Abstract
−1 Programmed ribosomal frameshifting (PRF) in synthesizing the gag-pro precursor polyprotein of Simian retrovirus type-1 (SRV-1) is stimulated by a classical H-type pseudoknot which forms an extended triple helix involving base–base and base–sugar interactions between loop and stem nucleotides. Recently, we showed that mutation of bases involved in triple helix formation affected frameshifting, again emphasizing the role of the triple helix in −1 PRF. Here, we investigated the efficiency of hairpins of similar base pair composition as the SRV-1 gag-pro pseudoknot. Although not capable of triple helix formation they proved worthy stimulators of frameshifting. Subsequent investigation of ∼30 different hairpin constructs revealed that next to thermodynamic stability, loop size and composition and stem irregularities can influence frameshifting. Interestingly, hairpins carrying the stable GAAA tetraloop were significantly less shifty than other hairpins, including those with a UUCG motif. The data are discussed in relation to natural shifty hairpins.
INTRODUCTION
Ribosomal frameshifting is a translational recoding event in which a certain percentage of ribosomes are forced to shift to another reading frame in order to synthesize an alternative protein. This switch occurs at a specific position on the mRNA, called the slip site or slippery sequence, and can be either forwards (+1) or backwards (−1). The nature and efficiency of frameshifting depends on several factors, including tRNA availability and modifications, and mRNA primary and secondary structure (1,2).
The signals that are responsible for −1 frameshifting comprise two elements: a slippery sequence where the actual reading shift takes place, and a downstream located structural element which greatly stimulates the efficiency of frameshifting. Although the mechanism is still elusive, the present view is that the downstream structure forms a physical barrier that blocks EF-2 function and causes ribosomes to stall in their translocation step. This ‘roadblock’ puts tension on the mRNA–tRNA interaction. The tension can be relieved by the realigning of A-site and P-site tRNAs in the 5′-direction, whereafter EF-2 can do its work and the ribosome resumes translation in the −1 reading frame (3).
In general, a pseudoknot is more efficient in stimulating frameshifting than a hairpin of the same sequence composition. This difference is likely related to a higher thermodynamic stability of the pseudoknot. Indeed, from thermodynamic analysis it appears that pseudoknots are more stable than their hairpin counterparts (4–6). Recent studies employing mechanical ‘pulling’ of frameshifter pseudoknots have shown a correlation between the mechanical strength of a pseudoknot and its frameshifting capacity (7,8), and the influence of major groove and minor groove triplex structures (9). The higher strength of a pseudoknot can be primarily attributed to the formation of base triples between the lower stem S1 and loop L2 (Figure 1A), making it more resistant against unwinding by an elongating ribosome (8,10). Base triples in several pseudoknots, such as Beet western yellows virus (BWYV) p1–p2 (11), Pea enation mosaic virus type-1 (PEMV-1) p1–p2 (6), Sugarcane yellow leaf virus (ScYLV) p1–p2 (12) and Simian retrovirus type-1 gag-pro (SRV-1) (13,14) have been shown to play an essential role in frameshifting. For pseudoknots with a longer stem S1 of 10–11 bp, like that of Infectious Bronchitis Virus (IBV), base triples do not appear to contribute to frameshifting (15).
Figure 1.
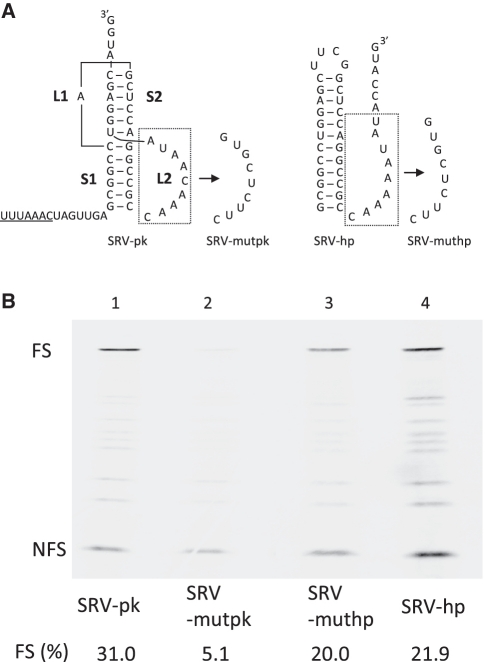
Hairpin derivative of the Simian retrovirus type-1 (SRV-1) gag-pro frameshift pseudoknot is an efficient frameshift stimulator. (A) Schematic representation of the SRV-1 pseudoknot (SRV-pk) and its hairpin derivative (SRV-hp). Mutations in SRV-pk loop L2 (SRV-mutpk) and SRV-hp (SRV-muthp) are indicated. The slippery sequence is underlined. (B) SDS–PAGE analysis of 35S-methionine-labeled translation products in rabbit reticulocyte lysate (RRL). −1 Ribosomal frameshifting is monitored by appearance of the 65-kD product (FS). The non-shifted zero-frame product is indicated by NFS. Quantitative analysis of frameshifting efficiency [FS (%)] is described in ‘Materials and Methods’ section.
