Abstract
The molecular basis of resistance to nucleotide excision repair (NER) of certain bulky DNA lesions is poorly understood. To address this issue, we have studied NER in human HeLa cell extracts of two topologically distinct lesions, one derived from benzo[a]pyrene (10R-(+)-cis-anti-B[a]P-N2-dG), and one from the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (C8-dG-PhIP), embedded in either full or ‘deletion’ duplexes (the partner nucleotide opposite the lesion is missing). All lesions adopt base-displaced intercalated conformations. Both full duplexes are thermodynamically destabilized and are excellent substrates of NER. However, the identical 10R-(+)-cis-anti-B[a]P-N2-dG adduct in the deletion duplex dramatically enhances the thermal stability of this duplex, and is completely resistant to NER. Molecular dynamics simulations show that B[a]P lesion-induced distortion/destabilization is compensated by stabilizing aromatic ring system–base stacking interactions. In the C8-dG-PhIP-deletion duplex, the smaller size of the aromatic ring system and the mobile phenyl ring are less stabilizing and yield moderate NER efficiency. Thus, a partner nucleotide opposite the lesion is not an absolute requirement for the successful initiation of NER. Our observations are consistent with the hypothesis that carcinogen–base stacking interactions, which contribute to the local DNA stability, can prevent the successful insertion of an XPC β-hairpin into the duplex and the normal recruitment of other downstream NER factors.
INTRODUCTION
Bulky DNA lesions are subject to removal by the nucleotide excision repair (NER) machinery in order to maintain the integrity of the genome. There are two NER pathways, transcription-coupled repair (TCR) and global genomic repair (GGR). In TCR (1,2), the lesions cause the RNA polymerase to stall, which results in the recruitment of other NER factors that remove the lesions. In GGR (3), the local distortion/destabilization of the DNA caused by the lesions is recognized by the NER factors XPC/Rad23B (5); these factors bind to the site of the lesion, and cause a local strand separation involving ∼6 bp in the case of benzo[a]pyrene (B[a]P)-derived adducts (4). Other factors are subsequently recruited to this complex and ultimately produce dual incisions and the excision of lesion-containing oligonucleotides that are 24–32 nt in length (4–6).
The structural properties of bulky DNA lesions that elicit NER are the subject of considerable interest, particularly since the relative incision efficiencies for structurally different adducts vary over several orders of magnitude (3,7,8). Moreover, the base sequence context in which a given lesion is embedded can modulate the repair efficiencies (9,10). A recently determined crystal structure of a truncated form of yeast Saccharomyces cerevisiae Rad4/Rad23 (11), an ortholog of the mammalian XPC/Rad23B, in a complex with an oligonucleotide containing a T<>T cyclobutane pyrimidine dimer (CPD) lesion, reveals that a β-hairpin is inserted into the DNA helix that stabilizes the separation between the damaged and complementary undamaged strand. The position of the lesion could not be determined, but the two mismatched thymine bases in the complementary strand opposite the CPD were flipped out of the duplex and in contact with amino acid residues in two of the three β-hairpins in this complex. In prokaryotic NER, a crystal structure of a complex of the Bacillus caldotenax UvrB bound to double-stranded damaged DNA (12) also shows β-hairpin intrusion between the two strands and partner base flipping as an element in lesion recognition. It has therefore been suggested that the flipped out bases in the complementary strand that are normally positioned opposite the lesion may play a role in the recognition step of both prokaryotic and eukaryotic NER (11–14). Indeed, base-displaced intercalated 10S-(−) and 10R-(+)-cis-anti-B[a]P-N2-dG (cis-B[a]P-dG) (Figure 1A) adducts in fully double-stranded DNA, in which both the damaged base and the partner base are extruded from the double helix, are excellent NER substrates (13,15). However, the same cis-B[a]P-dG adducts are completely resistant to NER in human cell extracts when the nucleotide dCp opposite the lesion is removed (13,15). Such deletion duplexes might be biologically significant since they could arise during DNA replication in vivo when DNA polymerases fail to insert a dNTP opposite bulky lesions, and skip instead to the next 5′-downstream unmodified template base (16–18).
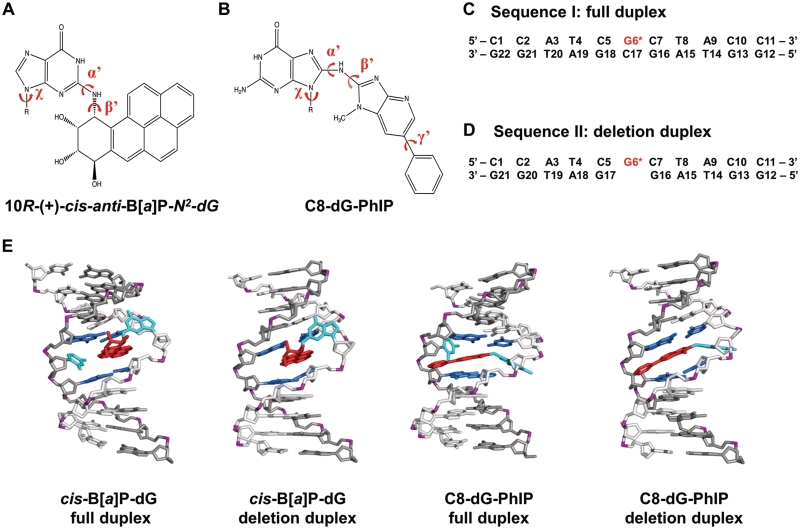
Figure 1.
