAGGTTCCATCTGCCAGGTA-3′ and 5′-CACACAGTTCGCCTCTTTGA-3′; mouse TATA-binding protein (mTBP), 5′-CAGCCTTCCACCTTATGCTC-3′ and 5′-TTGCTGCTGCTGTCTTTGTT-3′; mouse ribosomal protein L32 (mRPL32), 5′-AACCCAGAGGCATTGACAAC-3′ and 5′-CACCTCCAGCTCCTTGACAT-3′; rAANAT, 5′-TTTGAGATTGAGCGCGAAG-3′ and 5′-TCGAACCAGCCCAGTGAC-3′
Real-time bioluminescence monitoring
A total of 5.0 × 105 cells were plated into 35-mm culture dishes. When the cells reached confluence, the medium was exchanged with medium containing 100 nM dexamethasone. After 2 h, this medium was replaced with complete medium supplemented with 50 mM HEPES (pH 7.2) and 0.1 mM luciferin (Promega). Cultures were maintained at 37°C and bioluminescence was monitored continuously using an AB-2500 Kronos luminometer (Atto Corporation). Photon counts were integrated over 1-min intervals.
RNA interference
The sequences of synthesized siRNAs were as follows. siCon: 5′-UUCUCCGAACGUGUCACGUTT-3′ (Samchully Pharm.), sihnRNP Q: 5′-AGACAGUGAUCUCUCUCAUTT-3′ (Dharmacon Research). For siRNA transfection into NIH3T3 cell lines, a microporator (Digital-Bio) was used, according to the manufacturer's instructions.
Protein preparation and immunoblot analysis
For immunoblotting, cells were disrupted with complete protein solubilizing buffer containing 1% SDS and M urea in PBS, followed by sonication. Fractionation of NIH3T3 cells into cytosolic extracts was performed as described (30). Immunoblot analyses were performed with monoclonal anti-14-3-3ξ (Santa Cruz), polyclonal antibody against hnRNP Q (Sigma-Aldrich). HRP-conjugated mouse, rabbit (KPL) or rat (Santa Cruz) secondary antibodies were detected with SUPEX ECL reagent (Neuronex) and a LAS-4000 system (FUJI FILM), according to the manufacturer's instructions.
In vitro binding assay and immunoprecipitation
In vitro binding assay was performed as described previously (30). In brief, XbaI-linearized pSK′-5′-UTR constructs were transcribed using T7 RNA polymerase (Promega) in the presence of [α-32P] UTP. Twenty micrograms of nuclear extracts or 40 μg of cytosolic extracts were incubated with labeled RNAs at 30°C. After 30 min of incubation, the mixtures were UV-irradiated on ice for 15 min with a CL-1000 UV-crosslinker (UVP). The samples were detected with autoradiography after SDS–PAGE. To confirm the identity of the UV cross-linked protein, 3 μg of a polyclonal antibody against hnRNP Q or pre-immune serum were added to RNase-digested reaction mixtures. After 16 h, Protein G agarose beads (Amersham bioscience) were added. After a further incubation for 3 h, precipitates were detected with autoradiography after SDS–PAGE.
RESULTS
mPer3 mRNA stability is regulated in a circadian phase-dependent manner
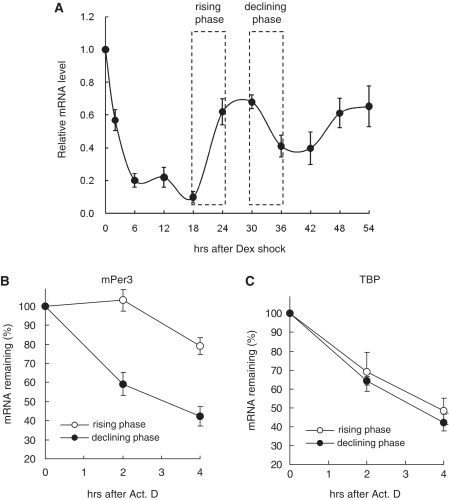
In circadian phase-synchronized NIH3T3 fibroblasts, endogenous mPer3 mRNA levels began to increase after 18 h of dexamethasone treatment and decreased after reaching their peak levels between 24 and 30 h (Figure 1A). To examine whether mPer3 mRNA stability was regulated by circadian rhythm, we analyzed the mRNA decay kinetics in the circadian phase-synchronized NIH3T3 cells. To compare the decay kinetics of the rising phase or the declining phase, transcription was blocked with actinomycin D at 18 or 30 h after dexamethasone shock, respectively. As shown in Figure 1B and Supplementary Figure S1, mPer3 mRNA stability was dramatically regulated in a circadian phase-dependent manner. mPer3 mRNA was very stable in the rising phase (especially during the first 2 h), whereas it was rapidly degraded in the declining phase. To explore the specificity of these results, we then analyzed the decay kinetics of TBP mRNA, whose levels are constant, irrespective of circadian rhythm (27). As expected, TBP mRNA decay kinetics was not altered during circadian rhythm (Figure 1C) in contrast to the mPer3 mRNA. Based on these results, we concluded that mPer3 mRNA stability is regulated by circadian rhythm and that the circadian control of mRNA stability is a specific phenomenon that is restricted to a subset of mRNAs.
Figure 1.