Although a hairpin is considered to be a less efficient frameshift-inducing secondary structure than a pseudoknot, some viruses like Human immunodeficiency virus (HIV) (16), Human T-lymphotropic virus type-2 (HTLV-2) (17) and Cocksfoot mottle virus (CfMV) (18) make use of a simple hairpin to stimulate substantial levels of frameshifting. In addition, frameshifting in the prokaryotic dnaX gene requires, next to an upstream enhancer, the presence of a hairpin as well (19). A few studies have investigated a correlation between hairpin stability and frameshift efficiency of natural shifty hairpins (19,20). Nonetheless, certain studies have shown that a hairpin composed of the same base pairs as a frameshifter pseudoknot is not very efficient in inducing frameshifting in mammalian cells and lysates (21–23) but is in other systems (24).
Here, we have carried out a systematic analysis of the frameshift-inducing efficiency of hairpins derived from the SRV-1 gag-pro frameshifter pseudoknot. Investigation of about 30 different hairpin constructs revealed that next to thermodynamic stability, also loop size and composition, and stem irregularities can significantly influence frameshifting. Our data showed that there exists no base specific contacts between the hairpin and the ribosome during frameshifting and suggests that the hairpin primarily serves as a barrier to allow repositioning of tRNAs at the slippery site.
MATERIALS AND METHODS
Mutations in the SRV-1 gag-pro frameshifting signal were made in an abridged version of plasmid SF2 (25) which is derivative of pSFCASS5 (26), a frameshift reporter construct. In this version, the entire BglII–NcoI fragment of pSF2 was replaced by a synthetic dsDNA fragment (5′-GATCTTAATACGACTCACTATAGGGCTCATTTAAACTAGTTGAGGGGCCATATTTCGC-3′, a SpeI restriction site is underlined). This yielded plasmid pSF208 in which the original GGGAAAC slippery sequence has been replaced by the more slippery UUUAAAC sequence (26). pSF208 was digested with SpeI and NcoI, and sets of complementary oligonucleotides corresponding to the various mutants were inserted. A list of oligonucleotides is available upon request. All constructs were verified by automated dideoxy sequencing using chain terminator dyes (LGTC, Leiden).
In vitro transcription
DNA templates were linearized by BamHI digestion and purified by successive phenol/chloroform extraction and column filtration (Qiagen, Benelux). SP6 polymerase directed transcriptions were carried out in 50 µl reactions containing ∼2 µg linearized DNA, 10 mM NTPs, 40 mM Tris–HCl (pH 7.9), 10 mM NaCl, 10 mM DTT, 6 mM MgCl2, 2 mM spermidine, 6 U of RNase inhibitor (RNAsin, Promega, Benelux) and 15 U of SP6 polymerase (Promega, Benelux). After an incubation period of 2 h at 37°C, samples were taken and run on agarose gels to determine the quality and quantity of the transcripts. Appropriate dilutions of the reaction mix in water were directly used for in vitro translations. Alternatively, transcripts were purified by phenol/chloroform extraction and isopropanol precipitation and quantified by UV absorption as described previously (14).
In vitro translation
Experiments were carried out in duplicate using serially—in water—diluted mRNAs with final concentrations of 5 nM. Reactions contained 4 µl of an RNA solution, 4.5 µl of rabbit reticulocyte lysate (RRL, Promega), 0.25–1 µl of 35S methionine (Amersham, in vitro translation grade), 0.5 µl of 1 mM amino acids lacking methionine and were incubated for 60 min at 28°C. Samples were boiled for 3 min in 2× Laemmli buffer and loaded onto 12% SDS polyacrylamide gels. Gels were dried and exposed to phosphoimager screens. Band intensity of 0-frame and −1 frameshift products was measured using a Molecular Imager FX and Quantity One software (Biorad). Frameshift percentages were calculated as the amount of −1 frameshift product divided by the sum of 0 and −1 frame products, corrected for the number of methionines (10 in the 0-frame product and 28 in the fusion product), multiplied by 100.