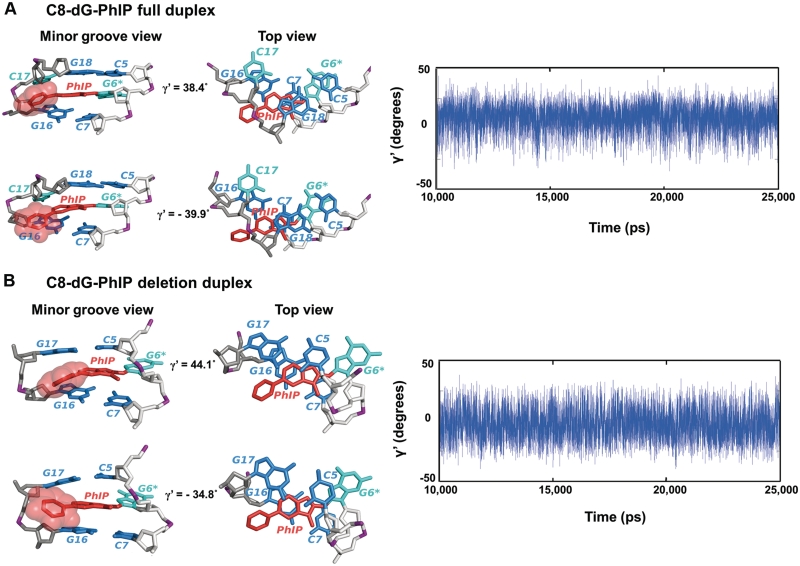
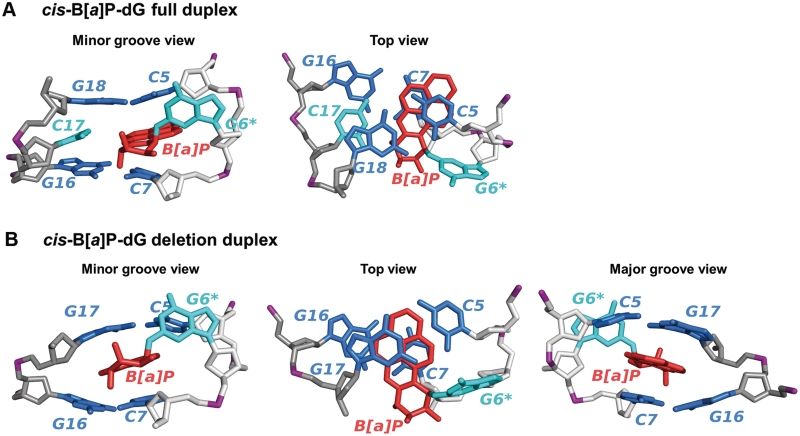
Structures and sequences of cis-B[a]P-dG and C8-dG-PhIP-modified duplexes. (A) Chemical structure of the cis-B[a]P-dG adduct. The glycosidic torsion angle, χ for both modified duplexes is defined as O4′−C1′−N9−C4. The B[a]P-dG linkage site torsion angles, α′ and β′, are defined as follows: α′ = N1(G)−C2(G)−N2(G)−C10(B[a]P) and β′ = C2(G)−N2(G)−C10(B[a]P)−C9(B[a]P). (B) Chemical structure of the C8-dG-PhIP adduct. The PhIP-dG linkage site torsion angles, α′ and β′, are defined as follows: α′ = N9(G)−C8(G)−N(PhIP)−C2(PhIP) and β′ = C8(G)−N(PhIP)−C2(PhIP)−N1(PhIP). The torsion angle γ′ is defined as C7(PhIP)−C6(PhIP)−C1′(PhIP)−C2′(PhIP). (C) Sequence I. Sequence context of the 11/11-mer full duplex. (D) Sequence II. Sequence context of the 11/10-mer deletion duplex. G6*, colored in red, represents the lesion-modified guanine. (E) NMR solution structures for the cis-B[a]P-dG-modified 11/11mer full and 11/10mer deletion duplexes (34,35) and C8-dG-PhIP 11/11mer full duplex (33) utilized as initial structures for the MD simulations. The C8-dG-PhIP deletion duplex was modeled based on the cis-B[a]P-dG deletion duplex NMR structure (35), as described in ‘Materials and Methods’ section. The duplexes are viewed from the minor groove side. Hydrogen atoms and phosphate oxygen atoms are deleted for clarity. In the modified full duplexes, the lesion strand backbone is colored in light gray and the complementary strand is colored in dark gray. Phosphate atoms are colored purple. The adduct is colored red, the modified guanine and its partner base are colored cyan, and the neighboring bases are colored blue.
In this work we explored the molecular basis of the resistance of bulky lesions to NER and the role of a flipped-out base, by studying the repair efficiencies of a DNA adduct derived from the heterocyclic aromatic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a member of a different class of bulky carcinogenic compounds than polycyclic aromatic hydrocarbons (PAHs). Such heterocyclic aromatic amines (HAAs) are highly potent mutagens and suspected human carcinogens, and are present in cigarette smoke and in meats and fish cooked at high temperatures (19–24). Following metabolic activation (25–30), PhIP forms several covalent adducts with DNA (31,32), of which the C8-dG-PhIP adduct (Figure 1B) is the most prevalent and only well characterized lesion (31,32). An NMR solution structure of the C8-dG-PhIP adduct (G*) has revealed that ∼90% of the population of this adduct in sequence I (Figure 1C) adopts a base-displaced intercalated conformation (33), which is in the same family of adduct conformations as the cis-B[a]P-dG adduct (34,35) (Figure 1A). However it is noteworthy that the topology of the C8-dG-PhIP adduct with its fewer aromatic rings, elongated ring system, bulky methyl group, and mobile phenyl ring (Figure 1B) is very different from that of the cis-B[a]P-dG adduct (Figure 1A).
