Abstract
Telomere length/DNA content has been measured in epidemiological/clinical settings with the goal of testing a host of hypotheses related to the biology of human aging, but often the conclusions of these studies have been inconsistent. These inconsistencies may stem from various reasons, including the use of different telomere length measurement techniques. Here, we report the first impartial evaluation of measurements of leukocyte telomere length by Southern blot of the terminal restriction fragments and quantitative PCR (qPCR) of telomere DNA content, expressed as the ratio of telomeric product (T)/single copy gene (S) product. Blind measurements on the same samples from 50 donors were performed in two independent laboratories on two different occasions. Both the qPCR and Southern blots displayed highly reproducible results as shown by r values > 0.9 for the correlations between results obtained by either method on two occasions. The inter-assay CV measurement for the qPCR was 6.45%, while that of the Southern blots was 1.74%. The relation between the results generated by Southern blots versus those generated by qPCR deviated from linearity. We discuss the ramifications of these findings with regard to measurements of telomere length/DNA content in epidemiological/clinical circumstances.
INTRODUCTION
Many independent studies have related telomere length in human cells (as most commonly measured in peripheral leukocytes) to risks, incidence and mortality for a variety of diseases. However, these studies variously use one of three different methods to assess telomere length properties: Southern blotting (1), quantitative PCR (qPCR) (2,3) or flow-FISH (4). The Southern blot analysis of the length of the terminal restriction fragments (TRFs) of chromosomes measures [in kilobases (kb) or nucleotides (nt)] the mean length and length distributions of not only the canonical telomeric region (strictly TTAGGG repeats). However, it also adds in the non-canonical region of the telomeres up to the nearest restriction site that is the target of a given set of enzymes used to fragment the DNA prior to Southern blot analysis (1). In contrast, the qPCR (2,3) and Flow-fluorescence in situ hybridization (FISH) (4) techniques measure only the canonical component of telomeres. While the qPCR method ‘normalizes’ the quantity of telomere product to a single gene to provide a mean telomere length for the cell population, the FISH method quantifies the telomere florescence signal by using cells of known telomere length as controls, and can provide information on the distribution of telomeric DNA content in a cell population.
Because each of these three methods employs different laboratory-based tools and methodologies and, furthermore, generates distinct telomere parameters, comparisons between studies have often been difficult. Moreover, current telomere length measurement methods require expertise, which is not uniform across laboratories, and this can further complicate the interpretation and comparison of different studies. For instance, the reported measurement error of telomere length or DNA content, expressed in the inter-assay coefficient of variation (CV), has ranged from 2.27% to 28% for the qPCR method that measures telomere DNA content (5–8) and from 1.5% to 12% for the Southern blot analysis of the mean length of the TRFs (9,10). When reported, the inter-assay CV of telomere signal by FISH amounts to >5% (11–13). However, none of the reported ‘low’ CV of telomere length measurements by various methods has been impartially verified.
The central aim of this report was to compare the inter-assay CV of telomere length/DNA content, measured on two occasions by Southern blots of the TRFs or by qPCR. The FISH method requires intact nuclei and prompt processing of samples. For this reason, in epidemiological research, the Southern blot analysis and qPCR methods, which are the focus of this impartial evaluation, have been typically used rather than FISH. In addition, we discuss several other relevant features of the two methods and their potential ramifications for epidemiological and other research.
MATERIALS AND METHODS
Overall design
Two laboratories independently measured telomere length by Southern blot analysis of the TRFs (Aviv's Lab) or telomere DNA content by qPCR (Blackburn's Lab). These measurements of leukocyte telomere length (LTL) were performed on 50 blinded DNA samples from white donors, ages 41–70 (mean 55.3 ± 9.6), BMI 21–35 (mean 27.9 ± 3.5) and 50% women, whose blood was collected from 2004 to 2008. DNA was extracted by QIAamp DNA blood kits in Hunt's Lab.