Frameshift assays in mammalian cells
Candidates of interest were constructed in a dual luciferase vector, pDUAL-HIV(0), essentially as described previously (14,27). In short, pDUAL-HIV(0) was digested by KpnI and BamHI, followed by insertion of complementary oligonucleotides to clone the SRV-1 gag-pro pseudoknot, various hairpins as shown in Figures 2C and 5, and a negative control (NC) which formed no apparent secondary structure downstream of the slippery sequence. An in-frame control was constructed by inserting an A-residue upstream of the cytosine in the UUUAAAC slippery sequence of a 12 bp hairpin frameshift construct. HeLa cells were cultured in DMEM/high glucose/stable glutamine (PAA Laboratories GmbH, Germany) and supplemented with 10% fetal calf serum and 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were kept in a humidified atmosphere containing 5% CO2 at 37°C. Assay protocols were described previously (14). Briefly, cells were transfected with 300 ng of plasmid using 1 µl of lipofectamine-2000 (Invitrogen) in a 24-well plate. Cells were lysed 24 h after transfection and luciferase activities were quantified by Glomax-multidetector (Promega, Benelux) according to manufacturer's protocol. Frameshifting efficiency was calculated by dividing the ratio of Renilla luciferase (RL) over Firefly luciferase (FL) activity of the mutant by the RL/FL ratio of the in-frame control, multiplied by 100.
Figure 2.
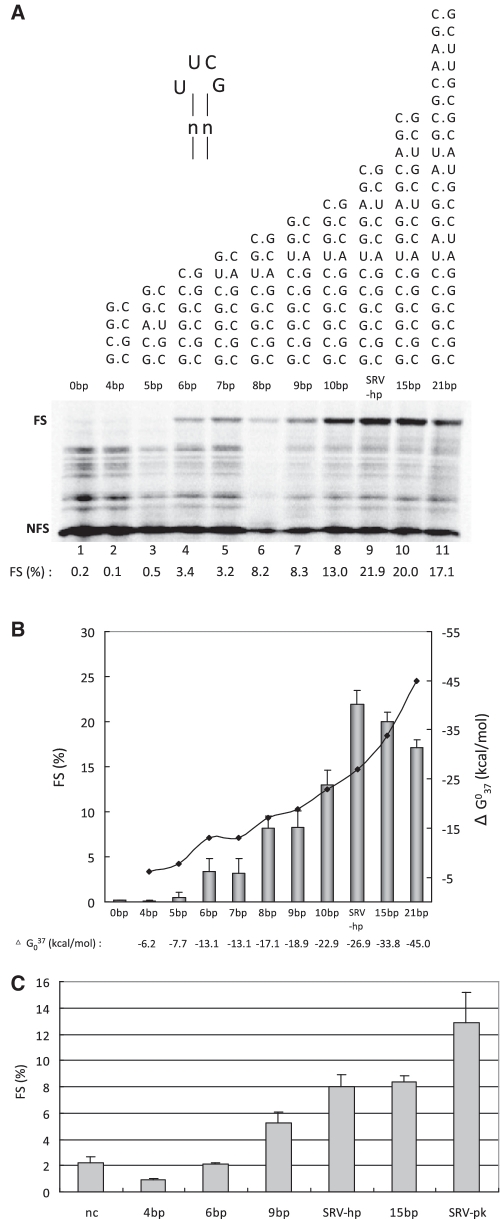
Influence of stem length of UUCG-capped hairpins on −1 ribosomal frameshifting efficiency in vitro and in vivo. (A) SDS–PAGE analysis of 35S-methionine-labeled translation products in RRL using mRNAs with hairpins of various stem lengths. See legend to Figure 1B for more details. The base composition of the various stems is shown. The 0 bp is the control without a hairpin. (B) Graph showing the relation between frameshifting efficiency (indicated by bars) on the left y-axis and predicted thermodynamic stability by MFOLD [indicated by a solid diamond (filled diamond) on the right y-axis]. The average FS (%) and error bars were from at least three independent experiments. (C) Selected hairpins with different stem lengths were assayed for their ability to induce −1 ribosomal frameshifting in HeLa cells. The frameshifting efficiency was obtained by measuring dual-luciferase activity of a frameshift reporter construct (see ‘Materials and Methods’ section). The in vivo experiments were done at least three times in triplicate.
Figure 5.
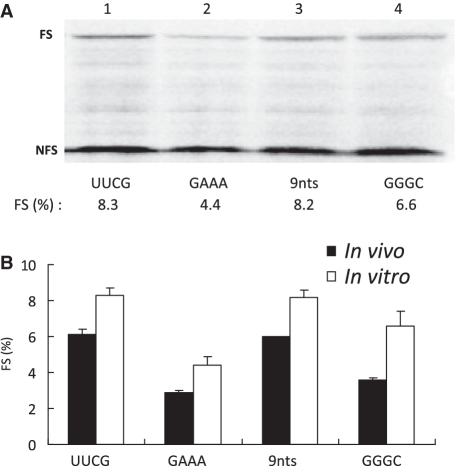
Comparison of in vitro and in vivo frameshifting efficiencies induced by four selected 9 bp hairpins with different loops. (A) SDS–PAGE analysis of 35S-methionine-labeled translation products in RRL of mRNAs containing the 9 bp hairpin with the indicated loop sequence. See legend to Figure 1B for more details. (B) Comparison of −1 ribosomal frameshifting in vitro and in vivo. The in vitro efficiency (white bar) was obtained by quantifying autoradiograms and averaging of at least three independent experiments. In vivo frameshifting efficiency (black bar) was obtained by measuring dual-luciferase activity of a frameshift reporter construct in HeLa cells (see ‘Materials and Methods’ section). The in vivo experiments were done at least three times in triplicate.