We compare here the NER of the C8-dG-PhIP and the cis-B[a]P-dG adducts in full and deletion duplexes in the sequences defined in Figure 1C and D. We show that both of these lesions, positioned in fully complementary duplexes of the identical sequence context, are excised with similar high efficiencies in human NER in HeLa cell extracts. While the cis-B[a]P-dG adduct is completely resistant to NER in the deletion duplex (Figure 1D), the PhIP adduct positioned in the identical deletion duplex is excised, but with an efficiency that is reduced to 15–20% of that in the full duplex. In order to examine the origins of the differences in the NER response, we determined the impact of these lesions on the thermal stabilities of the double-stranded DNA. We find that both lesions destabilize the full duplexes to near-similar extents; however, the two lesions stabilize the deletion duplexes to significantly different extents, with the cis-B[a]P-dG adduct being more stabilizing. To gain molecular insights on the structural and dynamic properties of the lesions that govern their relative NER susceptibilities, we used molecular modeling and molecular dynamics (MD) simulations for the full and deletion duplexes (Figure 1E). These simulations pinpoint differences in structural distortions and helix-stabilizing/destabilizing properties, and thereby delineate the characteristics of the lesions that elicit resistance to NER.
MATERIALS AND METHODS
Synthesis of modified oligonucleotide sequences and preparation of NER-substrates
The synthesis of the site-specifically modified 11-mer 5′-CCATCGCTACC to generate the centrally positioned C8-dG-PhIP adduct was accomplished by the methods described by Brown et al. (31). Briefly, N-acetoxy-PhIP was dissolved in methanol (1 mg/ml) and added gradually (aliquots added over a 5 h period) to a solution containing the 11-mer oligonucleotide, in 10 mM sodium citrate buffer solution containing 1 mM EDTA and 0.1 M NaCl at pH 7.0 for 60 min at 37°C. The desired modified oligonucleotides were separated and purified by reversed phase HPLC methods utilizing a Microsorb-MV 100-5 C18 250 × 4.6 mm column. The characteristics of the C8-dG-PhIP adducts embedded in the 11-mer oligonucleotides are described in Supplementary Data (Supplementary Figures S1–S4). The cis-B[a]P-dG adducts in the same 11-mer oligonucleotides were prepared as described in detail elsewhere (36). For the thermal melting experiments, unmodified or carcinogen-modified 11-mers were annealed with their fully complementary 11-mer strand 5′-GGTAGCGATGG, or the 10-mer strand 5′-GGTAGGATGG to generate the full and deletion duplexes, respectively. The 135-mer full duplexes (the full sequence is depicted in Supplementary Figure S5) used in the NER experiments were prepared by annealing—ligation techniques, using two 67-mer oligonucleotides and the centrally positioned, modified and 32P-labeled 11-mer 5′-32p-CCATCG*CTACC (where G* denotes the site of the lesion) and a fully complementary 135-mer duplex as described in detail by Kropachev et al. (37). The analogous deletion duplexes were generated in the same manner, except that the dCp nucleotides in the complementary strands opposite the lesion were missing. In all cases, the adducts were positioned at the 67th nucleotide counted from the 5′-side.
Thermal stabilities of oligonucleotide duplexes
Melting curves of the modified duplexes were measured using UV absorbance methods based on Kolbanovskiy et al. (38). Briefly, the unmodified 5′-CCATCGCTACC and modified 5′-CCATCG*CTACC 11-mers were annealed to the complementary strand 5′-GGTAGCGATGG-3′ in a 1:1 ratio of modified:complementary strand DNA, in 20 mM phosphate buffer (pH 7.0) containing 100 mM NaCl. Melting curves were generated using the Cary 100 Bio UV/Vis spectrophotometer. The temperature was increased by 0.1°C increment per minute, followed by 2 s average reading times of the absorbance at 260 nm with the buffer solution being used as the reference.
Human NER assays in HeLa cell extracts
These NER assays were conducted as described by Kropachev et al. (37). Briefly, each reaction contained 1 pmol of P32-internally labeled 135mer duplex with and/or without adduct, in an aqueous mixture generated by mixing 17.5 µl of a 1 M KCl solution with 20 µl of a 10 mM Tris–ATP (pH 7.9) and 10 µl freshly prepared cell extract solution containing 40–45 µg of protein), and sufficient dialysis buffer (12 mM MgCl2, 25 mM HEPES–KOH pH 7.9, 2.5 mM Dithiothreitol, 1 mM EDTA 10% glycerol) to produce a final volume of 50 µl. After appropriate incubation times and subsequent preparation of the samples (37), the reaction products were loaded onto a 12% polyacrylamide, 8 M urea denaturing gel. The gels were dried, exposed, and analyzed using a Storm 840 phosphoimager and Image-Quant software.
Molecular modeling and MD simulations
We performed 25 ns of MD simulations for the C8-dG-PhIP and the cis-B[a]P-dG 11/11-mer full and 11/10-mer deletion duplexes, utilizing NMR solution structures of these adducts as the basis for the initial models (33–35) (Figure 1E). The C8-dG-PhIP deletion duplex (Figure 1E) was modeled based on the NMR structure of the cis-B[a]P-dG deletion duplex (35), since no NMR structure was available. Full details of the molecular modeling, force fields and MD protocols, are given in the Supplementary Data (Supplementary Tables S1 and S2). For all cases, the MD simulations achieved good stability, fluctuating around the mean, after 10.0 ns. Structural ensembles from 10.0 to 25.0 ns were employed for analyses. Details of the analyses are provided in the Supplementary Data.