Circadian rhythm regulates mPer3 mRNA stability. (A) Endogenous mPer3 mRNA oscillation pattern was confirmed. For the decay kinetics of the rising or declining phases, transcription was blocked after 18 or 30 h of dexamethasone treatment, respectively. Both of rising and declining phases were indicated. (B) mRNA decay analyses were performed as mentioned in Materials and methods section. Decay kinetics of endogenous mPer3 mRNA from circadian phase-synchronized NIH3T3 fibroblasts with dexamethasone treatment are shown. The mPer3 mRNA levels were determined by quantitative real-time RT–PCR and normalized to RPL32 mRNA levels. (C) Decay kinetics of TBP mRNA from circadian phase-synchronized NIH3T3 fibroblasts with dexamethasone treatment are shown. The TBP mRNA levels were determined by quantitative real-time RT–PCR and normalized to RPL32 mRNA levels. All results are presented as the mean ± SD of three experiments.
3′-UTR-mediated mRNA decay is necessary but not sufficient for the circadian control of mPer3 mRNA stability
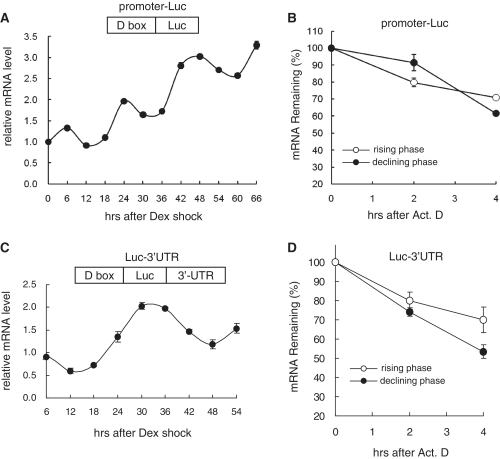
To examine whether the mPer3 mRNA oscillation pattern was regulated with only transcriptional control, we established NIH-mPer3 promoter-Luc stable cell lines that express the luciferase gene under the control of the wild-type mPer3 promoter without any UTRs. Surprisingly, the total amount of reporter mRNA increased gradually after phase-synchronization with dexamethasone. Although it seemed to show a 24-h period oscillation pattern to some degree, a breakdown of the mRNA level balancing mechanism could be a potential obstacle in cellular homeostasis (Figure 2A). In the mPer3 promoter-Luc stable cell lines, the reporter mRNA was also transcribed by a D box element within the wild-type mPer3 promoter; nevertheless, the reporter mRNA showed a continuously accumulating pattern. In that case, it is possible that the decay of the reporter mRNA was less active in the NIH-mPer3 promoter-Luc stable cell lines. As expected, the mRNA degradation kinetics of the UTR-free reporter was much slower than that of UTR-bearing endogenous mPer3 mRNA (Figure 2B). Moreover, we did not observe a phase-dependency of mRNA decay.
Figure 2.
The mPer3 3′-UTR is necessary but not sufficient for the circadian control of mRNA stability. (A and C) Temporal expression profiles of luciferase (Luc) mRNA without a UTR (A) or with the mPer3 3′-UTR (C) are shown from NIH-mPer3 promoter-Luc stable cells (A) or NIH-Luc-WT 3′-UTR stable cells (C) after circadian phase-synchronization with dexamethasone treatment. Schematic diagrams of reporter genes expressed in each cell lines are shown. When dexamethasone was added, the time was set to 0. Reporter mRNA levels were determined by quantitative real-time RT–PCR and normalized to TBP mRNA levels. The initial level of reporter mRNA was arbitrarily set to 1.0. (B and D) Decay kinetics of Luc mRNA without a UTR (B) or with the mPer3 3′-UTR (D) are shown. The reporter mRNA levels were determined by quantitative real-time RT–PCR and normalized to RPL32 mRNA levels. All results are presented as the mean ± SD of three experiments.
Previously, we established NIH-Luc-WT 3′-UTR stable cell lines that produce mRNA of the Luc coding region followed by the 3′-UTR of the wild-type mPer3, and showed that the mPer3 3′-UTR is necessary for circadian mRNA oscillations (16). To examine whether mPer3 3′-UTR-mediated mRNA decay is sufficient for the circadian control of mRNA stability, we analyzed the decay kinetics of Luc mRNA containing the mPer3 3′-UTR after blocking transcription. As shown in Figure 2D, although the reporter mRNA was a little more stable in the rising phase than in the declining phase, the mPer3 3′-UTR alone could not reproduce the dramatic changes of mRNA decay kinetics as shown for the endogenous mPer3 mRNA. The reporter mRNA oscillation profile showed an ∼6-h phase delay compared to endogenous mPer3 mRNA (Figure 2C) (16). According to these results, we concluded that the contribution of the mPer3 3′-UTR alone is not sufficient to replicate the endogenous oscillation pattern of mPer3 mRNA. Therefore, we needed to analyze the role of the mPer3 5′-UTR in mRNA decay regulation.