RESULTS
Hairpin derived from the SRV-1 gag-pro pseudoknot is an efficient frameshift stimulator
In contrast to earlier reports involving the IBV frameshifting pseudoknot (21,22), we found that in the case of the SRV-1 gag-pro frameshift inducing pseudoknot a hairpin of similar composition as the pseudoknot did stimulate frameshifting in vitro (Figure 1A and B). The 12 bp hairpin derivative of the SRV-1 pseudoknot (SRV-hp) showed 22% frameshifting efficiency, whereas the SRV-1 pseudoknot (SRV-pk) in this context yielded 31%. The pseudoknot in these experiments is a modified version of the wild-type SRV-1 pseudoknot previously used for NMR and functional analysis (14). We note that the UUUAAAC slippery sequence was used to enhance the sensitivity of the in vitro frameshifting assay. This sequence is ∼1.5-fold more slippery than the wild-type GGGAAAC slippery sequence (28). In the latter context, the hairpin was indeed less efficient (data not shown) while a non-slippery variant, GGGAAGC, was not effective at all (<0.2%, data not shown). Two other known efficient slip sites, AAAAAAC and UUUUUUA, caused 23 and 27%, respectively, of ribosomes to switch frame in the presence of the 12 bp hairpin (data not shown). These data showed that the 12 bp hairpin is a genuine stimulator of frameshifting.
Since the hairpin construct also contained sequences resembling those of L2 of the pseudoknot construct, it was theoretically possible that these nucleotides could take part in the same base triples. To investigate this possibility, we replaced the downstream sequence in the hairpin construct (SRV-muthp). This did not affect the frameshift efficiency of the hairpin construct. In contrast, the same mutations in the pseudoknot context (SRV-mutpk) reduced its activity about 6-fold (Figure 1B). Thus, it is unlikely that triple helix formation or other tertiary interactions contribute to hairpin-dependent frameshifting; the hairpin as such seems to be sufficient.
Effect of hairpin stem size on frameshifting efficiency
Next, we investigated the role of stem length on frameshifting efficiency. Increasing stem size from 12 to 15 or 21 bp did not significantly alter frameshifting (Figure 2A). On the other hand, decreasing stem size led to a steady decrease in frameshifting efficiency which seemed to vanish around a stem size of 5 bp or ΔG37 of −7.7 kcal/mol (Figure 2B). Thermodynamic stabilities were calculated at the MFOLD website using version 2.3 parameters (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3), as these were previously shown to better fit in vivo hairpin stabilities (29). These data support the notion that downstream structures serve as barriers to stall translating ribosomes to stimulate frameshifting, and demonstrate that there is a correlation between the thermodynamic stability of a hairpin and its frameshift inducing capacity.
A selection of above hairpins was cloned into a dual-luciferase reporter plasmid and their frameshifting efficiency assayed in mammalian cells (Figure 2C). Although the absolute level of frameshifting was lower than in vitro, the trend was similar and showed maximal frameshifting of ∼8% around 12–15 bp. The pseudoknot in these assays was 1.6 times more efficient than the 12 and 15 bp hairpins, close to the in vitro ratio of 1.4 (see above). Thus, the hairpin derivative can effectively substitute for the SRV-1 pseudoknot in −1 ribosomal frameshifting.