Two sets of aliquots were prepared in Hunt's Lab from these samples. The two sets had different randomly assigned ID numbers. The first set was shipped in parallel to Blackburn's lab and Aviv's Lab. The second set was shipped approximately 2 months later only after LTL measurements for the first set had been completed and results transmitted electronically to Hunt for statistical analysis. In this way, both labs ‘blindly’ performed the telomere length/DNA content measurements on two occasions.
Telomere DNA content by qPCR
The telomere length measurement assay was adapted from the published original method by Cawthon (2,3). The telomere thermal cycling profile consisted of: Cycling for T (telomeric) PCR: denature at 96°C for 1 s, anneal at 54°C for 60 s, with fluorescence data collection, 30 cycles. Cycling for S (single copy gene) PCR: denature at 95°C for 15 s, anneal at 58°C for 1 s, extend at 72°C for 20 s, eight cycles; followed by denature at 96°C for 1 s, anneal at 58°C for 1 s, extend at 72°C for 20 s, hold at 83°C for 5 s with data collection, 35 cycles.
The primers for the telomere PCR were tel1b [5′-CGGTTT(GTTTGG)5GTT-3′], used at a final concentration of 100 nM, and tel2b [5′-GGCTTG(CCTTAC)5CCT-3′], used at a final concentration of 900 nM. The primers for the single-copy gene (human beta-globin) PCR were hbg1 [5′ GCTTCTGACACAACTGTGTTCACTAGC-3′], used at a final concentration of 300 nM, and hbg2 [5′-CACCAACTTCATCCACGTTCACC-3′], used at a final concentration of 700 nM. The final reaction mix contained 20 mM Tris–HCl, pH 8.4; 50 mM KCl; 200 µM each dNTP; 1% DMSO; 0.4× Syber Green I; 22 ng Escherichia coli DNA per reaction; 0.4 U of Platinum Taq DNA polymerase (Invitrogen Inc.) per 11 µl reaction; 0.5–10 ng of genomic DNA. Tubes containing 26, 8.75, 2.9, 0.97, 0.324 and 0.108 ng of a reference DNA (from Hela cancer cells) were included in each PCR run so that the quantity of targeted templates in each sample was determined relative to the reference DNA sample by the standard curve method. Each concentration of the reference DNA was run as quadruplets and samples were run as triplicates.
To control for inter-assay variability, eight control DNA samples from cancer cell lines were included in each run. The cell lines included 293T, H1299, UMUC3 and UMUC3 cells infected with a lentiviral construct containing the telomerase RNA gene to extend telomeres harvested at various population doublings after infection. In each assay batch, the T/S ratio of each control DNA was divided by the average T/S for the same DNA from 10 runs to obtain a normalizing factor. This was done for all eight samples and the average normalizing factor for all eight samples was used to correct the participant DNA samples to obtain the final T/S ratio.
The T/S ratio for each sample was measured twice. In this procedure, when the duplicate T/S value and the initial value varied by >7%, the sample was run a third time and the average of two closest values was reported. Typically, in cohorts from human clinical studies, ∼15% of samples have needed to be assayed the third time.
Telomere length measurement by Southern blot analysis of the TRFs
This measurement was performed as described previously (1). First DNA integrity was evaluated by SYBR Green I, after resolving each sample (10 ng) on 1% agarose gel at 200 V for 60 min. Second, samples were digested with restriction enzymes Hinf I (10 U) and Rsa I (10 U; Roche). This cocktail is commonly used in epidemiological studies to generate the TRFs. Digested DNA samples (3 µg each), and DNA ladders (1-kb DNA ladder plus λ DNA/Hind III fragments; Invitrogen, Carlsbad, CA) were resolved on 0.5% agarose gels. After 16 h, the DNA was depurinated for 15 min in 0.25 N HCl, denatured 30 min in 0.5 M NaOH/1.5 mol/l NaCl and neutralized for 30 min in 0.5 mol/l Tris, pH 8/1.5 M NaCl. The DNA was transferred for 1 h to a positively charged nylon membrane (Roche) using a vacuum blotter (Boeckel Scientific, Feasterville, PA). Membranes were hybridized at 65°C with the DIG labeled telomeric probe overnight in 5× SSC, 0.1% Sarkosyl, 0.02% SDS and 1% blocking reagent (Roche). They were washed three times at room temperature in 2× SSC, 0.1% SDS each for 15 min and once in 2× SSC for 15 min. The DIG-labeled probe was detected by the DIG luminescent (Roche) and exposed on X-ray film. All autoradiographs were scanned, and the TRF signal was digitized. The optical density values versus DNA migration distances were converted to optical density (adjusted for background)/molecular weight versus molecular weight [for more details, see (1)]. Each set of samples was run on duplicates resolved on different gels.