The mPer3 5′-UTR mediates translational regulation-coupled mRNA decay
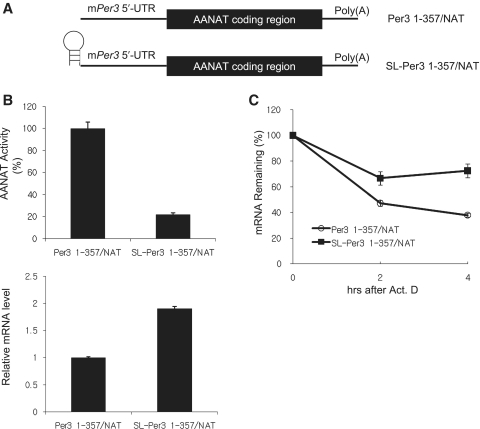
Although a large number of studies have demonstrated 5′-UTR-mediated translational regulation (33,34), several studies also reported that mRNA stability could be regulated by the 5′-UTR, a mechanism called translational regulation-coupled mRNA decay. In such cases, translational inhibition caused mRNA stabilization (35–39). To test whether this mechanism also exists in the regulation of the levels of mPer3 mRNA, we cloned a reporter construct by inserting the wild-type mPer3 5′-UTR upstream of serotonin N-acetyltransferase (AANAT) coding region (Per3 1–357/NAT) (32). We also prepared the other construct that had a stem-loop structure at the upstream of the mPer3 5′-UTR in 5UTR-NAT reporter to block the ribosome scanning (SL-Per3 1–357/NAT) (Figure 3A). The effect of this structure on translation was analyzed by transient transfection followed by measurement of AANAT activity. As expected, the stem-loop-harboring construct showed lower AANAT activity than the wild-type construct, even though reporter mRNA level was elevated due to higher transfection efficiency (Figure 3B). This result indicated that the stem–loop structure could inhibit the translation of downstream gene efficiently. Interestingly, the construct with a lower translation rate showed a slower mRNA decay rate (Figure 3C). This reciprocal relationship between translation efficiency and mRNA stability suggests that the mPer3 5′-UTR mediates a coupling between translation and mRNA decay.
Figure 3.
The mPer3 5′-UTR mediates translation-mRNA decay coupling. (A) Schematic diagrams of pcNAT reporters. mPer3 5′-UTR with (SL-Per3 1–357/NAT) or without (Per3 1–357/NAT) stem–loop structure was fused to the AANAT coding region. (B) Translation efficiency of each pcNAT reporter was measured by the AANAT assay. An amount of 0.5 μg of each construct was transiently transfected into HEK293A cells. The AANAT assay was performed 24 h after transfection. The AANAT level in Per3 1–357/NAT reporter transfected cells was arbitrarily set to 100 (upper panel). The reporter transcript level of each construct was determined using quantitative RT–PCR, and normalized to RPL32 mRNA levels (lower panel). (C) The decay kinetics of AANAT mRNA expressed from each reporter are shown. Cells were treated with 5 μg/ml actinomycin D, 24 h after transfection of each reporter. All results are representative of at least three independent experiments. The error bars represent the mean ± SEM of duplicate measurements.
Increasing evidence has suggested that upstream open reading frames (uORFs) can regulate their mRNA translation. mPer3 has four uORFs within the mPer3 5′-UTR, including one overlapping uORF. To determine the potential role of these uORFs, we generated four single-nucleotide mutant constructs in which AUG was changed to AAG (Per3 uAUG-1 mut/NAT, Per3 uAUG-2 mut/NAT, Per3 uAUG-3 mut/NAT, Per3 uAUG-4 mut/NAT). The effects of these mutations on translation were analyzed by transient transfection and the reporter, AANAT assay. However, we could not observe any differences in AANAT level between WT and mutant constructs (Supplementary Figure S4A). In addition, mRNA decay kinetics of WT and mutant reporters were comparable (Supplementary Figure S4B). Therefore, we concluded that uORF-mediated translation regulation is not crucial step in the case of mPer3.
Circadian control of mPer3 mRNA stability requires the cooperative function of the 5′- and 3′-UTRs
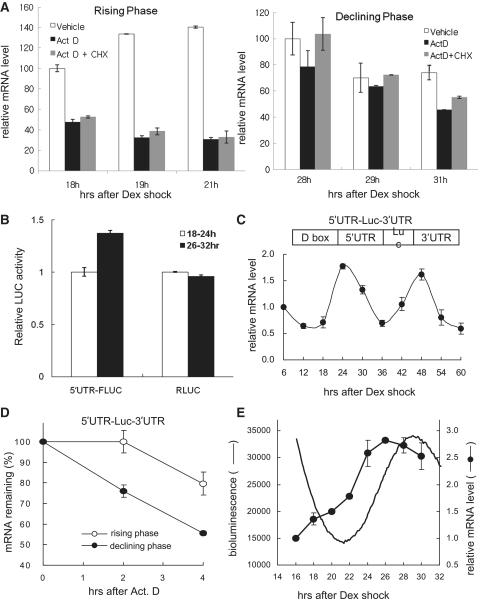
Since mPer3 mRNA stability was regulated in a circadian phase-dependent manner, we hypothesized that mPer3 translational regulation-coupled mRNA decay would also be rhythmically regulated. To confirm the hypothesis, we compared the decay rate of endogenous mPer3 mRNA between the translation-inhibited condition and the normal condition in both the rising and declining phases in phase-synchronized NIH3T3 mouse fibroblasts. Interestingly, mPer3 mRNA was more stable when both transcription and translation were blocked by actinomycin D and cycloheximide treatment, respectively, than only when transcription was blocked. Moreover, this phenomenon was more dramatic in the declining phase when mPer3 mRNA decay was accelerated (Figure 4A). This result suggests that translation-mRNA decay coupling may occur differentially, dependent on the mRNA oscillation phase.