Bulges and mismatches decrease frameshifting efficiency
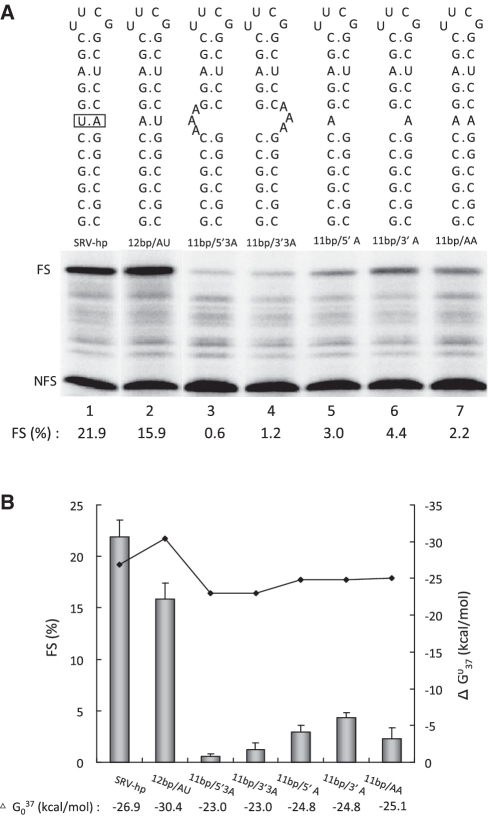
Bulges and mismatches are known to change twisting and bending of a regular stem and are thus expected to influence the way in which a ribosome encounters a hairpin structure (11,30). To investigate a possible effect of helical twisting and bending on frameshifting, we introduced mismatches and bulges in the 12 bp stem at a position corresponding to the junction in the SRV pseudoknot (Figure 3A). Introduction of an A·A mismatch halfway through the stem (11 bp/AA) decreased frameshifting about 10 fold, although its predicted thermodynamic stability of −25.1 kcal/mol is comparable to that of a regular hairpin of 10 bp, yielding 13% frameshifting (Figure 2B). The frameshift inducing ability was recovered when the base pair was restored to A–U (12 bp/AU).
Figure 3.
Influence of bulges and mismatches in the middle part of the hairpin on frameshifting efficiency. (A) SDS–PAGE analysis of 35S-methionine-labeled translation products in RRL using mRNAs containing the indicated hairpins. See legend to Figure 1b for more details. (B) Graph showing the relation between the predicted thermodynamic stability and frameshift efficiency. See legend to Figure 2B for more details.
We also introduced a single or triple adenosine bulge at either side of the stem, to investigate potential bending effects on frameshifting. Figure 3A and B show that the single adenosine bulge mutant decreased frameshifting, depending on the location of the bulge, five to seven fold compared to the 12 bp hairpin construct. When the bulge was enlarged to three adenosines the frameshifting was almost abolished. Interestingly, the effect of bulges at the 3′ side of the stem was less dramatic than those at the 5′ side.
Loop composition affects frameshifting efficiency
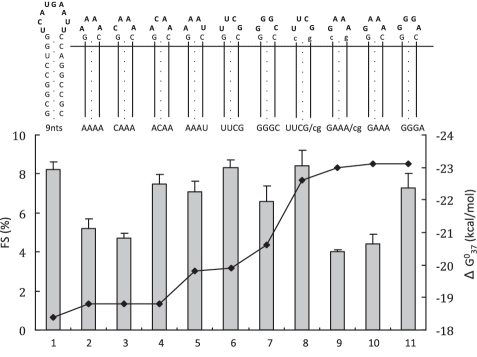
The loop composition plays a major role in hairpin stability, RNA/RNA and RNA/protein interaction. These factors may directly influence hairpin-induced ribosomal frameshifting efficiency. To explore the correlation between loop composition and frameshifting efficiency, a number of loop mutations were introduced in the context of a 9 bp stem (Figure 4). We note that the UUCG tetraloop with a CG closing base pair (cbp) has higher stability (∼2 kcal/mol) than that with a GC cbp (31). Therefore, we first tested if this different cbp affected frameshifting efficiency. Our results showed that there is no difference in frameshifting efficiency between UUCG and UUCG/cg constructs (Figure 4, bars 6 and 8). Replacing the UUCG tetraloop by GGGC which, due to its high content of purines, is among the most disfavored tetraloops (32) had only a marginal effect on frameshifting (Figure 4, compare bars 6 and 7 and Figure 5A, lanes 1 and 4). Interestingly, increasing the loop size to 9 nt, which is predicted to lower the stability of stem did not affect frameshifting (Figure 4, bar 1; Figure 5A, lane 3).
Figure 4.
Influence of loop sequence and closing base pair (cbp) on −1 ribosomal frameshifting efficiency. The composition of various loops capping a 9 bp stem is shown in bold, and CG-cbps are shown in lower case. The constructs are named after their loop sequence followed by the ‘/cg’ extension when the cbp was changed from G–C to C–G. Slippery sequence and spacer are the same as in the construct shown in Figure 1A. Graph is similar to that of Figure 2B except that on the right y-axis ΔG starts from −18 kcal/mol.
Substituting UUCG by another stable tetraloop sequence (GAAA) resulted in a 2-fold decrease in frameshifting (Figure 5A, lanes 1 and 2) either with GC (Figure 4, bar 10) or CG cbp (Figure 4, bar 9). We designed another five loop mutants to try to explain the low efficiency of the GAAA tetraloop constructs. Constructs AAAA and CAAA induced 5.2% and 4.7% frameshifting, respectively (Figure 4, bars 2 and 3), which is close to that of the GAAA constructs. The efficiency of two other A-rich loop mutants, ACAA and AAAU, was 7.5% and 7.1%, respectively (Figure 4, bars 4 and 5), thereby closely matching that of the UUCG constructs. Finally, the GGGA tetraloop construct, belonging to the stable GNRA tetraloop family, induced 1.7-times more frameshifting than its GAAA sibling (Figure 4, bar 11). These data suggest that the presence of 3 or 4 adenines at the 3′ side of a tetraloop is unfavorable for frameshifting.