RESULTS
The C8-dG-PhIP and cis-B[a]P-dG adducts destabilize the full duplexes but stabilize the deletion duplexes to different extents
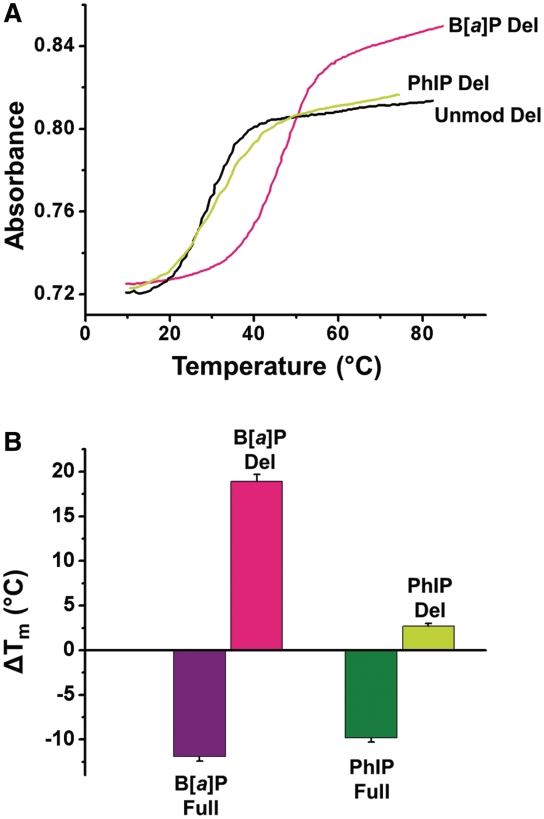
The impacts of these lesions on the duplex thermodynamic properties are reflected in the melting points, Tm, of relatively short oligonucleotide duplexes (39–41). In addition, hyperchromicities (percentage of difference in the absorbance measured at 260 nm of single- and double-stranded oligonucleotides) are indicative of the extent of base stacking interactions in double-stranded DNA. Typical melting curves of the unmodified and modified 11/10-mer deletion duplexes with and without C8-dG-PhIP or cis-B[a]P-dG adducts are depicted in Figure 2A. The melting points of these duplexes specify the temperature at which 50% of the single-stranded molecules are in the double-stranded and single-stranded forms, and are summarized in Table 1. The changes in the melting points relative to the unmodified duplexes are defined by ΔTm = Tm (modified) – Tm (unmodified) and are summarized in Figure 2B and Table 2. The Tm-values of the C8-dG-PhIP and the cis-B[a]P-dG-modified 11/11-mer full duplexes are lower by ∼10° and ∼12°, respectively, than those of the unmodified duplexes. Thus, both lesions destabilize the full duplexes to almost the same extent. The hyperchromicity values of the C8-dG-PhIP-modified (15%), cis-B[a]P-dG-modified (12%) and unmodified (14%) full duplexes measured at 260 nm are similar (Table 1), indicating that the overall impact of both lesions on the DNA base stacking interactions in these 11/11-mer full duplexes is not large, except at the site of the lesion.
Figure 2.
Thermal melting properties of cis-B[a]P-dG and C8-dG-PhIP-modified duplexes. (A) UV melting profiles of unmodified (Unmod), cis-B[a]P-dG (B[a]P) and C8-dG-PhIP (PhIP) modified deletion duplexes measured at 260 nm. The full (Full) and deletion (Del) duplexes are defined in Figure 1 and the Tm values and hyperchromicity values are summarized in Table 1. (B) Comparisons of the ΔTm = Tm (modified) – Tm (unmodified values) based on thermal melting data of Table 1. The melting points of the duplexes, Tm, are averages of 2–3 experiments.
Table 1.
Tm and hyperchromicity values of unmodified, C8-dG-PhIP and cis-B[a]P-dG-modified full (Full) and deletion duplexes (Del)
| Duplexes | Tm (°C) | Hyperchromicity (%) |
|---|---|---|
| Unmodified Full | 51.7 ± 1.0 | 14 |
| B[a]P Full | 39.4 ± 0.8 | 12 |
| PhIP Full | 41.9 ± 0.8 | 15 |
| Unmodified Del | 27.3 ± 0.5 | 11 |
| B[a]P Del | 46.2 ± 0.8 | 14 |
| PhIP Del | 30.0 ± 0.7 | 11 |
Table 2.
Summary of experimental NER efficiencies, ΔTm and structural/energetic properties of the C8-dG-PhIP and cis-B[a]P-dG adducts in full (Full) and deletion (Del) duplexesa
| Lesion-containing duplex | Relative incision efficiency | ΔTm (°C) | Stacking interaction (kcal/mol) | Unpaired partner base | ΔIH (flanking base pairs) | Adduct topology |
|---|---|---|---|---|---|---|
| B[a]P Full | 111 ± 4 | −11.9 | −25.5 | Yes | 784 | Rigid, more aromatic rings |
| PhIP Full | 100 ± 5 | −10.6 | −19.4 | Yes | 209 | Mobile phenyl and methyl groups |
| PhIP Del | 16 ± 5 | 2.7 | −22.4 | No | 257 | Mobile phenyl and methyl groups |
| B[a]P Del | 0 | 18.9 | −27.9 | No | 41 | Rigid, more aromatic rings |
aThe relative incision efficiencies are the same as shown in Figure 4 at 30 min of incubation. ΔTm values are the same as shown in Figure 2B. For each modified DNA duplex, ΔIH is the trajectory summed IH values of both G:C base pairs flanking the lesion minus the trajectory summed IH values of the G16:C7 and G18:C5 base pairs in the unmodified duplex, and was obtained as described in the Supplementary Data. Stacking interaction energies are the mean values of the vdW interaction energies shown in Figure 7A.
