Abstract
In the Drosophila germline, retrotransposons are silenced by the PIWI-interacting RNA (piRNA) pathway. Telomeric retroelements HeT-A, TART and TAHRE, which are involved in telomere maintenance in Drosophila, are also the targets of piRNA-mediated silencing. We have demonstrated that expression of reporter genes driven by the HeT-A promoter is under the control of the piRNA silencing pathway independent of the transgene location. In order to test directly whether piRNAs affect the transcriptional state of retrotransposons we performed a nuclear run-on (NRO) assay and revealed increased density of the active RNA polymerase complexes at the sequences of endogenous HeT-A and TART telomeric retroelements as well as HeT-A-containing constructs in the ovaries of spn-E mutants and in flies with piwi knockdown. This strongly correlates with enrichment of two histone H3 modifications (dimethylation of lysine 79 and dimethylation of lysine 4), which mark transcriptionally active chromatin, on the same sequences in the piRNA pathway mutants. spn-E mutation and piwi knockdown results in transcriptional activation of some other non-telomeric retrotransposons in the ovaries, such as I-element and HMS Beagle. Therefore piRNA-mediated transcriptional mode of silencing is involved in the control of retrotransposon expression in the Drosophila germline.
INTRODUCTION
Silencing of mobile elements in germ cells depends on a distinct class of 24- to 30-nt long RNAs called PIWI-interacting RNAs (piRNAs) associated with Argonaute proteins from the PIWI subfamily (1,2). piRNA pathway mutations lead to overexpression and mobilization of retrotransposons in the germline (1,3,4). However, the mechanism of the piRNA-mediated gene silencing in Drosophila remains obscure. Short RNAs were shown to target the associated protein complex to degrade complementary mRNA (5). Short-RNA-mediated heterochromatin assembly was described in fission yeast, plants, and ciliates (6–8). In this case, short RNAs guide histone methyltransferase to the target locus to methylate lysine 9 of histone H3 (H3K9) with subsequent binding of heterochromatic protein 1 (HP1) or its homolog (Swi6 in fission yeast). Heterochromatization diminishes the transcriptional capacity of the target locus resulting in transcriptional silencing. How do piRNAs maintain transposon silencing in the germline? In mice, transposon-specific piRNAs drive transposon promoter methylation in the male germline (2,9). The three Drosophila PIWI proteins, PIWI, Aubergine (Aub) and Argonaute3 (Ago3) which are found in the germline, are associated with short RNAs in vivo and demonstrate cleavage activity of the complementary RNA in vitro, implicating their involvement in the post-transcriptional degradation of mRNA (1,10,11). Despite numerous studies describing the piRNA-mediated repression of retrotransposons in the Drosophila germline, only a few of them suggest mechanism of repression. piRNA-mediated post-transcriptional destabilization of retroelement transcripts was shown (12). A post-transcriptional mechanism was proposed to be involved in the piRNA-mediated silencing of testis-expressed Stellate repeats (13) and the non-LTR retrotransposon I-element (14). At the same time, the piRNA-mediated silencing by transgenes located at subtelomeric regions was shown to be dependent on such heterochromatic components as HP1 or its partner, Su(var)3–7 (15). Changes of the chromatin structure of retroelements caused by piRNA pathway mutations in spn-E gene (16) also indicate that piRNAs may target chromatin modifications. Most probably, both transcriptional and post-transcriptional pathways may be involved in the piRNA-mediated silencing of the same target gene. Klattenhoff and Theurkauf (4) proposed a model of compartmentalization of the piRNA pathway. According to this model, cytoplasmic proteins Aub and Ago3 catalyze the RNA cleavage in the cytoplasm, most likely in the perinuclear structure nuage operating at the post-transcriptional level. Being localized in the nucleus, PIWI might be involved in the transcriptional silencing of its target locus. Based on the well-known scheme of the siRNA-mediated heterochromatin formation in yeast, there were attempts to directly connect piRNA pathway components with heterochromatin marks such as HP1 and histone H3 modifications (H3K9Me2 and H3K9Me3) in Drosophila. Such approach results in the appearance of several contradicting facts. PIWI was shown to be a component of chromatin (17) which mediates HP1 recruitment to the heterochromatic loci (18,19). On the other hand, PIWI was described as a nucleoplasmic protein which is not associated with chromatin (16,20). Formation of heterochromatin and its association with HP1 was shown to be independent of the piRNA pathway (21). A reasonable explanation for this apparent contradiction is that different short RNA targets are regulated by distinct mechanisms depending of their genomic location. In our study we decided to directly estimate the transcriptional status of retroelements in the germline of the piRNA pathway mutants rather than to look for the vestiges of known chromatin modifying pathways in attempt to connect them with piRNA pathway. The main objects of this study are the Drosophila telomeric retrotransposons.