Loop composition affects frameshifting efficiency in vivo
To further examine the role of the loop identity or size in ribosomal frameshifting, we cloned some of the above loop mutants into a dual-luciferase reporter plasmid and assayed their frameshifting efficiency in mammalian cells (Figure 5B). Our data show that the effects of loop nucleotides are comparable in vitro and in vivo. The stable GAAA tetraloop construct again had the lowest frameshifting efficiency (Figure 5B, 2.9%), which was half that of the UUCG construct (Figure 5B, 6.1%).
DISCUSSION
Most RNA viruses that make use of ribosomal frameshifting employ pseudoknot structures instead of simple hairpins for this job. The reason for this may be the presence of a triple helix interaction between S1 and L2 in most frameshifter pseudoknots, which has been suggested to be a poor substrate for the ribosomal helicase (13,33) and hence increases ribosomal pausing and the time window for slippage. Although pausing is critical, it is not sufficient for efficient frameshifting (34). Previously, it was shown that a 17 bp hairpin with a calculated stability of −31.2 kcal/mol derived from the minimal IBV pseudoknot induced 5- to 10-fold less frameshifting in RRL (22) than its parent pseudoknot even though both the hairpin and the pseudoknot can pause ribosomes at the same position and to a similar extent (34). In the present study, a 12 bp hairpin derivative of the SRV-1 gag-pro pseudoknot with a calculated stability of −26.9 kcal/mol was capable of inducing 22% of frameshifting, which is only 1.4-fold less than its pseudoknotted counterpart. This indicated that a non-natural hairpin can be an efficient frameshift stimulator, at least in the SRV-1 model. Furthermore, our results showed that the frameshifting efficiency increased upon elongation of the length of the hairpin up to 12–15 bp, which is consistent with our previous data using antisense oligonucleotides of 12–15 nt to induce ribosomal frameshifting (35). More importantly, the frameshift inducing ability of these hairpin constructs with a perfect stem linearly correlated with the calculated thermodynamic stability, in agreement with two previous reports (19,20).
In the experiments of Bidou et al. (20) studying the HIV-1 gag-pol frameshift hairpin the stem-length was kept at 11 bp, while its stability was varied between −3.4 and −22.1 kcal/mol (recalculated using MFOLD 2.3) by changing the number of AU and GC base pairs in a small set of six hairpins. In the case of the dnaX gene of Escherichia coli 22 variants of the wild-type 11 bp hairpin were tested for their ability to stimulate −1 PRF at the AAAAAAG slippery sequence. Hairpin stabilities varied between −10.4 and −27 kcal/mol and a positive correlation between frameshifting efficiency and calculated stability was observed both in the presence (R2 = 0.62) and absence (R2 = 0.72) of upstream enhancer (19). The dnaX gene with the highly efficient (prokaryotic) AAAAAAG slippery sequence is not directly comparable to our in vitro system; a 6 bp hairpin in the dnaX gene displayed 17% of frameshifting without upstream enhancer, whereas a 6 bp hairpin in our system induced only 3.5% of frameshifting.
In the HIV-1 gag-pol gene Bidou et al. (20) observed a 15–20% decrease in frameshifting in vivo with their most stable hairpin, similar to our results with the 21 bp hairpin. However, in our case, the stability at which this happened was −45 kcal/mol much higher than their most stable hairpin of −22.1 kcal/mol. It is possible that this difference is due to the different experimental systems. Although it has been suggested that too stable stems increase the time for tRNAs to shift back into the 0-frame again (20) we believe that our 21 bp hairpin is less efficient because it has more AU bps in the middle of the stem compared to the 12 and 15 bp hairpins (Figure 2A). The experiments with hairpins harboring bulges or mismatches halfway through the stem demonstrated that this region is quite important for frameshifting (Figure 3A and B). Even though the overall stability of these constructs was comparable to that of a hairpin of 9 or 10 bp, their frameshift activity was equal or lower than that of a 6 bp hairpin of −13.1 kcal/mol: as if the mismatch or bulge after the 6th base pair disconnected the upper part of the stem. This observation is reminiscent of the overall destabilizing effect of mismatches in DNA hairpins. In a pioneer single-molecule pulling study, it was shown that introducing a mismatch in a 20 bp DNA hairpin shifted its transition state close to the location of the mismatch (36). Our data also comply with this mechanical study and suggest that mechanical stability may be a better parameter than thermodynamic stability to describe the frameshift efficiency of hairpins.