In contrast to the destabilizing impact of the lesions on the thermodynamic stabilities that are reflected in the Tm-values of the modified 11/11-mer full duplexes, the modified 11/10-mer deletion duplexes are stabilized by the same lesions (Figure 2). While the Tm of the unmodified deletion duplex is 24°C lower than that of the unmodified 11/11-mer full duplex (Table 1), both kinds of lesions stabilize the deletion duplexes. The C8-dG-PhIP lesion stabilizes the deletion duplex modestly by ΔTm ∼ +3°C (Figure 2B and Table 2), and the cis-B[a]P-dG lesion more prominently by ΔTm ∼ +19°C (Figure 2B and Table 2). The hyperchromicity of the unmodified 11/10-mer deletion duplex (11%) is 3% points lower than that of the full duplex (Table 1), as expected from the loss of base-base stacking interactions in the deletion duplex. Remarkably, the hyperchromicity rises to 14% in the cis-B[a]P-dG deletion duplex (Table 1), which is the same as the value for the full unmodified duplex; this suggests an increase in stacking due to interactions between the B[a]P residue and the neighboring bases, which dramatically stabilize the cis-B[a]P-dG deletion duplex. The enhanced stability of the deletion duplex is indicated by the prominent increase in the Tm-value (Table 1). Overall, these results clearly point out that bulky polycyclic aromatic DNA lesions exert not only destabilizing effects by distorting the normal B-DNA helical structure, but are also capable of stabilizing the same duplexes via intermolecular van der Waals (vdW) stacking and other interactions with DNA residues.
Full duplexes are excellent substrates of NER while in deletion duplexes the B[a]P adduct is fully resistant and the PhIP adduct elicits only moderate repair
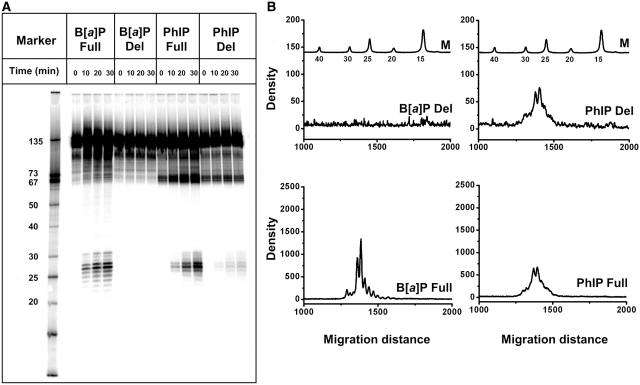
The incubation of 135-mer duplexes (Supplementary Figure S5) containing single C8-dG-PhIP and cis-B[a]P-dG lesions in HeLa cell extracts gives rise to the appearance of radioactively labeled oligonucleotide dual incision fragments 24–32 nt in length that can be resolved from the 135-mer duplexes by denaturing gel electrophoresis (Figures 3A and B). A small fraction (<10%) of the C8-dG-PhIP adducts, but not the cis-B[a]P-dG adducts, decompose during the workup of the samples subsequent to the incubation reaction. This is evident from the appearance of 67-mer oligonucleotide cleavage fragments in denaturing gels that appear even in the case of untreated control samples (Figure 3A). Furthermore, the fractions of these decomposition products are independent of incubation time in NER-viable cell extracts, while the fractions of NER incision products increase linearly with incubation time in the same extracts (Supplementary Figures S6 and S7).
Figure 3.
NER assays in Human HeLa cell extracts. (A) Autoradiogram of a representative denaturing 12% polyacrylamide gel showing incisions of full (Full) and deletion (Del) 135-mer duplexes containing single cis-B[a]P-dG (B[a]P) and C8-dG-PhIP (PhIP) adducts in HeLa cell extracts at 10, 20 and 30 min of incubation. The zero time point samples serve as controls (these samples were incubated in heat-deactivated cell extracts, but otherwise underwent the same treatment). Lane Marker: oligonucleotide markers of different lengths. (B) Densitometry tracings of the 30 min lanes shown in the autoradiogram in Figure 3A. M: markers.
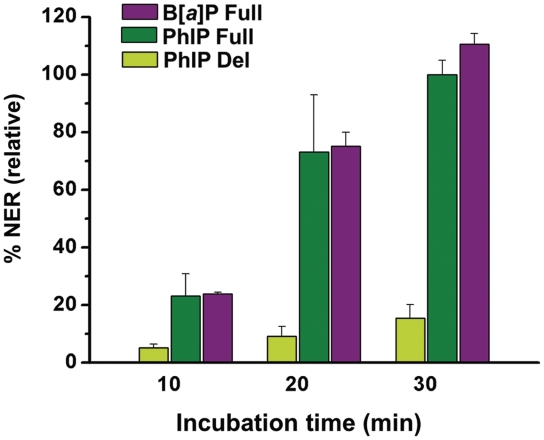
Examples of typical densitometry tracings of the 30 min gel electrophoresis lanes are depicted in Figure 3B. Comparisons of the relative extents of NER were calculated from the integration of the radioactivity in the 25–32 oligonucleotide dual excision fragments divided by the total radioactivity in each lane of the gel autoradiogram (Figure 3A). The NER dual incision efficiencies varied from 0% to ∼5–8% in the experiments reported here. In order to achieve reproducibility and to take account of varying activities of different cell extracts, the NER efficiencies in each gel autoradiogram were normalized to the value obtained with the C8-dG-PhIP-modified 135/135-mer full duplexes after an incubation time of 30 min. These values were arbitrarily assigned a value of 100, and all other NER efficiencies are reported relative to this value for each gel. The relative efficiencies of dual incisions of the C8-dG-PhIP and cis-B[a]P-dG-modified 135/135-mer full duplexes increase as a function of incubation time within the range of 0–30 min (Figure 4). The resistance of the cis-B[a]P-dG-modified 135/134-mer deletion duplex to NER is evident from the flat densitometry trace shown in Figure 3B, which is indistinguishable from the base line. Thus, NER is completely abolished for this adduct (13,15). In contrast, the C8-dG-PhIP-modified deletion duplex is incised in the cell extracts (Figures 3A and 4). However, at all incubation times the incision efficiencies are uniformly 5–6 times lower in the C8-dG-PhIP-modified deletion duplexes than in the C8-dG-PhIP-modified full duplexes (Figure 4).