Figure 4.
Circadian control of mPer3 mRNA stability requires the cooperative function of the 5′- and 3′-UTRs. (A) Phase-synchronized NIH3T3 fibroblasts were treated with vehicle, actinomycin D, or actinomycin D with 50 μg/ml cycloheximide at 17 h (rising phase) or 27 h (declining phase) after dexamethasone treatment. The endogenous mPer3 mRNA levels were determined by quantitative real-time RT–PCR and normalized to RPL32 mRNA levels at the indicated time points. The initial level of mPer3 mRNA (at 18 h in rising phase, at 28 h in declining phase) was arbitrarily set to 100. The results are presented as the mean ± SD of three experiments. (B) An amount of 2 μg of in vitro transcribed two mRNA reporters, 5′-capped 5′-UTR-FLUC or 5′-capped RLUC, were co-transfected at 18 or 28 h after synchronization. After 6 h, the luciferase assay was performed. The FLUC and RLUC level during 18–26 h after synchronization were set to 1.0. The error bars represent the mean ± SEM of duplicate measurements. (C) Temporal expression profiles of luciferase (Luc) mRNA with both the mPer3 5′- and 3′-UTRs are shown from NIH-mPer3 promoter-5′-UTR-Luc-3′-UTR stable cells after circadian phase-synchronization with dexamethasone (Dex) treatment. Reporter mRNA levels were determined by quantitative real-time RT–PCR and normalized to TBP mRNA levels. The initial level of reporter mRNA was arbitrarily set to 1.0. (D) The decay kinetics of Luc mRNA with both the mPer3 5′- and 3′-UTRs are shown. The reporter mRNA levels were determined by quantitative real-time RT–PCR and normalized to the RPL32 mRNA levels. In (C) and (D), each point presented is the mean ± SD of three experiments. (E) Comparison of LUC mRNA and protein profiles. Real-time bioluminescence of NIH-mPer3 promoter-5′-UTR-Luc-3′-UTR stable cells was monitored. The unit of bioluminescence (y-axis) is photons/min. Reporter mRNA levels were determined by quantitative real-time RT–PCR and normalized to the TBP mRNA levels.
However, upregulation of mPer3 mRNA stability by cycloheximide treatment could result from the synthesis blockade of proteins that promote the clearance of mPer3 mRNA. To exclude this possibility, two cap-harboring mRNA reporters were generated by in vitro transcription: the 5′-UTR-FLUC (firefly luciferase) reporter, composed of the mPer3 5′-UTR followed by FLUC, and the RLUC reporter which contains only RLUC (Renilla luciferase) coding sequence. Two mRNA reporters were transiently co-transfected into phase-synchronized NIH3T3 fibroblasts at 18 or 28 h after dexamethasone shock. Six hours later, translated FLUC and RLUC levels were analyzed by the luciferase assay. RLUC level which determined the transfection efficiency was slightly decreased in 28–34 h after synchronization than in 18–24 h. Nevertheless, the 5′-UTR-FLUC transcript was ∼1.5-fold more translated in the mPer3 mRNA decay-favoring declining phase than in the rising phase (Figure 4B). This result suggests that mPer3 5′-UTR-mediated translation is also rhythmically regulated.
Since 3′-UTR-mediated mRNA decay was not sufficient to reproduce the phase-dependent mRNA stability changes of mPer3 and we identified that the 5′-UTR was also involved in the mRNA decay process, we established NIH-mPer3 promoter-5′-UTR-Luc-3′-UTR stable cell lines that express Luc mRNA containing both the 5′- and 3′-UTRs of mPer3 mRNA upstream and downstream of the Luc coding sequences, respectively. Interestingly, the Luc mRNA with both the 5′- and 3′-UTRs of mPer3 (i.e. 5′-UTR-Luc-3′-UTR) oscillated similarly to the endogenous mPer3 mRNA (Figure 4C). The notable thing is that the decay kinetics of this reporter mRNA were nearly the same as the endogenous mPer3 mRNA (Figure 4D). This result suggests that the cooperative functions of the 5′- and 3′-UTRs are necessary for the circadian control of the decay rate of mPer3 mRNA.
As shown in Figure 4E, the mRNA level was in the rising phase at 16 h after dexamethasone treatment and increased continuously to 26 h. Interestingly, the oscillation profile of Luc activity was very different from that of the reporter mRNA. Especially, in spite of rising mRNA levels, the protein levels decreased until ∼21.5 h after dexamethasone treatment. As a result, a distinct phase-lag between the mRNA and protein levels was observed. This result supported our hypothesis that translation was inhibited during the early rising phase of mRNA levels and translational inhibition may induce the stabilization of the mRNA in the rising phase.