Telomeres in Drosophila are maintained by transpositions of specialized telomeric retroelements rather than by the telomerase activity that adds short DNA repeats to chromosome ends in other eukaryotes. HeT-A, TART and TAHRE non-LTR retrotransposons are found at Drosophila telomeres in mixed tandem head-to-tail arrays; their 3′-ends are orientated towards the centromere (22–24). HeT-A and TART elements were shown to be transcribed bidirectionally from internal sense and antisense promoters (25–27). The abundance of telomeric retroelement transcripts, both sense and antisense, as well as the frequency of their transpositions on the chromosome ends, is controlled by a piRNA-mediated mechanism (28,29). The mechanism of piRNA-mediated telomeric retrotransposon silencing in the Drosophila germline remains elusive. The question about the mechanism of the telomeric retroelement silencing is of great interest because, in the case of transcriptional silencing, it might concern such an important process as formation of the telomeric chromatin which protects chromosome ends from fusion and is involved in meiotic and mitotic telomere behavior in different organisms. In Schizosaccharomyces pombe, centromeric and subtelomeric repeats of the transposon origin impact the siRNA-mediated spreading of heterochromatin through the activity of HP1 family proteins, histone methyltransferases, and the histone deacetylase complex (30–32).
Here, we have demonstrated that expression of the reporter genes driven by the HeT-A promoter is activated in piRNA mutants independently of where a reporter is located, in euchromatin or at the chromosome end. Furthermore, nuclear run-on analysis (NRO) was applied to determine directly whether the piRNAs induce transcriptional silencing of the telomeric retrotransposons. We have demonstrated piRNA-mediated transcriptional repression of endogenous HeT-A and TART retroelements, HeT-A-containing constructs located out of natural telomeres as well as some other non-telomeric retrotransposons. The data of NRO assay are in accord with chromatin immunoprecipitation (ChIP) analysis of histone H3 modifications which are characteristics of transcriptionally active loci. Although the protein machinery that transmits a signal from piRNA to chromatin complexes remains elusive, these data provide strong evidence of the piRNA-mediated transcriptional silencing in the Drosophila germline.
MATERIALS AND METHODS
Drosophila strains
Strains bearing spindle-E (spn-E) mutations were ru1 st1 spn-E1 e1 ca1/TM3, Sb1 es and ru1 st1 spn-Ehls3987 e1 ca1/TM3, Sb1 es. aub mutants were aubQC42/CyO and aubHN/CyO. vasa locus mutations were vigEP812 and vasPH165. These mutations affect two genes, vig and vasa, located at the same locus (33,34). We used piwi2 and piwi3 mutations. P-element-mediated germ line transformation of Df (1)w67c23(2), y embryos was performed according to standard protocol (35). Multiple transgenic Drosophila lines were established (Supplementary ‘Materials and Methods’ section). si_piwi flies were F1 of the crossing between #101658 line bearing piwi hairpin construct (VDRC Stock Center) and #25751 line (P{UAS-Dcr-2.D}1, w1118, P{GAL4-nos.NGT}40, Bloomington Stock Center) providing GAL4 expression under the control of the germline-specific promoter of nanos gene. Control flies were F1 of the crossing of #60000 (w1118, VDRC Stock Center) and #25751 (Bloomington Stock Center).
Reporter construct design
Constructs were made in the CaSpeR-AUG-β-gal vector. The HeT-A sequence was derived from clone z2 containing the fragment of the HeT-A element attached to a terminal deleted X chromosome (29). Clone z2 was used as a template to amplify 434 bp of the HeT-A promoter region. HeT-A PCR fragment obtained with 5′-ATGAATTCATCCATCGCCCGCAACATG-3′ and 5′-ATGGATCCTTTGCTGGTGGAGGTACGGAGAC-3′ primers was inserted into the EcoRI and BamHI polylinker sites of CaSpeR-AUG-β-gal.
In situ RNA hybridization, DNA FISH and immunohistochemistry
In situ RNA analysis was carried out according to the previously described procedure (36) using DIG-labeled strand-specific HeT-A riboprobes and alkaline phosphatase (AP)-conjugated (diluted at 1/2000) anti-digoxigenin (DIG) antibodies (Roche). The HeT-A probe contained a fragment of the ORF (nucleotides 4330–4690 of the GenBank sequence DMU06920). The combination of protein and DNA localization was done according to the previously described procedure (37). Primary polyclonal rabbit anti-HP1 antibodies (Covance, diluted at 1:1000) were used. The probes used for DNA FISH analysis were: yellow, PCR amplified fragment using primers 5′-CTGATCACCTTGGTGACGCCG-3′ and 5′-AATTCGCGTATCCGTGGTCAAGTC-3′; TART, cloned fragment of TART ORF2, corresponding to 434–2683 nt in GenBank sequence DMU02279; HeT-A, cloned fragment of HeT-A ORF, corresponding to 1746–4421 nt in GenBank sequence DMU 06920. To stain DNA, ovaries were incubated in PBS containing 0.5 µg/ml DAPI. The confocal microscope, Zeiss LSM 510 Meta, was used to visualize the ovaries.