Figure 4.
Incision kinetics of the internally labeled 135-mer modified duplexes in HeLa cell extracts. The incision efficiencies were normalized in each of the four independent experiments to the value obtained with the C8-dG-PhIP full duplex (relative value of 100) at the 30 min incubation time point. The averages and standard deviations shown were obtained from these normalized values. Note that the cis-B[a]P-dG deletion duplex is not incised.
Molecular modeling and MD simulations
In order to obtain structural, dynamic and energetic understanding of the experimental thermal melting and NER studies, we performed molecular modeling and 25 ns MD simulations for the C8-dG-PhIP and cis-B[a]P-dG adducts, in 11/11-mer full and 11/10-mer deletion duplexes. These were based on their base-displaced intercalated solution structures (Figure 1E) that were determined by NMR methods (33–35). There is no experimental NMR data for the C8-dG-PhIP deletion duplex. As detailed in the Supplementary Data, the known cis-B[a]P-dG deletion duplex NMR solution structure (35) was utilized to obtain an initial structure for MD simulation of the C8-dG-PhIP deletion duplex, by replacing the B[a]P-modified guanine with the PhIP-modified guanine (Figure 1E). We carried out detailed analyses of the MD trajectories between 10 and 25 ns during which the simulations were stable, as shown by the RMSD data for each 25 ns MD simulation (Supplementary Figure S8).
Base-displaced intercalated conformations affect duplex structures and stabilities differently depending on adduct topology and type of duplex
MD simulations show base-displaced intercalated conformations of the C8-dG-PhIP and cis-B[a]P-dG adducts in full and deletion duplexes (Figures 5 and 6). These simulations indicate that there are structural features that distinguish the properties of the PhIP- and B[a]P-derived duplexes although both are intercalated with the displacement of the damaged bases and their partners from their normal intra-helical positions. The topological differences between the lesions are notable: the PhIP is more elongated, contains only three aromatic rings, one of which is the mobile phenyl ring, while the B[a]P is rigid and planar, and contains four fully aromatic rings. While the overall features of the C8-dG-PhIP and cis-B[a]P-dG-modified full and deletion duplexes shown in the ensembles of structures derived from the MD simulations are similar to those of their respective NMR solution structures (33–35), the simulations provide important new insights into the lesion dynamics.
Figure 5.
Molecular dynamics of the C8-dG-PhIP-modified full and deletion duplexes. Hydrogen atoms and phosphate oxygen atoms are deleted for clarity. The lesion strand backbone is in light gray and the complementary strand is in dark gray. Phosphate atoms are purple. The adduct is red, the modified guanine and its partner base are cyan, and the neighboring bases are blue. (A) Dynamics of phenyl ring of the C8-dG-PhIP-modified full duplex. The structures with the highest and lowest γ′ values (38.4° and −39.9°) are shown on the left to emphasize the dynamics of the phenyl ring. The central trimer is viewed from the minor groove side (left) and along the helix axis (Top view, right). The time dependence of the γ′ torsion angle is shown on the right. (B) Dynamics of phenyl ring of the C8-dG-PhIP-modified deletion duplex. The structures with the highest and the lowest γ′ values (44.1° and −34.8°) are shown on the left to emphasize the dynamics of the phenyl ring. The central trimer is viewed from the minor groove side (left) and along the helix axis (Top view, right). The time dependence of the γ′ torsion angle is shown on the right.
Figure 6.
Representative structures of the cis-B[a]P-dG-modified full and deletion duplexes. Hydrogen atoms and phosphate oxygen atoms are deleted for clarity. The lesion strand backbone is in light gray and the complementary strand is in dark gray. Phosphate atoms are purple. The adduct is red, the modified guanine and its partner base are cyan, and the neighboring bases are blue. (A) Best representative structure of the cis-B[a]P-dG-modified full duplex viewed from the minor groove and along the helix axis. The best representative structure viewed along the helix axis (Top view) is shown on the right to emphasize stacking interactions with neighboring bases. (B) Best representative structure of the cis-B[a]P-dG-modified deletion duplex viewed from the minor groove, along the helix axis and from the major groove. The best representative structure viewed along the helix axis (Top view) is shown in the middle to emphasize stacking interactions with neighboring bases. The best representative structure viewed from the major groove side is shown on the right to emphasize the wedge-like shape induced by the deletion.
C8-PhIP-dG-modified duplexes: the dynamic phenyl ring
In the full duplex, the PhIP phenyl ring protrudes into the minor groove towards the complementary strand and rotates rapidly. This rapid rotation is reflected by the dynamic oscillation, between 38.4° and −39.9°, of the γ′ torsion angle (Figure 1B) shown in Figure 5A and Movie S1 in Supplementary Data. Both G6* and its partner base C17 are displaced into the major groove of the full duplex. G6* stacks with base C5 on its 5′-side, while the inserted imidazopyridine (IP) ring of PhIP stacks mainly with G18 in the helix. In the deletion duplex, the phenyl ring also rotates rapidly, with the γ′ torsion angle (Figure 1B) adopting values between 44.1° and −34.8°. The dynamics of this rotation is shown in Figure 5B and Movie S2 in Supplementary Data. Furthermore, the PhIP phenyl ring protrudes more into the minor groove than in the full duplex, because the absence of dC17 compresses the partner strand. In concert, G6* protrudes further into the major groove away from C5 and the PhIP IP ring now stacks with C5, as well as with G16 (Figure 5B Top view). Modest stacking interactions between the IP ring and other adjacent bases are equivalent in the full and the deletion duplexes (Supplementary Table S3). The increased stacking interactions involving C5 and G16 provide stabilization to the deletion duplex containing this adduct, as revealed in the thermal melting data (Figure 2 and Table 1).