Downregulation of hnRNP Q not only reduces the translation efficiency but also increases the mRNA stability of mPer3
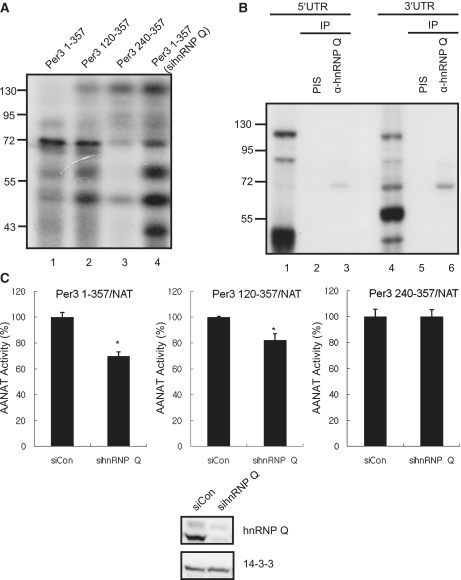
To identify the cis-acting region and trans-acting factors which are in charge of translation initiation and mRNA decay regulation, we performed in vitro binding assay followed by UV crosslinking with deletion constructs. While there was little change in protein binding pattern when the 5′ proximal 119 nt within the 5′-UTR of the mPer3 mRNA were deleted (Per3 120–357), several proteins were dramatically dissociated when additional 120 nt were deleted (Per3 240–357) (Figure 5A). Previously, we reported that 68-kDa protein hnRNP Q bound to 5′-UTR of AANAT, the key enzyme in the melatonin biogenesis, and enhanced its translation kinetics (30). Also, we already identified that hnRNP Q could control the translation initiation of mRev-erb α in phase-dependent manner (40). In line with such findings, we discovered that 68-kDa protein showed dramatically reduced binding to the Per3 240–357 construct. As a result of in vitro binding assay with whole cell lysates from NIH3T3 cell lines that hnRNP Q was downregulated with siRNA, the binding of 68-kDa protein was markedly reduced and this protein was exactly the same size of hnRNP Q (Figure 5A). To confirm the interaction between hnRNP Q and mPer3 mRNA, UV crosslinking followed by immunoprecipitation with antibodies against hnRNP Q was performed. Among several cellular proteins bound to radioisotope-labeled UTRs of mPer3, we confirmed that hnRNP Q was clearly bound to both 5′- and 3′-UTR of mPer3. In contrast, we could not detect any bands in precipitates using pre-immune serum (Figure 5B). The association between 5′-UTR of mPer3 and hnRNP Q was confirmed by RNA affinity purification using biotin–streptavidin interaction followed by immunoblot analysis (Supplementary Figure S2).
Figure 5.
hnRNP Q regulates the translation-coupled mRNA decay of mPer3. (A) Cellular proteins that bound to the wild-type or truncated forms of mPer3 5′-UTR were analyzed with the in vitro binding followed by UV-crosslinking assay. Whole lysates were extracted from NIH3T3 cells (lane 1–3). Cellular proteins that bound to the full length of the mPer3 5′-UTR were analyzed. Whole lysates were extracted from NIH3T3 cells which hnRNP Q was downregulated (lane 4). The sizes of proteins are indicated to the left of the panels. (B) Identification the interaction between hnRNP Q and both UTRs of mPer3, using UV crosslinking followed by immunoprecipitation. In lane 1 and 4, cellular proteins that bound to the mPer3 5′-UTR (lane 1) or 3′-UTR (lane 4) were analyzed with the in vitro binding followed by UV-crosslinking assay. Cytoplasmic lysates were extracted from NIH3T3 cells. The sizes of proteins are indicated to the left of the panels. PIS, pre-immune serum. (C) Translation initiation controlled by wild-type or truncated forms of mPer3 5′-UTR is analyzed when the hnRNP Q is downregulated. Per3 120–357/NAT and Per3 240–357/NAT are deletion constructs of Per3 1–357/NAT. 0.5 μg of reporter plasmid was transiently transfected into NIH3T3 cell lines which were pre-transfected with either control siRNA (siCon) or siRNA against hnRNP Q (sihnRNP Q). Translation efficiency of AANAT reporter was measured by the AANAT assay. For transfection control, 0.1 μg of beta-galactosidase plasmid was cotransfected. The AANAT assay and beta-galactosidase assay were performed at 36 h after transfection. The normalized AANAT level in siCon-transfected cells was arbitrarily set to 100. The significance of differences of AANAT activities was determined by Student's t-test. *P < 0.005. (D) After co-transfection of 0.05 μg of AANAT reporter with either siCon or sihnRNP Q, the decay kinetics of the reporter mRNA was analyzed. Cells were treated with 5 μg/ml actinomycin D, at 18 h after co-transfection. Knockdown efficiency was confirmed by western blot. (E) After transfection with either siCon or sihnRNP Q, the decay kinetics of endogenous mPer3 was analyzed. Cells were treated with 5 μg/ml actinomycin D, 24 h after siRNA-transfection. (D and E) Decay kinetics of TBP mRNA was analyzed for negative control. Each mRNA levels was determined by quantitative real-time RT–PCR and normalized to RPL32 mRNA levels. In (C) to (E), all results are representative of at least three independent experiments. The error bars represent the mean ± SD of duplicate measurements.