NRO assay
Ovaries from ∼300 females were dissected and homogenized with a Dounce homogenizer (Sigma) in 300 µl of buffer HB [15 mM HEPES, pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.05% NP40, 1× complete protease inhibitor (Roche)] containing 0.35 M sucrose. All steps of nuclei isolation and washing were done on ice with ice-cold solutions. Homogenized solution was filtered through Miracloth membrane (Calbiochem). The filtrate was overlaid on a cushion containing 0.8 ml of 0.8 M sucrose-HB as the bottom phase and 0.3 ml of 0.35 M sucrose-HB as upper phase and centrifuged at 10 000 rpm for 10 min in Eppendorf centrifuge at 4°C. The supernatant was discarded and nuclei pellet was resuspended in 0.5 ml of RB buffer (5 mМ Tris, pH 8.0, 5 mМ MgCl2,150 mМ KCl, 1 mМ DTT, 1× complete protease inhibitor (Roche) and centrifuged at 6000 rpm for 10 min at 4°C. The pellet was resuspended in 100 µl RB containing 0.5 mM of each rATP, rGTP, rCTP, 5-bromouridine 5′-triphosphate (BrUTP, Sigma) and 1 U/µl of RNasin (Promega) and was incubated at 25°C for 30 min. After the run-on reaction RNA was isolated using TRIzol LS reagent (Invitrogen) according to the manufacturer’s guidelines. NRO RNA fraction was immuno-purified using anti-Bromodeoxyuridine agarose conjugate IgG (Santa Cruz Biotechnology). Agarose beads were blocked in IP buffer (150 mМ NaCl, 50 mМ Tris–HCl, pH 8.0, 0.05% NP40, 1 mМ EDTA, 1× complete protease inhibitor) containing 0.1% polyvinylpyrrolidone and 1 mg/ml ultrapure BSA (Ambion). NRO-RNAs were added to 50 ml beads in 500 ml of IP buffer containing 1 U/µl of RNasin (Promega), and allowed to bind 1 h. The beads were washed in IP buffer five times. RNA was extracted using TRIzol reagent (Invitrogen) and precipitated. Descriptions of the steps of NRO assay as well as estimation of purity of NRO RNA fraction are done in Supplementary Figures S1–S3 and Supplementary ‘Materials and Methods’ section. Three to six independent NRO experiments were done for every genotype.
ChIP
A total of ∼100 pairs of ovaries were dissected for every IP reaction. ChIP was performed according to the published procedure (38). Chromatin was immunoprecipitated with the following antibodies: anti-dimethyl-histone H3 Lys4 (Upstate) and anti-dimethyl-histone H3 Lys79 (Millipore). Quantitative PCR was conducted on DT-96 machine from DNA Technology, Russia. Quantities of target genomic regions precipitated by different antibodies were normalized to a fragment of transcriptionally inactive intergenic spacer in the 60D region. The ratio of these values calculated for homozygous and heterozygous spn-E mutants or si_piwi and control w1118 flies represents at the histograms. Eight serial 3-fold dilutions of input DNA of corresponding line were amplified in duplicates with each primer pair to make a standard curves. Standard deviation of duplicate PCR measurements for two independent ChIP experiments was calculated to determine the error of measurement. rp49 gene was included in the analysis as positive control for transcriptionally active chromatin. Levels of H3K79me2 and H3K4me2 marks at rp49 are similar in spn-E/+ and spn-E/spn-E mutants as well as in w1118 and si_piwi flies and display 30- to 80-fold enrichment compared to 60D intergenic spacer in different ChIP assays (data not shown). Normalization of ChIP and NRO data on the retrotransposon copy number was done using ChIP input data for each genotype. No substantial differences in the retrotransposon copy number were observed between spn-E/+ and spn-E/spn-E or w1118 and si_ piwi flies.
RESULTS
HeT-A/yellow endogenous reporter gene is a target for the piRNA-mediated silencing
Broken chromosome ends, as well as normal chromosomes, can be elongated by attachments of telomeric retroelements. We used Drosophila lines with HeT-A attachments to a truncated X chromosome with a break in the yellow locus to study expression of an endogenous reporter yellow gene in the piRNA pathway mutants. If the HeT-A attachment occurred after degradation of the yellow promoter, the yellow gene is transcribed from the HeT-A promoter located at its 3′ region (39,40). This results in a yellow-to-black change in aristae pigmentation which serves as a marker of terminal attachment events.