To compute the mean TRF length, we used the following equation:
where ODi = optical density at position i and Li is TRF length at position i.
Statistical analysis
Coefficients of variation were calculated for the duplicate samples from each of the 50 subjects for each method using the pooled standard deviation of the duplicates divided by the overall mean of the 100 measurements. Linear regression was used to estimate the r between the two methods and to estimate the slopes of LTL with age. A quadratic term was included in the linear models to test whether or not the relationship between results generated by the two methods was linear. No a priori assumptions were made regarding the behavior of the curve beyond the data points.
RESULTS
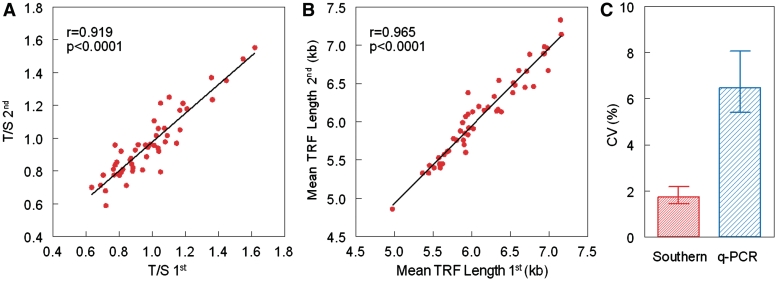
Figure 1 displays correlations between the means of the replicates of the LTL measurements for the 50 samples in the second versus the first set of measurements (qPCR results, Figure 1A; Southern blot results of mean TRF length, Figure 1B). Clearly, both the qPCR and Southern blots display highly reproducible results, as expressed in r > 0.9. On the two occasions, the inter-assay CV measurement for the qPCR was 6.45%, while that of the Southern blots was 1.74% (Figure 1C).
Figure 1.
Correlations between the first set of LTL measurements and second set of measurements by qPCR (A) and Southern blot analysis (B) and the inter-assay variation ±95% confidence interval of the two methods (C).
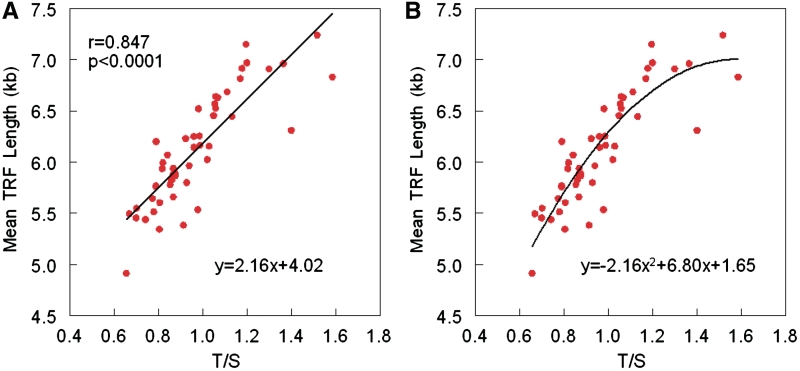
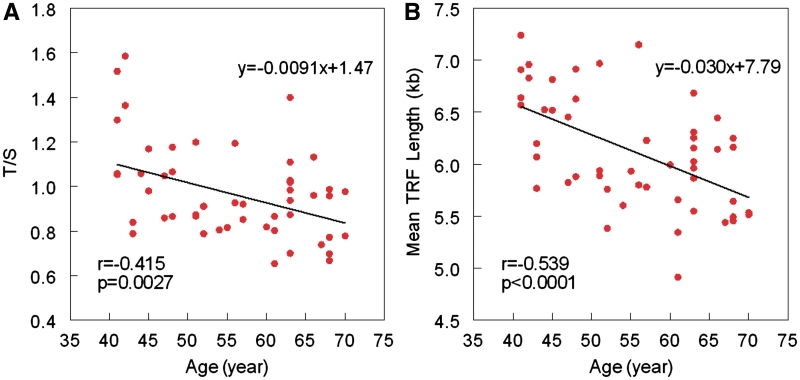
Figure 2 displays the relation between the mean TRF length versus the T/S ratio. The Figure 2A shows the linear regression of this relation. However, a quadratic term that was added to the linear model was statistically significant (P = 0.003) (Figure 2B). The adjusted R2 for the model improved slightly from 0.703 to 0.748. Figure 3 shows the age-dependency of telomere lengths across the age range of the donors based on results generated by the qPCR (Figure 3A) and those generated by the Southern blot method (Figure 3B). Age explained 17.2% of the inter-individual variation in the T/S ratio and 29% of the inter-individual variation in the mean TRF length.