To test whether a deficiency in hnRNP Q also affects the translation efficiency of mPer3, we transiently transfected either control siRNA or the siRNA against hnRNP Q (sihnRNP Q) into NIH3T3 cells. After 12 h, we additionally transfected the mPer3 5′-UTR-inserted pcNAT reporter (Per3 1–357/NAT) into siRNA-transfected NIH3T3 cells. The amount of translated proteins was measured by AANAT assay. Interestingly, translated protein level showed ∼20% decrease in sihnRNP Q-transfected cells, compared to siCon-transfected cells (Figure 5C). Translational reduction in hnRNP Q-downregulated cells was also observed when 5′ proximal 119 nt within the 5′-UTR of the mPer3 mRNA were deleted (Per3 120–357/NAT). However, there was little reduction in the AANAT level in hnRNP Q-knockdowned cells when 120 nt within the 5′-UTR of the mPer3 mRNA were additionally removed (Per3 240–357/NAT). Reduction of hnRNP Q level was confirmed by western blotting (Figure 5C).
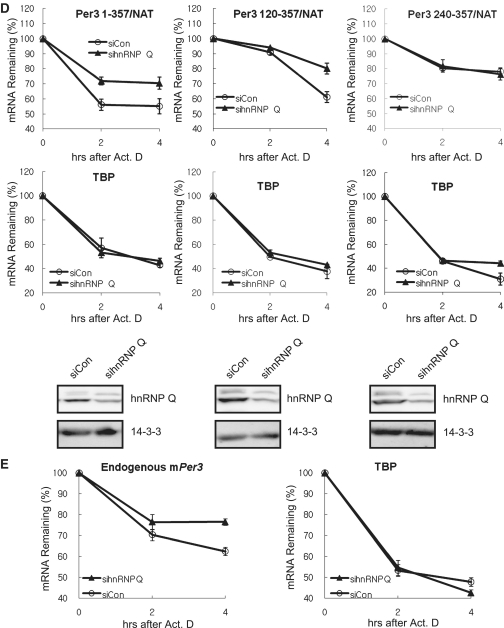
Next, we sought to determine whether the decrease in reporter translation by downregulation of hnRNP Q could influence the stability of reporter mRNA. Consistent with the result of Figure 3, Per3 1–357/NAT and Per3 120–357/NAT transcripts were more stable when translation initiation was less active by sihnRNP Q-transfection. However, we could not observe any differences in the stability of Per3 240–357/NAT transcripts when hnRNP Q level was downregulated (Figure 5D), mirroring the results obtained from in vitro binding assay and AANAT assay. Furthermore, the degradation kinetics of endogenous mPer3 was also slower in sihnRNP Q-transfected condition than in siCon-transfected condition (Figure 5E). To determine the specificity of hnRNP Q downregulation effect, we exogenously expressed hnRNP Q in endogenous hnRNP Q-downregulated cells. As a result, altered mRNA decay kinetics of mPer3 by hnRNP Q knockdown was rescued by exogenous hnRNP Q expression (Supplementary Figure S3A). However, the degradation kinetics of endogenous TBP mRNA was not altered in all conditions (Figure 5D, E and Supplementary Figure S3B). These results supported that the mPer3 mRNA decay was connected to its translation kinetics and the central region of mPer3 5′-UTR (120–239 nt) was responsible for coupling of translation and mRNA decay. Moreover, hnRNP Q was strong candidate in this regulatory mechanism.
hnRNP Q binds to 5′-UTR of mPer3 in a phase-dependent manner, and maintains the robust mRNA oscillation
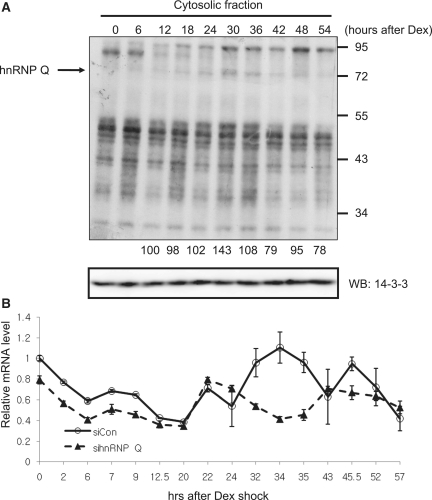
Since both translation and mRNA decay are more active in the declining phase and hnRNP Q was concerned with those events, we hypothesized that the binding affinity of hnRNP Q to 5′-UTR of mPer3 would be higher in the declining phase than in the rising phase. Based on the general knowledge that a translation occurs in the cytoplasm, we analyzed the phase-dependent interaction between cytosolic hnRNP Q and 5′-UTR of mPer3. Overall, even though hnRNP Q did not bind to 5′-UTR of mPer3 transcript in abundance, this interaction was highly enhanced 30–36 h after phase-synchronization with dexamethasone treatment (Figure 6A). Moreover, this time period was coincident with the declining phase that we already defined. To clarify the role of hnRNP Q in the oscillation of mPer3 mRNA, we tranfected either a control siRNA or hnRNP Q siRNA into NIH3T3 fibroblasts, and synchronized a circadian phase of cells with dexamethasone treatment. Interestingly, we observed that overall oscillation phase of mPer3 mRNA was advanced in hnRNP Q-downregulated cells. Moreover, the amplitude was evidently decreased in hnRNP Q-downregulated environment (Figure 6B) However, the oscillation pattern of mCry1 mRNA, another clock gene, was comparable in hnRNP Q-knockdowned condition (unpublished data). We also confirmed that oscillation phase of mPer3 mRNA was unchanged when hnRNP R was downregulated (unpublished data). These results suggested that the precise regulation of the translation-mRNA decay coupling through hnRNP Q was necessary for the robust and scheduled oscillation of mPer3.