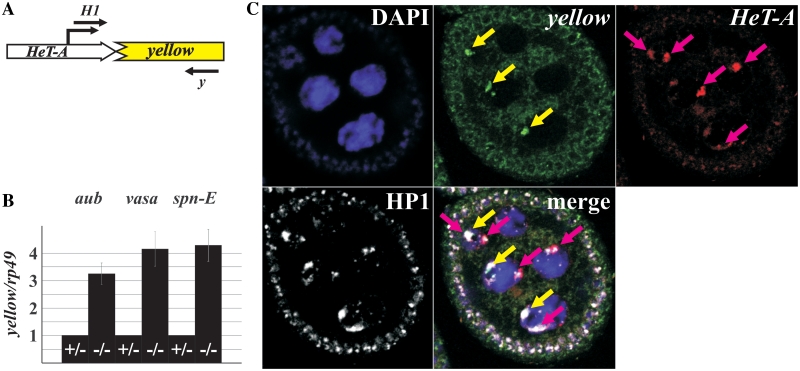
Yellow expression was analyzed in three independent lines with HeT-A attachments at different positions of the degraded yellow promoter region in the presence of piRNA pathway gene mutations. In such lines, a HeT-A/yellow fusion transcript is formed. Here, we present data for z2 line, in which HeT-A attachment has occurred 50-bp downstream of the yellow transcription start site. The 5′ RACE analysis using yellow-specific primers revealed HeT-A/yellow 5′-ends at positions −30- and −95-bp upstream of the HeT-A polyadenylation site in the z2 line. A major transcription site was mapped to position −30 because transcription at the −30 position is initiated about ten times more efficiently than that at the −95 position (Supplementary Figure S4). The RT–PCR analysis revealed that the abundance of HeT-A/yellow transcripts is under control of the piRNA silencing pathway components Spn-E, Aub and a product of the vasa locus (Figure 1); it is several times more abundant in the ovaries of transheterozygous mutants as compared with heterozygous flies. It should be noted, that we obtained similar effects for all studied lines with the HeT-A attachment to the truncated yellow (data not shown).
Figure 1.
HeT-A/yellow reporter gene expression in the piRNA pathway mutants. (A) A schematic representation of the HeT-A/yellow reporter construct. Position of the transcription start site is indicated by broken arrow. Primers used in the RT–PCR analysis are shown. (B) RT–qPCR analysis of the HeT-A/yellow transcript amount in the ovaries of spn-E, aub, and vasa mutants. Bars of histograms represent a ratio of HeT-A/yellow to rp49 transcript abundance in the ovaries of transheterozygous aubQC42/aubHN (–/–), spn-E1/spn-Ehls3987 (–/–), or vigEP812/vasPH165 (–/–) flies related to this ratio in aubQC42/CyO (+/−), spn-E1/TM3 (+/−), or vigEP812/CyO (+/−) females. (C) Terminally deleted chromosome recruits the telomere capping protein HP1 in ovaries. Immunostaining of HP1 (white) reveals numerous signals in the ovarian nuclei. DNA FISH was performed with yellow (green signals indicated by yellow arrows) or HeT-A (red signals indicated by pink arrows) DNA probes. HP1 locates at the ends of normal chromosomes (HeT-A staining) as well as of terminally deleted chromosome (yellow staining) of the z2 line. DNA is stained with DAPI (blue).
In Drosophila, chromosome ends are capped by a protein complex including HP1 (41,42). Despite the absence of telomeric retrotransposon sequences at terminally deleted chromosomes, HP1 binds the terminus of these chromosomes in salivary glands, thus providing the chromosome capping (41,43). An extensive co-localization between a signal from anti-HP1 antibodies with the yellow and HeT-A staining is seen on ovarian nurse cell chromosomes of the z2 line. Thus, HP1, a main component of the cap, is associated with the normal chromosome ends as well as with broken X chromosome in ovaries (Figure 1C). This fact allows us to consider that HeT-A/yellow reporter localizes in a protein environment similar to that of endogenous chromosome ends.
Expression of the HeT-A/lacZ transgenic construct is upregulated in the ovaries of piRNA pathway mutants
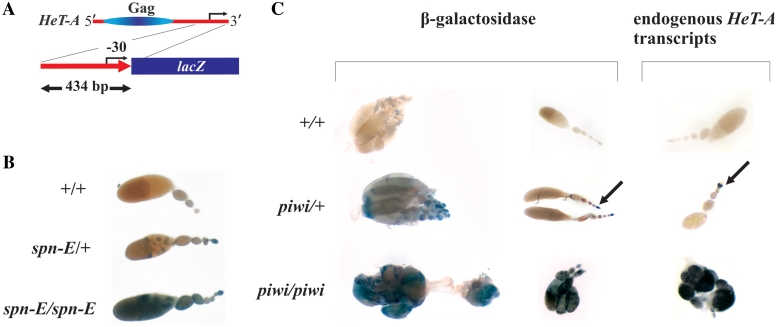
To address how the chromatin context affects the piRNA-mediated transgene silencing, we have studied the expression of a reporter gene driven by the HeT-A promoter and located in euchromatin, in the ovaries of piRNA pathway mutants. A construct containing a 3′-UTR fragment of the HeT-A copy derived from the z2 line, fused to lacZ was used to generate transgenic flies (Supplementary ‘Materials and Methods’ section). The expression of transgenic β-galactosidase was studied in the ovaries of piRNA pathway mutants. We observed derepression of the transgene in the ovaries of spn-E and piwi mutants in three independent lines containing the transgene at different euchromatic sites (Figure 2B and C). Increase of the β-galactosidase activity in spn-E mutants correlates with an elevated level of HeT-A/lacZ RNA according to the RT–PCR analysis (Supplementary Figure S5).