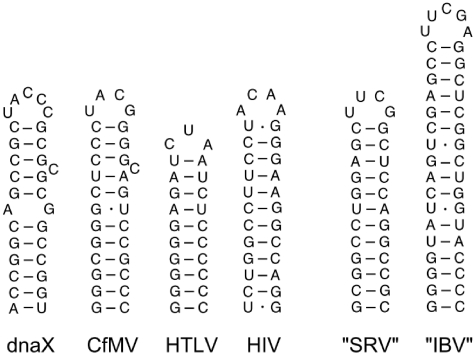
In addition to the mentioned dnaX and HIV-1 gag-pol hairpins, other examples of frameshifter hairpins are found in HTLV-2 and CfMV (Figure 6). HTLV-2 gag-pro features a perfect 10 bp hairpin with CUA tri-loop which induces 9% frameshifting in RRL (16). The CfMV 2a–2b frameshifting hairpin consists of 12 bp, one cytidine bulge close to the top, and a stable UACG tetraloop and is capable of inducing 11% of frameshifting in a wheat germ cell-free system (WGE) (17). What these hairpins have in common is their length of 10–12 bp, their relatively low number of mismatches and bulges, their small loops and their high GC content, especially in the bottom 6 bp. These features are also applicable to the good frameshifters from our dataset. Interestingly, these features do not all apply to the minimal IBV hairpin (Figure 6) that is derived from the so-called minimal IBV pseudoknot. Despite its large size of 17 bp, absence of mismatches and bulges, presence of a small loop, the stability of the middle part of the hairpin, i.e. bp 5–9, is not very high. This could be the reason why its activity in RRL is 5-10 fold lower (22) than of its parent pseudoknot, whose activity is 42% (25). Surprisingly, in our assays the frameshift-inducing efficiency of the IBV hairpin was 26% (data not shown), which is in stark contrast to the 4–8% reported by Brierley et al. (22). This discrepancy may be due to experimental conditions: in our experiments we used non-capped transcripts, a 7-nt spacer and RRL from Promega whereas the Brierley's lab used capped transcripts, a 6-nt spacer and in-house prepared RRL. On the other hand, the 26% we obtained for the IBV hairpin would be a factor of 1.6 lower than the 42% reported for the IBV pseudoknot (25), and is similar to the ratio of 1.4 and 1.6 we obtained for SRV in vitro and in vivo, respectively.
Figure 6.
Overview of naturally occurring hairpins involved in −1 ribosomal frameshifting and two synthetic pseudoknot-derived hairpins (SRV and IBV). HIV, Human immunodeficiency virus type 1 gag-pol; HTLV, Human T-lymphotropic virus type 2 gag-pro; CfMV, Cocksfoot mottle virus; IBV, Infectious bronchitis virus; SRV, Simian retrovirus type 1.
Remarkably, in WGE the IBV hairpin has been reported to induce high levels (34%) of frameshifting versus 51% for the IBV pseudoknot (24). In that study modified extracts were used that are somewhat more frameshift-prone than the standard wheat-germ extracts. Nevertheless, the ratio between pseudoknot and hairpin-induced frameshifting in this system is also 1.5. This number may reflect the additional interactions, like base triples, in a pseudoknot that make it a better frameshift stimulator than a hairpin.
In addition to stem size, loop composition is another determinant of hairpin stability. An important subgroup of hairpin loops is the tetraloop, which is the most common loop size in 16S and 23S ribosomal RNAs (37). The tetraloops with consensus UNCG, GNRA, or CUUG loop sequence form stable loop conformations (38,39). As opposed to the mentioned stable tetraloops, purine-rich (32) and larger loops (40) are considered to be less favorable for hairpin formation. Our results showed that the GGGC loop is indeed less efficient in inducing frameshifting but the larger loop construct (9 bp/9 nt), although having a lower thermodynamic stability, showed comparable frameshifting efficiency to the stable UUCG tetraloop hairpin. This is consistent with previous studies that showed that increasing the size of the loop in a hairpin or pseudoknot can increase frameshift-inducing ability to a certain extent (21,41). Although larger loops seem efficient in inducing frameshifting, in known examples of frameshifter hairpins, there are no loop sizes of more than 5 nt. This could relate to hairpin folding kinetics (40) or to nuclease sensitivity.
Intriguingly, we found that a 9 bp stem capped with a GAAA tetraloop is 2-fold less efficient in inducing frameshifting than its UUCG counterpart in vitro and in vivo. It has been reported that GAAA tetraloops are frequently involved in RNA tertiary interactions (42). We hypothesize that the GAAA tetraloop may be involved in an unknown RNA tertiary structure with ribosomal RNA, thereby interfering with frameshifting. The fact that in the known natural examples of frameshifter hairpins, the GAAA tetraloop, despite its high stability, is absent can be taken as support for this hypothesis (Olsthoorn, unpublished data). Further investigation of this observation may lead to new insights in ribosomal frameshifting.