Figure 6.
hnRNP Q interacts with mPer3 5′-UTR dynamically and is necessary for the robust oscillation mPer3 mRNA. (A) Dynamics of the binding affinity of several proteins to mPer3 5′-UTR mRNA after synchronization was analyzed with the UV crosslinking assay (upper panel). The loading amounts of cytosolic extracts were confirmed by western blotting of the 14-3-3 level (lower panel). The normalized band intensities of hnRNP Q are shown below the upper panel. The intensity at 12 h after synchronization was arbitrarily set as 100. (B) The endogenous mPer3 mRNA levels were determined using quantitative real-time RT–PCR and normalized to RPL32 mRNA levels at the indicated time points in phase-synchronized control siRNA or hnRNP Q siRNA-transfected NIH3T3 cells. Initial levels of mPer3 mRNA in control siRNA-transfected cells were arbitrarily set as 1.0. These results represent the mean ± SD of three experiments.
DISCUSSION
The regulation of mRNA decay rate, which is accomplished through changes in mRNA half-life, is an important step in the control of gene expression, as changes in mRNA abundance may alter the amount of the corresponding protein. For oscillating mRNAs, precise control of mRNA stability is even more critical because alterations to mRNA stability could affect the overall mRNA oscillation profile (16). A number of studies have shown that diverse stimuli have effects on the half-lives of mRNA (17,22–24). As for the circadian control of mRNA stability, the temporal regulation of Drosophila period (per) mRNA stability was previously predicted by computer modeling based on the comparison of mRNA levels and transcription profiles (13). Given that circadian rhythm is a fundamental biological phenomenon (2), it is not surprising that the circadian rhythm regulates mRNA stability; however, little is known about the circadian regulation of mRNA stability.
In this study, we demonstrated that the turnover rate of mPer3 mRNA was regulated by circadian rhythm, providing the first example of the circadian control of a clock gene whose mRNA stability is regulated by both of the 5′- and 3′-UTRs. This result implies that the molecular circadian rhythm could regulate the biological clock by controlling the mRNA stability of core clock genes. Given that the stability of TBP mRNA was independent of circadian rhythm (Figure 1B), the circadian regulation of the mRNA decay rate seems to be a specific phenomenon restricted to a subset of genes. In this regard, it will be interesting to identify the common features of those mRNAs whose stability is regulated according to circadian rhythm.
The UTRs of mRNA are known to play essential roles in post-transcriptional control, including the regulation of mRNA stability, subcellular localization, and translation efficiency (16,20,34,41). Many studies have focused on the independent functions of either the 5′-UTR or the 3′-UTR, nevertheless, the combined functions of the 5′- and 3′-UTRs were reported in some studies; the hypoxia-induced stabilization of VEGF mRNA was dependent on the cooperation of the 5′-UTR, 3′-UTR, and coding region (42); Her-2 uORF-mediated translational inhibition was reversed by the Her-2 3′-UTR in cancer cells (43). Here, we showed that the circadian control of mPer3 mRNA stability was also accomplished by the cooperative function of the 5′- and 3′-UTRs. The circadian regulation of mRNA stability was not mimicked by the 3′-UTR-mediated mRNA decay alone (Figure 2D) and required the additional function of the 5′-UTR.
As shown in Figure 4D, the cooperative function of the 5′- and 3′-UTRs reproduced the decay kinetics of endogenous mPer3 mRNA in the rising phase. Nevertheless, since the Luc mRNA with both the 5′- and 3′-UTRs (i.e. 5′-UTR-Luc-3′-UTR) was a little more stable than the endogenous mPer3 mRNA in the declining phase (Figures 1B and 4D), it should be noted that an additional region of mPer3 mRNA seems to be needed to fully reproduce the decay kinetics observed in the declining phase. The missing region may be located in the coding region of mPer3 mRNA, like VEGF mRNA (42) or c-fos mRNA (39).
Even though the 5′-UTR alone can mediate translational regulation-coupled mRNA decay (Figure 3B and C), circadian control of mPer3 mRNA stability was not sufficiently regulated with only the 5′-UTR, given that the Luc mRNA with the mPer3 5′-UTR was degraded with almost the same rate in the rising and declining phases (unpublished data). Our results suggest that the functional role of the mPer3 3′-UTR in mRNA decay process is necessary in addition to the 5′-UTR.
We already verified that hnRNP Q could control the translation initiation of AANAT via an internal ribosomal entry site-mediated translation. However, we could not figure out any internal ribosomal entry site in 5′-UTR of mPer3 (unpublished data) by using a dicistronic vector system (30). In this study, it is likely that hnRNP Q controls the translation of mPer3 in a cap-dependent manner rather than in a cap-independent manner. Although knockdown of hnRNP Q resulted in inefficient mRNA decay of mPer3, overall mPer3 mRNA level was less in sihnRNP Q-transfected cells than in siCon-transfected cells. This result remains a possibility that hnRNP Q is also involved in the transcriptional regulation of mPer3. Indeed, it has been reported that several hnRNP proteins participate in transcription regulation (44,45). In addition, it was recently identified that hnRNP Q has a critical role in splicing events (46). Indeed, there are four mPer3 transcripts mediated by alternative splicing or alternative promoter, according to ASTD database (47). Therefore, it will be worthwhile to explore whether hnRNP Q can function as a transcriptional (co)factor or splicing factor on mPer3.