Figure 2.
HeT-A/lacZ reporter gene expression in the piRNA mutants. (A) A schematic representation of the HeT-A/lacZ reporter construct. Position of the transcription start site is indicated by arrow. (B) X-gal staining of the β-galactosidase in the ovaries of Df (1)w67c23(2), y (+/+), spn-E1/TM3 (spn-E/+) and spn-E1/spn-Ehls3987 (spn-E/spn-E) flies bearing HeT-A/lacZ transgene (H95 line). Identical results were obtained for H12 and H124 transgenic lines. (C) Comparison of the endogenous HeT-A transcript localization (on the right) and β-galactosidase staining (on the left) in the ovaries of Df (1)w67c23(2), y (+/+), piwi2/CyO (piwi/+), piwi2/piwi3 (piwi/piwi) flies bearing HeT-A/lacZ transgene (H12 line). Arrows indicate staining in the germarium.
It is noteworthy, that expression pattern of the reporter gene reproduces the expression patterns of endogenous telomeric HeT-A elements in the ovaries. Figure 2 shows distribution of endogenous HeT-A transcripts in the ovaries of wild-type flies, piwi/+ and piwi/piwi females in comparison with the transgenic β-galactosidase staining in the flies of the same genotypes. No staining was observed in the wild-type ovaries. In heterozygous piwi mutants, lacZ expression is detected in the germarium, which is similar to the distribution of endogenous HeT-A transcripts (Figure 2C). Immunostaining of β-galactosidase detects it in the germinal cysts of piwi/+ females (Supplementary Figure S6). Thus, the tissue-specific expression of the transgene in ovaries is regulated by the HeT-A promoter region which is present in the construct. Using 5′ RACE analysis, we have shown that a fused HeT-A/lacZ transcript contains ∼30 nt of the HeT-A 3′ region as a result of the presence of transcription initiation site in the HeT-A 3′-UTR (Supplementary Figure S4). We failed to detect a second transcription start site, possibly due to the absence of some regulatory elements in the construct which provide a proper initiation of transcription. Thus, expression of the reporter genes driven by the HeT-A promoter is affected by the piRNA silencing pathway independent of the reporter gene location, in euchromatin or in telomeric heterochromatin.
Transcriptional analysis
Previously, we demonstrated that abundance of telomeric element transcripts of both directions is regulated by the piRNA pathway. HeT-A and TART transcripts of both polarities accumulate in the germ cell nuclei of homozygous piRNA pathway mutants (27). HeT-A and TART transcript hybridization signals in nuclei look like distinct bright dots, which may reflect the accumulation of nascent transcripts at the sites of transcription. To verify this assumption we performed a combined RNA and DNA localization in ovaries using RNA and DNA FISH analysis (Supplementary ‘Materials and Methods’ section). HeT-A sense and TART antisense transcripts were shown to be accumulated at the telomeric loci in spn-E mutants (Supplementary Figure S7). This observation argues in favor of a transcriptional or co-transcriptional mechanism of the piRNA-based expression regulation of endogenous telomeric elements.
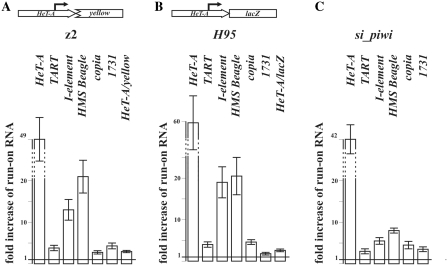
To directly determine whether the piRNAs induce transcriptional silencing of the telomeric retrotransposons, we performed NRO assay. This method is capable of estimating the density of transcriptionally active RNA–polymerase complexes at the locus of interest (44,45). Retrotransposons were shown to be transcribed by RNA polymerase II (46). To isolate fractions of NRO RNA, we performed run-on reaction in the presence of BrUTP followed by immunoprecipitation and purification of BrUTP-labeled nascent RNA (Supplementary Figure S3). NRO RNA was reverse transcribed and used for qPCR analysis. rp49 served as an internal control for the normalization of transcription level values. We estimated transcription levels of endogenous and transgenic telomeric elements in piRNA pathway gene spn-E mutants. The H95 transgenic line and z2 line bearing HeT-A attachment to the broken X-chromosome were used for analysis. For both lines, transcription level of endogenous HeT-A and TART elements was significantly higher in the spn-E/spn-E ovaries than in spn-E/+ ones (Figure 3A and B). HeT-A/lacZ and HeT-A/yellow transcription levels also increased in the ovaries of spn-E mutants (Figure 3A and B). These data indicate that telomeric retrotransposons and reporter constructs bearing HeT-A promoter undergo piRNA-mediated transcriptional silencing.