In conclusion, our data show that hairpins of various base composition in stem and loop can act as efficient frameshift stimulators. Combined with previous studies on antisense-induced frameshifting (43,44), these data support the notion that downstream structures primarily serve as barriers to stall translating ribosomes to stimulate frameshifting. Although there exists a linear relationship between calculated stability and frameshifting, local destabilizing elements like bulges or mismatches in a hairpin can greatly influence frameshift-inducing activity. Future experiments addressing the mechanical strength of these hairpins (7–9) may help to improve our understanding of the basics of ribosomal frameshifting.
FUNDING
Funding for open access charge: Leiden Institute of Chemistry, Leiden University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Fabien Sohet and Maarten Laurs for their initial contributions to this subject.
REFERENCES
- 1.Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 2.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nixon PL, Giedroc DP. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- 5.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nixon PL, Rangan A, Kim YG, Rich A, Hoffman DW, Henning M, Giedroc DP. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TM, Reihani SN, Oddershede LB, Sørensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I., Jr Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plant EP, Dinman JD. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YG, Su L, Maas S, O'Neill A, Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl Acad. Sci. USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish PV, Hennig M, Giedroc DP. A loop 2 cytidine-stem1 minor groove interaction as a positive determinant for pseudoknot-stimulated -1 ribosomal frameshifting. Proc. Natl Acad Sci. USA. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michiels PJ, Versleijen AA, Verlaan PW, Pleij CW, Hilbers CW, Heus HA. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsthoorn RC, Reumerman R, Hilbers CW, Pleij CW, Heus HA. Functional analysis of the SRV-1 RNA frameshifting pseudoknot. Nucleic Acids Res. 2010;38:7665–7672. doi: 10.1093/nar/gkq629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient -1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudin C, Mazauric MH, Traïkia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 17.Kim YG, Maas S, Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchesi J, Mäkeläinen K, Merits A, Tamm T, Mäkinen K. Regulation of -1 ribosomal frameshifting directed by cocksfoot mottle sobemovirus genome. Eur. J. Biochem. 2000;267:3523–3529. doi: 10.1046/j.1432-1327.2000.01379.x. [DOI] [PubMed] [Google Scholar]
- 19.Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli DNAX ribosomal frameshifting: programmed efficiency of 50% J. Mol. Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset JP. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 21.Brierley I, Rolley NJ, Jenner AJ, Inglis SC. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marczinke B, Hagervall T, Brierley I. The Q-base of asparaginyl-tRNA is dispensable for efficient -1 ribosomal frameshifting in eukaryotes. J. Mol. Biol. 2000;295:179–191. doi: 10.1006/jmbi.1999.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napthine S, Vidakovic M, Girnary R, Namy O, Brierley I. Prokaryotic-style frameshifting in a plant translation system: conservation of an unusual single-tRNA slippage event. EMBO J. 2003;22:3941–3950. doi: 10.1093/emboj/cdg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the ‘slippery-sequence’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ten Dam EB, Brierley I, Inglis S, Pleij CW. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 28.ten Dam EB, Verlaan PW, Pleij CW. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 29.de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Kang H, Shen LX, Chamorro M, Varmus HE, Tinoco I., Jr A characteristic bent conformation of RNA pseudoknots promotes -1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- 31.Blose JM, Proctor DJ, Veeraraghavan N, Misra VK, Bevilacqua PC. Contribution of the closing base pair to exceptional stability in RNA tetraloops: roles for molecular mimicry and electrostatic factors. J. Am. Chem. Soc. 2009;131:8474–8484. doi: 10.1021/ja900065e. [DOI] [PubMed] [Google Scholar]
- 32.Nagel JH, Flamm C, Hofacker IL, Franke K, de Smit MH, Schuster P, Pleij CW. Structural parameters affecting the kinetics of RNA hairpin formation. Nucleic Acids Res. 2006;34:3568–3576. doi: 10.1093/nar/gkl445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu CH, Noteborn MH, Olsthoorn RC. Stimulation of ribosomal frameshifting by antisense LNA. Nucleic Acids Res. 2010;38:8277–8283. doi: 10.1093/nar/gkq650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 38.Cheong C, Varani G, Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 39.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 40.Kuznetsov SV, Ren CC, Woodson SA, Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamm T, Suurväli J, Lucchesi J, Olspert A, Truve E. Stem-loop structure of Cocksfoot mottle virus RNA is indispensable for programmed -1 ribosomal frameshifting. Virus Res. 2009;146:73–80. doi: 10.1016/j.virusres.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 43.Olsthoorn RC, Laurs M, Sohet F, Hilbers CW, Heus HA, Pleij CW. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:1702–1703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard MT, Gesteland RF, Atkins JF. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]