cis-B[a]P-dG-modified duplexes: planar, rigid and bulky B[a]P ring
The cis-B[a]P-dG adduct contains more aromatic rings, is less elongated than the PhIP residue, and is planar and rigid (Figure 1). In the cis-B[a]P-dG-modified full duplex, the G6* base is displaced into the minor groove, while C17 is displaced into the major groove; however, this displacement is less pronounced than in the case of the C8-dG-PhIP-modified full duplex (Figures 5A and 6A). In the deletion duplex, due to the compression of the backbone, G16 and G17 collapse onto each other and form a wedge-like shape around the B[a]P rings (Figure 6B). The B[a]P aromatic ring system is shifted more towards the complementary strand and more into the minor groove than in the full duplex (Figure 6A and B). These conformational changes provide better aromatic ring overlap between B[a]P rings and neighboring bases, primarily G17 and the periphery of G16, in the deletion duplex than in the full duplex (Figure 6A and B and Supplementary Table S3); this results in stabilization that is manifested in the thermal melting points (Figure 2 and Table 1).
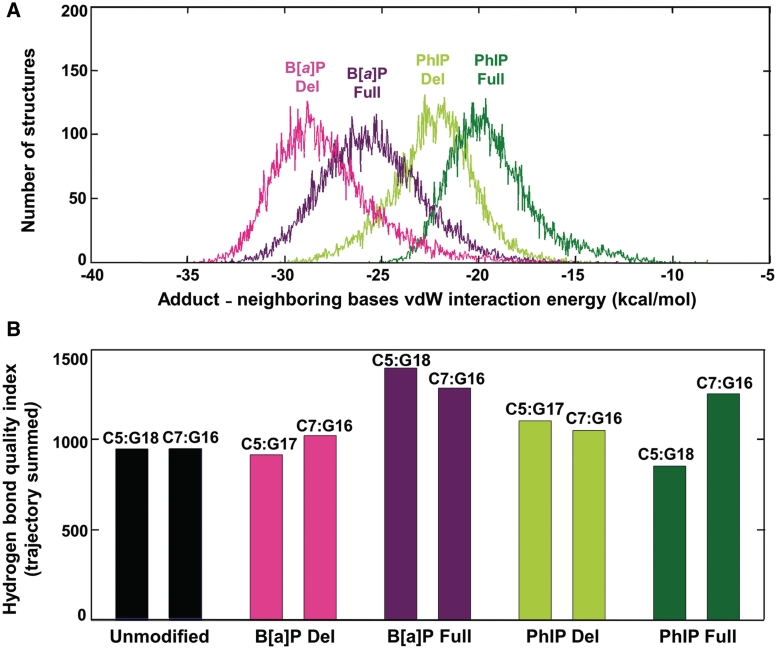
Stacking interaction energies between the aromatic ring systems of the lesions and adjacent base pairs differ in full and deletion duplexes
In order to gain deeper insights into the effects of PAH adduct ring system topology on stacking interactions between the adduct and the adjacent base pairs, we computed the vdW interaction energy (EvdW) between the adduct aromatic rings and all the adjacent bases; this energy was computed for each of the distinct structures (total of 15 000) that evolve during the MD simulations from 10 to 25 ns, as detailed in Supplementary Data. Figure 7A shows the number of structures from the total ensemble at each given vdW interaction energy value, and is called a population distribution. As expected, vdW interaction energies are weaker for the elongated PhIP with its dynamically mobile phenyl ring than for the rigid, planar and more aromatic B[a]P ring system. This dynamic, non-planar ring protrudes into the minor groove, and forces the PhIP IP rings toward this groove; consequently, stacking interactions between the PhIP ring system and neighboring bases is diminished. It is evident that the vdW interaction energies between the lesion aromatic rings and the adjacent bases are stronger in the deletion duplexes than the full duplexes in each case. This stems from the compressed backbone of the partner strand in the deletion duplex, which allows better aromatic ring system overlap between the lesion and adjacent bases in both cases, as detailed above. The lack of the dCp nucleotide opposite the lesion allows for better B[a]P – base stacking interactions, thus pointing to the destabilizing influence of the partner dCp nucleotide opposite the lesion in the full duplex. The contributions to the stacking energy of each base are provided in Supplementary Table S3.
Watson–Crick base pairing is impacted differently depending on the lesion and type of duplex
The distortion of normal Watson–Crick hydrogen bonding is an important NER recognition signal (37,42). Therefore, for both PhIP and B[a]P adducts, in full and deletion duplexes, we have evaluated the impact of the lesion on the local hydrogen bond quality for the base pairs flanking the site of the lesion, namely C5:G18 and C7:G16 in full duplexes and C5:G17 and C7:G16 in deletion duplexes. We utilized the hydrogen bond quality index (IH) developed by us previously (43), to evaluate the deviation of the hydrogen bonds for a selected base pair from ideal Watson–Crick hydrogen bonding; we computed the IH sum over the 15 000 structures collected between 10 and 25 ns of MD simulation (see Supplementary Data). Ideal Watson–Crick hydrogen bonds have an IH-value of zero. The more the hydrogen bond distances and angles differ from those in ideal Watson–Crick hydrogen bonds, the larger the value of IH. Such deviations in distances and angles are produced by various combinations of disturbances to the global base–base parameters, such as shear, stretch, stagger, buckle, propeller and opening, which can vary in concert (44) (Supplementary Figure S9).