Figure 3.
NRO analysis of the retrotransposon transcription in the spn-E mutants and in the piwi-knockdown flies. NRO analysis of transcription of the endogenous retrotransposons and reporter genes in the ovaries of z2 line (A) and H95 line (B) bearing spn-E mutations. A schematic representation of the reporter construct is given above the diagram. NRO analysis of the endogenous retrotransposon transcription in the ovaries of si_piwi flies (C). Bars of histograms represent a ratio of genes to be tested to rp49 transcript abundance in the spn-E1/spn-Ehls3987 or si_piwi flies related to this ratio in spn-E1/TM3 or w1118, respectively.
PIWI is a crucial component of piRNA pathway. Its mutations lead to a drastic ovary degeneration impeding experiments with piwi mutants. We applied RNAi approach to repress piwi expression in the germline (see ‘Materials and Methods’ section). In si_piwi flies PIWI was detected only in the follicular cells whereas ovarian germ cells lacked PIWI (Supplementary Figure S8). These flies are sterile in spite of morphologically normal ovary development. RT–PCR analysis demonstrates derepression of different retrotransposons in the si_piwi ovaries (Supplementary Figure S9). piwi knockdown results in the HeT-A and TART retrotransposon transcript accumulation in the ovaries (Supplementary Figure S10). The pattern of their distribution is similar to that in the other piRNA pathway mutants (28,34). HeT-A piRNAs are not detected in the ovaries of si_piwi flies (Supplementary Figure S10). NRO assay of the si_piwi ovaries reveals activation of endogenous HeT-A and TART transcription (Figure 3C).
Thus, piRNA-mediated telomeric retrotransposon silencing is in part transcriptional. Is this phenomenon universal for the retrotransposon silencing in the germline? NRO assay of spn-E mutants and si_piwi ovaries reveals accumulation of run-on transcripts of I-element, HMS Beagle, 1731 and copia retrotransposons, which were described previously as targets of the piRNA-mediated silencing (14,34,47) (Supplementary Figure S9 and Figure 3).
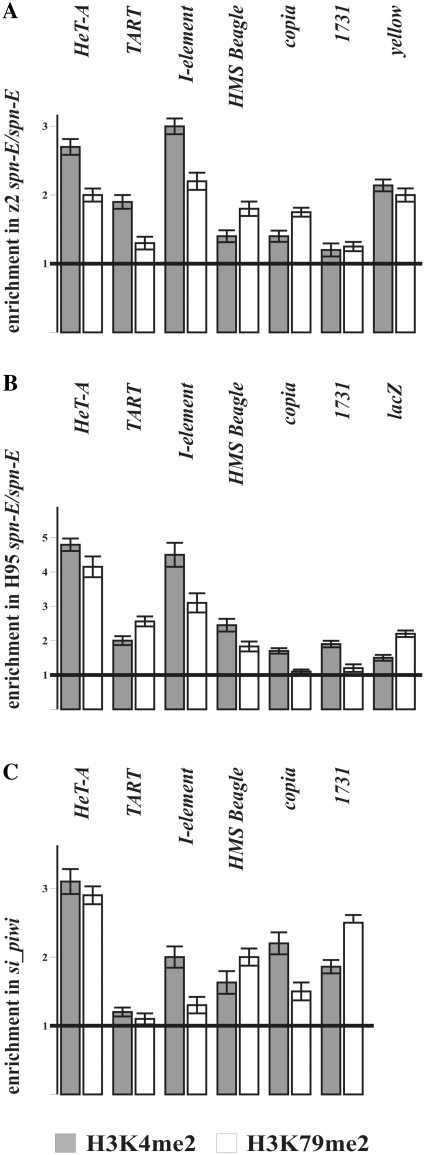
Taking into account the result of NRO assay, we examined the presence of transcriptionally active chromatin marks on the retrotransposon sequences in the piRNA pathway mutants. We focused on the two modifications of histone H3, dimethylation of lysine 79 (H3K79me2) and dimethylation of lysine 4 (H3K4me2). These two histone modifications were previously reported to be tightly linked to RNA polymerase II activity and correlate with the level of transcription (48). We performed ChIP experiments of chromatin of ovarian tissues comparing spn-E1/spn-Ehls3987 mutants to spn-E1/+ heterozygous flies. We observed enrichment of H3K79me2 and H3K4me2 transcription elongation marks within HeT-A, TART, HMS Beagle and I-element sequences in the ovaries of spn-E1/spn-Ehls3987 (Figure 4A and B). Data concerning copia and 1731 expression are ambiguous. These retrotransposons express not only in the germline but also in follicular cells at a substantial level (data not shown). This could mask transcriptional effects (if any) occurring in the germline. ChIP experiments performed in ovaries of si_ piwi flies show increase of both histone marks at the retrotransposon chromatin compared to control w1118 line (Figure 4C). HeT-A/lacZ and HeT-A/yellow transgenes are also enriched by H3K79me2 and H3K4me2 marks in the H95 and z2 lines, respectively, bearing spn-E mutations (Figure 4A and B). ChIP analysis was done for T6 line bearing spn-E mutations. In this line, HeT-A attachment to the truncated X chromosome has occurred 24-bp upstream of the yellow transcription start site just downstream of the TATA-box. The data obtained are in accord with the results obtaining for the z2 line corroborating the transcriptional activation of the endogenous retrotransposons and HeT-A/yellow reporter in the ovaries of the spn-E mutants (data not shown).