Figure 2.
Linear (A) and curvilinear (B) models depicting the relation between the mean TRF length and the T/S ratio.
Figure 3.
Cross sectional analysis of age-dependent telomere shortening based on the qPCR (A) and Southern blots (B).
DISCUSSION
To our knowledge, this is the first study that has compared, as impartially as possible, measurements of LTL by qPCR versus Southern blot analysis in two laboratories with considerable experience in performing these two methods. In comparing the methods one must balance precision and other features of a given telomere length measurement method with its practicality.
It is important to point out that the pre-analytical factors that impact the Southern Blot analysis and qPCR for telomere length measurement could be very different. Whereas the integrity of genomic DNA is crucial for Southern blots, it is less critical for qPCR. However, residual PCR inhibitors that may still be present even after DNA purification could contribute to variability in qPCR. The influences of different types of blood collection tubes, as well as individual donor differences have been observed in a separate study (J. Lin, J. Cheon, E. Epel and E. Blackburn, manuscript in preparation). The present study used DNA samples that have been examined for their DNA length (lack of detectable low-molecular weight DNA fragments, as assayed on agarose gels) as a quality control for sample integrity, but we did not address pre-analytical conditions, including blood collection and storage, DNA purification and storage, that might contribute to qPCR variability.
As shown in Figure 1, we observed high correlations between the two sets of measurements of LTL performed by either the Southern blots of the TRFs or telomere DNA content generated by the qPCR. The measurement error, defined by the inter-assay CV, which on the two occasions tested here, were 6.45% for the qPCR, and 1.74% for the Southern blots. It is useful to regard this error in the context of the relation between LTL and age in the present study, and specifically with respect to epidemiological research and ultimately clinical practice.
The larger error of the qPCR relative to the Southern blots could explain the findings that age, across 30 years, accounts for 17.2% of the inter-individual variation in the mean T/S (qPCR method), while it explains 29% of this variation in the mean TRF length (Southern blot method) (Figure 3). Thus, we suggest that measurement error should be a primary consideration, as well as any unique demographic or biological features, in large-scale cross-sectional studies that show little or no LTL shortening over a wide age range.
As an illustrative example, consider an adult whose LTL = 6 kb. Measurement errors of 2 and 6% of 6 kb amount to 120 and 360 nt, respectively. Although the average rate of LTL shortening in adults is highly variable, in cross-sectional studies it on average amounts to ∼30 nt/year, as also shown in Figure 3. In equivalence of age-dependent LTL attrition, the error of the measurement by the Southern blot amounts to 4 years, while that of the qPCR amounts to 13 years. However, it is also important to note that the rate of LTL change over time has been shown in longitudinal studies to have a strong dependence upon baseline LTL, and thus base-line TL should also be co-varied in longitudinal analyses of telomere shortening (14–16). Thus, in cross-sectional studies the shortening rate is only inferred indirectly based on group analysis, and masks the findings that the rate of LTL shortening in individual adults measured longitudinally is highly variable.