Our previous report demonstrated that the functional role of the mPer3 3′-UTR in the mRNA decay process contributed to the oscillation pattern of mRNA. In addition, the present study revealed that mPer3 mRNA stability is also regulated in a circadian phase-dependent manner. It is the first evidence that the cooperative function of the 5′- and 3′-UTRs is necessary for the circadian control of the mPer3 mRNA profile and decay rate. Results from the transfection of mRNA and the comparison of mRNA levels with Luc activity suggested that translational regulation-coupled mRNA decay is the underlying mechanism controlling mPer3 mRNA stability. Although the mechanism of translation inhibition leading to the upregulation of mRNA stability still need to be determined, we suggest that translational regulation-coupled mRNA decay could be one of the possible mechanisms, which explains the distinct phase-lag between mRNA and protein levels.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Core Research Program/Anti-aging and Well-being Research Center; the Brain Korea 21 program; World Class University program (R31-10105) of the Ministry of Education, Science and Technology; National Research Foundation of Korea (NRF; 20100002146, 20100019706 and 20100030089).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ms Jiwon Lee for technical assistance. The authors also thank Dr Kiyoung Paek for experimental comments.
REFERENCES
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 4.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 5.Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J. Biol. Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Comparing clockworks: mouse versus fly. J. Biol. Rhythms. 2000;15:357–364. doi: 10.1177/074873000129001459. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clck genes in mouse peripheral tissues. BMC Mol. Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc. Natl Acad. Sci. USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darlington TK, Lyons LC, Hardin PE, Kay SA. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J. Biol. Rhythms. 2000;15:462–471. doi: 10.1177/074873040001500603. [DOI] [PubMed] [Google Scholar]
- 10.Wang GK, Ousley A, Darlington TK, Chen D, Chen Y, Fu W, Hickman LJ, Kay SA, Sehgal A. Regulation of the cycling of timeless (tim) RNA. J. Neurobiol. 2001;47:161–175. doi: 10.1002/neu.1024. [DOI] [PubMed] [Google Scholar]
- 11.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Hunter-Ensor M, Schotland P, Sehgal A. Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J. Biol. Rhythms. 1998;13:364–379. doi: 10.1177/074873098129000192. [DOI] [PubMed] [Google Scholar]
- 13.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 15.Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak E, Kim TD, Kim KT. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- 17.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 19.Tourriere H, Chebli K, Tazi J. mRNA degradation machines in eukaryotic cells. Biochimie. 2002;84:821–837. doi: 10.1016/s0300-9084(02)01445-1. [DOI] [PubMed] [Google Scholar]
- 20.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 21.Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulding WR, Czyzyk-Krzeska MF. Hypoxia-induced regulation of mRNA stability. Adv. Exp. Med. Biol. 2000;475:111–121. doi: 10.1007/0-306-46825-5_11. [DOI] [PubMed] [Google Scholar]
- 23.Staton JM, Thomson AM, Leedman PJ. Hormonal regulation of mRNA stability and RNA-protein interactions in the pituitary. J. Mol. Endocrinol. 2000;25:17–34. doi: 10.1677/jme.0.0250017. [DOI] [PubMed] [Google Scholar]
- 24.Gorospe M, Kumar S, Baglioni C. Tumor necrosis factor increases stability of interleukin-1 mRNA by activating protein kinase C. J. Biol. Chem. 1993;268:6214–6220. [PubMed] [Google Scholar]
- 25.Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 27.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 28.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 29.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganguly S, Coon SL, Klein DC. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–137. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- 32.Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray NK, Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 34.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 35.Savant-Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 36.Aharon T, Schneider RJ. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winstall E, Gamache M, Raymond V. Rapid mRNA degradation mediated by the c-fos 3′ AU-rich element and that mediated by the granulocyte-macrophage colony-stimulating factor 3′ AU-rich element occur through similar polysome-associated mechanisms. Mol. Cell Biol. 1995;15:3796–3804. doi: 10.1128/mcb.15.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curatola AM, Nadal MS, Schneider RJ. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol. Cell Biol. 1995;15:6331–6340. doi: 10.1128/mcb.15.11.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiavi SC, Wellington CL, Shyu AB, Chen CY, Greenberg ME, Belasco JG. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J. Biol. Chem. 1994;269:3441–3448. [PubMed] [Google Scholar]
- 40.Kim DY, Woo KC, Lee KH, Kim TD, Kim KT. hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res. 2010;38:7068–7078. doi: 10.1093/nar/gkq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dibbens JA, Miller DL, Damert A, Risau W, Vadas MA, Goodall GJ. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta A, Trotta CR, Peltz SW. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes Dev. 2006;20:939–953. doi: 10.1101/gad.1388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rauch J, O'Neill E, Mack B, Matthias C, Munz M, Kolch W, Gires O. Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells by regulating A-Raf transcription. Cancer Res. 2010;70:1679–1688. doi: 10.1158/0008-5472.CAN-09-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Liu J. Identification of heterogeneous nuclear ribonucleoprotein K as a transactivator for human low density lipoprotein receptor gene transcription. J. Biol. Chem. 2010;285:17789–17797. doi: 10.1074/jbc.M109.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol. Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, Kull M, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.