Figure 4.
The level of transcriptionally active chromatin marks increases at the retrotransposon sequences in the ovaries of piRNA pathway mutants. ChIP of retrotransposon and transgene sequences in the z2 line (A) and H95 transgenic line (B) bearing spn-E mutations and in RNAi piwi ovaries (C) with antibodies specific to H3K79me2 (white) and H3K4me2 (gray). Amount of the PCR product was normalized to that of 60D intergenic spacer for each primer set. Ratio of these values for transheterozygous spn-E1/spn-Ehls3987 to heterozygous spn-E1/+ mutants (A and B) or si_piwi to w1118 flies (C) is shown on histograms. Error bars indicate standard deviation of duplicate PCR measurements for two independent ChIP experiments.
Taken together, these data strongly suggest a transcription activation of retrotransposons in the germline of piRNA pathway mutants.
DISCUSSION
In this study, we have explored different approaches to reach understanding of the mechanism of the piRNA-mediated silencing in the germline. The main objects of our study are telomeric retroelements which are located at the telomere ends in a special environment of the telomeric chromatin.
A remarkable feature of the HeT-A promoter is its ability to reproduce an expression pattern characteristic of the endogenous promoter while being located in ectopic positions of the genome. The HeT-A promoter constructs were shown to be active in the Drosophila somatic tissues (49). We have shown a similar expression pattern of endogenous telomeric HeT-A and euchromatic HeT-A/lacZ transgenes in the germline. Thus, ∼400 bp of the HeT-A promoter is enough to provide tissue specificity in its expression.
We have shown that both yellow and lacZ reporter genes, located in telomeric region or in euchromatin, respectively, demonstrated the dependence of their expression on the piRNA pathway being transcribed from the HeT-A promoter. Fusion HeT-A/lacZ and HeT-A/yellow transcripts contain 30–95 bp of the HeT-A sequence that may be enough for homologous recognition and post-transcriptional degradation the fusion transcripts by the piRNA-mediated machinery. HeT-A-specific piRNAs complementary to the 3′ HeT-A fragment cloned in the HeT-A/lacZ construct are represented in the piRNA libraries (47). Their amount is strongly drops in piwi/piwi and spn-E/spn-E mutant ovaries (Supplementary Figure S11A). More interesting is the presence of the pull of piRNAs complementary to the very 3′ HeT-A end which is a part of the HeT-A-lacZ and HeT-A/yellow fusion transcripts (downstream of −30 transcription start site) (Supplementary Figure S11B). These piRNAs may recognize the fusion transcript resulting in the post-transcriptional degradation. According to another scenario, the same piRNAs may target a nascent transcript and transmit a signal to chromatin exerting transcriptional silencing.
The NRO assay confirmed by ChIP analysis reveals activation of endogenous HeT-A and TART transcription in spn-E mutants and in flies with piwi knockdown in the female germline. Most pronounced effect was observed for HeT-A retrotransposon. The accumulation of nascent HeT-A and TART transcripts at the telomeres of piRNA mutants also indicates a transcriptional or co-transcriptional mode of the regulation of their expression. Both reporter constructs bearing HeT-A promoter undergo piRNA-mediated transcriptional silencing independent of the construct location, in euchromatin or at the chromosome end. Transcriptional mechanism seems to be universal for the piRNA-mediated retrotransposon silencing in the germline. We detect transcriptional activation of I-element and HMS Beagle retrotransposons in spn-E mutants and in flies with piwi RNAi knockdown. Most likely, both post-transcriptional and transcriptional pathways may be involved in the piRNA-mediated silencing of the same target gene, although it is difficult to estimate quantitatively contribution of each mechanism in the total silencing effect. Deep sequencing of the run-on cDNA library and ChIP-seq analysis may be helpful to obtain more detailed information about targets of the piRNA-mediated transcriptional silencing.