In addition, age-dependent LTL attrition is only one component of LTL dynamics; the other component is birth LTL. The range of inter-individual variation in LTL in adults after age adjustment, in cross-sectional studies, is ∼3–4 kb, and an important contributor to this variation might be not only the rate of age-dependent LTL shortening during adult life, but the variation in LTL across newborns, which is ∼4–6 kb (11,17–19). Another significant contributor to LTL in adults is likely to be the rate of telomere attrition in the period from birth to early adulthood, which is much higher than that seen in adults in both humans (11,17,20) and non-human primates (21). Given this wide inter-individual variation in LTL, a 6% error in telomere length measurement might be acceptable in epidemiological research and clinical practice, if the goal of LTL measurement is not to determine the rate of LTL shortening but to rank individuals for susceptibility to a given disease based on having relatively short, average or long telomeres for their age.
Figure 2 displays deviation from linearity of the relation between the mean length of the TRFs and the T/S ratio, which was observed in other studies (2,22,23). The addition of a quadratic term increased the fit of the data to the regression line. No deviation from linearity is observed between the mean TRFs and telomere DNA content generated by a non-PCR method of the strictly canonical telomeric repeats region of telomeres, suggesting that this phenomenon is PCR mediated (23).
It is important to regard the ramifications of this work within the broader framework of epidemiology and clinical practice. Precision alone might not always justify the use of Southern blot method of the TRFs, since it is labor-intensive, costly, requires a large amount of DNA, cannot be performed in DNA that is even modestly degraded and requires expertise that might be available only in a few laboratories (1). Furthermore, while the qPCR method measures strictly the canonical region of telomeres, the TRF length data include long stretches of the non-canonical region, which might be variable across the general population due to polymorphism in the restriction sites that are the targets of the enzymes generating the TRFs.
Epidemiologists have employed various methods to measure telomere length but there has been less discussion of the shortcomings of the methods used. Moreover, measurements errors of the same method often differ among laboratories. Therefore, the findings of the present study by two laboratories with considerable experience in qPCR and Southern blots might not represent those generated in less experienced laboratories. For these reasons, large-scale epidemiological studies comparing the Southern blot analysis versus qPCR and possibly other methods (measured in a number of laboratories for each method) will bolster understanding of aging-related maladies and gage the risk of developing them. This will more fully address matters related to the question: which is the optimal method to measure telomere length? Without such studies, identifying the optimal method for assessing telomere length unbiased by preconceived views will be hampered.
FUNDING
Funding for open access charge: National Institutes of Health (grant numbers AG18734, AG030678 and AG20132 to A.A.); Barbro and Bernard Fund grant (to E.H.B.).
Conflict of interest statement. Drs. Blackburn and Lin are founders of Telome health.
REFERENCES
- 1.Kimura M, Stone RC, Hunt SC, Skurnick J, Lu X, Cao X, Harley CB, Aviv A. Measurement of telomere length by the southern blot analysis of the terminal restriction fragment lengths. Nat. Protoc. 2010;5:1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 2.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat. Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 5.Willeit P, Willeit J, Brandstätter A, Ehrlenbach S, Mayr A, Gasperi A, Weger S, Oberhollenzer F, Reindl M, Kronenberg F, et al. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler. Thromb. Vasc. Biol. 2010;30:1649–1656. doi: 10.1161/ATVBAHA.110.205492. [DOI] [PubMed] [Google Scholar]
- 6.McGrath M, Wong JY, Michaud D, Hunter DJ, De Vivo I. Telomere length, cigarette smoking, and bladder cancer risk in men and women. Cancer Epidemiol. Biomarkers Prev. 2007;16:815–819. doi: 10.1158/1055-9965.EPI-06-0961. [DOI] [PubMed] [Google Scholar]