A siRNA-mediated spreading of heterochromatin was reported to be involved in the centromeric and telomeric repeat silencing in yeast. We tested the Drosophila lines in which the attachments of HeT-A/TART occurred upstream of the intact yellow regulatory region to the terminal deleted X chromosome, at a distance of ∼100–1000 bp from the yellow transcription start site. In these cases, no changes of the yellow expression were observed in spn-E, aub and vasa mutants (data not shown). This fact indicates that the inactive chromatin, if to assume its formation at the HeT-A/TART retrotransposon sequences, does not spread onto the adjacent yellow promoter region. Indeed, the terminal retrotransposon array exhibits euchromatic characteristics since P-element insertions into this retrotransposon array are not silenced (50). Telomeric retrotransposon repeats display the features of open chromatin, however, no actively elongating RNA polymerase isoforms were detected in this region (51). Components of the transcriptional initiation complex of the retrotransposons may be considered as a putative link between piRNAs and inhibition of the transcription.
The aub and armi piRNA pathway gene mutations were shown to affect telomere capping protein recruitment (52) which may result in the telomeric retrotransposon overexpression. However, we show that transcription of non-telomeric retrotransposons as well as HeT-A/lacZ euchromatic transgene is activated as a result of the piRNA pathway disruption. This suggests that piRNA-mediated transcriptional silencing is rather universal phenomenon and not restricted to the specific telomeric region which is affected by the telomere capping. We do not exclude the possibility that genomic context influences the piRNA-mediated chromatin protein recruitment to the target sequences. This may be the case in the telomeric retrotransposon array as well. Transcriptional silencing complexes may be distinct on the distal-most telomeric retroelements covered by the capping proteins and on the telomeric retroelements located proximally.
Transposable element families consist of heterogeneous copies some of which are active and located in euchromatin, others are represented by damaged heterochromatic elements. It is clear that such transposon subfamilies have principally different regulatory mechanisms and chromatin structure. Most experimental approaches fail to distinguish between different types of transposons which result in the complicated superposition of distinct effects. An important result of this study is that unique transgene containing HeT-A promoter exhibits expression pattern and piRNA-mediated transcriptional silencing similar to the endogenous telomeric HeT-A. The transgenic system might be an important tool in the understanding of the piRNA-mediated transcriptional silencing mechanism in the germline.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Academy of Sciences program for Molecular and Cell Biology (to A.K. and M.S.); Russian Foundation for Basic Researches (09-04-00305 to A.K.); Federal Program ‘Scientific and scientific-pedagogical personnel of innovative Russia 2009–2013’ (state contract 02.740.11.0288). Funding for open access charge: Russian Foundation for Basic Researches (09-04-00305 to A.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Vladimir Gvozdev for critical reading of this manuscript and Alexandre Webster for checking of English spelling.
REFERENCES
- 1.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 4.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl Acad. Sci. USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 11.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc. Natl Acad. Sci. USA. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabehere D, Ronsseray S. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–1643. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 18.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 19.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 21.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 23.Biessmann H, Champion LE, O’Hair M, Ikenaga K, Kasravi B, Mason JM. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 25.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell PH, Belote JM, Levis RW. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 2006;34:5498–5507. doi: 10.1093/nar/gkl709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- 30.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 31.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 32.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 34.Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 35.Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 36.Kogan GL, Tulin AV, Aravin AA, Abramov YA, Kalmykova AI, Maisonhaute C, Gvozdev VA. The GATE retrotransposon in Drosophila melanogaster: mobility in heterochromatin and aspects of its expression in germline tissues. Mol. Genet. Genomics. 2003;269:234–242. doi: 10.1007/s00438-003-0827-1. [DOI] [PubMed] [Google Scholar]
- 37.Lavrov S, Dejardin J, Cavalli G. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol. Biol. 2004;247:289–303. doi: 10.1385/1-59259-665-7:289. [DOI] [PubMed] [Google Scholar]
- 38.Chanas G, Lavrov S, Iral F, Cavalli G, Maschat F. Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 2004;272:522–535. doi: 10.1016/j.ydbio.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol. Cell. Biol. 2000;20:7634–7642. doi: 10.1128/mcb.20.20.7634-7642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell. Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fanti L, Dorer DR, Berloco M, Henikoff S, Pimpinelli S. Heterochromatin protein 1 binds transgene arrays. Chromosoma. 1998;107:286–292. doi: 10.1007/s004120050310. [DOI] [PubMed] [Google Scholar]
- 42.Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, Gatti M, Mason JM. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29:819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigova A, Vagin V, Zamore PD. Measuring the rates of transcriptional elongation in the female Drosophila melanogaster germ line by nuclear run-on. Cold Spring Harb. Symp. Quant. Biol. 2006;71:335–341. doi: 10.1101/sqb.2006.71.031. [DOI] [PubMed] [Google Scholar]
- 47.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.George JA, Pardue ML. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2003;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, Walter MF, Mason JM. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171:1767–1777. doi: 10.1534/genetics.105.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J. Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- 52.Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.