- 7.De Vivo I, Prescott J, Wong JY, Kraft P, Hankinson SE, Hunter DJ. A prospective study of relative telomere length and postmenopausal breast cancer risk Cancer Epidemiol. Biomarkers Prev. 2009;18:1152–1156. doi: 10.1158/1055-9965.EPI-08-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Terry MB, Gurvich I, Liao Y, Senie RT, Santella RM. Short telomere length and breast cancer risk: a study in sister sets. Cancer Res. 2007;67:5538–4554. doi: 10.1158/0008-5472.CAN-06-3490. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick AL, Kronmal RA, Gardner JP, Psaty BM, Jenny NS, Tracy RP, Walston J, Kimura M, Aviv A. Leukocyte telomere length and cardiovascular disease in the Cardiovascular Health Study. Am. J. Epidemiol. 2007;165:14–21. doi: 10.1093/aje/kwj346. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff C, Petersen HC, Graakjaer J, Andersen-Ranberg K, Vaupel JW, Bohr VA, Kølvraa S, Christensen K. No association between telomere length and survival among the elderly and oldest old. Epidemiology. 2006;17:190–194. doi: 10.1097/01.ede.0000199436.55248.10. [DOI] [PubMed] [Google Scholar]
- 11.Rufer N, Brümmendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L, Schulzer M, Lansdorp PM. Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. J. Exp. Med. 1999;190:157–167. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat. Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 13.Zheng YL, Loffredo CA, Shields PG, Selim SM. Chromosome 9 arm-specific telomere length and breast cancer risk. Carcinogenesis. 2009;30:1380–1386. doi: 10.1093/carcin/bgp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farzaneh-Far R, Lin J, Epel E, Lapham K, Blackburn E, Whooley MA. Telomere length trajectory and its determinants in persons with coronary artery disease: longitudinal findings from the heart and soul study. PLoS One. 2010;5:e8612. doi: 10.1371/journal.pone.0008612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordfjäll K, Svenson U, Norrback KF, Adolfsson R, Lenner P, Roos G. The individual blood cell telomere attrition rate is telomere length dependent. PLoS Genet. 2009;5:e1000375. doi: 10.1371/journal.pgen.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aviv A, Chen W, Gardner JP, Kimura M, Brimacombe M, Cao X, Srinivasan SR, Berenson GS. Leukocyte telomere dynamics: longitudinal findings among young adults in the Bogalusa Heart Study. Am. J. Epidemiol. 2009;169:323–329. doi: 10.1093/aje/kwn338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frenck RW, Jr, Blackburn EH, Shannon KM. The rate of telomere sequence loss in human leukocytes varies with age. Proc. Natl Acad. Sci. USA. 1995;95:5607–5610. doi: 10.1073/pnas.95.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuda K, Bardeguez A, Gardner JP, Rodriguez P, Ganesh V, Kimura M, Skurnick J, Awad G, Aviv A. Telomere length in the newborn. Pediatr. Res. 2002;52:377–381. doi: 10.1203/00006450-200209000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Akkad A, Hastings R, Konje JC, Bell SC, Thurston H, Williams B. Telomere length in small-for-gestational-age babies. BJOG. 2006;11:318–323. doi: 10.1111/j.1471-0528.2005.00839.x. [DOI] [PubMed] [Google Scholar]
- 20.Sidorov I, Kimura M, Yashin A, Aviv A. Leukocyte telomere dynamics and human hematopoietic stem cell kinetics during somatic growth. Exp. Hematol. 2009;37:514–524. doi: 10.1016/j.exphem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Baerlocher GM, Rice K, Vulto I, Lansdorp PM. Longitudinal data on telomere length in leukocytes from newborn baboons support a marked drop in stem cell turnover around 1 year of age. Aging Cell. 2007;6:121–123. doi: 10.1111/j.1474-9726.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlenbach S, Willeit P, Kiechl S, Willeit J, Reindl M, Schanda K, Kronenberg F, Brandstätter A. Influences on the reduction of relative telomere length over 10 years in the population-based Bruneck Study: introduction of awell-controlled high-throughput assay. Int. J. Epidemiol. 2009;38:1725–1734. doi: 10.1093/ije/dyp273. [DOI] [PubMed] [Google Scholar]
- 23.Kimura M, Aviv A. Measurement of telomere DNA content by dot blot analysis. Nucleic Acids Res. 2011;39:e84. doi: 10.1093/nar/gkr235. [DOI] [PMC free article] [PubMed] [Google Scholar]