The mobility of the PhIP phenyl ring disturbs the base pairs flanking the lesion
In the C8-dG-PhIP-modified full duplex, distortion of the C7:G16 base pair is indicated by a higher trajectory summed IH-value than that of the analogous C7:G16 base pair in the unmodified duplex (Figure 7B). The origin of these distortions is the rotational mobility of the phenyl ring (Figure 5A): this mobility causes episodic crowding between the hydrogen atoms of the phenyl ring and G16, causing disturbances to the C7:G16 base pair, especially in buckle and propeller twist (Supplementary Figure S9 and Movie S1 in Supplementary Data). The methyl group also contributes to the distortion of the C7:G16 hydrogen bonds through close interaction with the sugar ring of dC7, thus somewhat displacing this base from its normal position relative to its partner G16. However, the other flanking base pair, C5:G18, remains essentially undistorted (Figure 5A), with an IH value comparable to that of the unmodified duplex (Figure 7B) since both the phenyl and methyl groups are further away from this base pair. In the C8-dG-PhIP-modified deletion duplex, the IH values of the C5:G17 and C7:G16 base pairs are modestly higher than for the C5:G18 and C7:G16 base pairs in the unmodified duplex (Figure 7B); this suggests small distortions of Watson–Crick hydrogen bonding caused by the rapid rotation of the phenyl ring in the minor groove of the deletion duplex, which produces episodic crowding between hydrogen atoms of the phenyl ring and the guanine bases G16 and G17 (Movie S2 in Supplementary Data). However, the crowding is less severe than in the full duplex, because the phenyl ring protrudes more into the minor groove in the deletion duplex and therefore does not significantly perturb the normal position of G16. Due to the compression of the complementary strand, G17 partially collapses onto G16.
Figure 7.
Stabilizing and destabilizing properties of lesions in modified duplexes. (A) The population distribution of the vdW interaction energy between adduct aromatic rings and the neighboring bases. The vdW interaction energies were calculated as described in Supplementary Data. Energies for the cis-B[a]P-dG (B[a]P) and C8-dG-PhIP (PhIP) modified full duplexes (Full) are represented in dark green and purple, and in light green and magenta for the respective deletion duplexes (Del). Mean values and standard deviations (kcal/mol): B[a]P Del, −27.9 ± 2.6; B[a]P Full, −25.5 ± 2.6; PhIP Del, −22.4 ± 2.1; PhIP Full, −19.4 ± 2.2. (B) Trajectory summed hydrogen bond quality index for the C:G base pairs 5′ and 3′ to the lesion in the cis-B[a]P-dG (B[a]P) and C8-dG-PhIP (PhIP) modified deletion (Del) and full (Full) duplexes, and for the C:G base pairs at comparable positions in the unmodified duplex. The trajectory summed hydrogen bond quality indexes were calculated as described in the Supplementary Data. The specific base pairs are labeled at the top of the bars. The bars are color-coded as in Figure 7A.
Base pairs flanking the cis-B[a]P-dG adduct are disturbed in the full duplex but scarcely in the deletion duplex
In the cis-B[a]P-dG-modified full duplex, the IH values of the C5:G18 and C7:G16 base pairs flanking the adduct are the highest observed in our simulations and thus are the most perturbed (Figure 7B). The origin of the disturbance to the C5:G18 base pair is the steric crowding between G18 and the hydrogen atom at the N2-G6* linkage site, as well as the N1 hydrogen atom of G6* (Movie S3 in Supplementary Data). These close contacts cause the C5:G18 base pair to episodically buckle and propeller twist. The C7:G16 base pair also buckles and propeller twists episodically because of steric crowding between the amino group of G16 and the hydrogen atom on the 7-hydroxyl group of the benzylic ring of the B[a]P aromatic residue (Movie S3 in Supplementary Data). By contrast, the hydrogen bond quality index IH values of the C5:G17 and C7:G16 base pairs flanking the aromatic B[a]P ring system in the cis-B[a]P-dG-modified deletion duplex are not much different from the IH values of the C5:G18 and C7:G16 base pairs in the unmodified duplex: the C5:G17 base pair is slightly lower and the C7:G16 base pair is slightly higher (Figure 7B). This difference arises from the backbone compression in the deletion duplex, which forces the benzylic ring to protrude further into the minor groove than in the full duplex (Figure 6B); as a result, there is much less steric crowding in the deletion duplex than in the full duplex (Movie S4 in Supplementary Data). The collapse is greater than for the C8-dG-PhIP deletion duplex, described above, because the interaction of G16 and G17 with the mobile phenyl ring prevents the greater collapse seen in the cis-B[a]P-dG deletion duplex.
DISCUSSION
NER of base-displaced intercalated full and deletion duplexes
Certain bulky DNA adducts, including N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-C8-AAF) (45), N-(deoxyguanosin-8-yl)-1-aminopyrene (dG-C8-AP) (46), 10S-(−) (47) and 10R-(+)-cis-anti-B[a]P-N2-dG (34), which adopt base-displaced intercalated conformations in full duplexes have been shown to be well repaired in human HeLa cell extract assays (13,15,48,49) or in the prokaryotic UvrABC system (50). Although each adduct has its unique structural features, common structural properties of base-displaced intercalation appear to offer similar recognition elements to the NER system. Several deletion duplexes, which involve B[a]P-derived stereoisomeric adducts of identical topology, are repair-resistant, namely the 10S (+)-trans-anti-B[a]P-N2-dG and the 10S-(−)-cis-anti-B[a]P-N2-dG ((−)-cis-B[a]P) adducts (15). The former deletion duplex adopts a base-displaced intercalated conformation (51). While there is no NMR solution structure available for the (−)-cis-B[a]P adduct in the deletion duplex, its comparable repair inhibition and thermal stabilization relative to the unmodified deletion duplex (Table 1) (Tm = 44 ± 0.5°C, ΔTm = 16.7°C, unpublished data) to the (+)-cis-B[a]P-dG deletion duplex investigated here, indicate that their overall structural characteristics are most likely similar. Additionally, the NER efficiency in a dG-C8-AP deletion duplex with the prokaryotic UvrABC system is quite inefficient compared to the same adduct with a C opposite this lesion (50), and the deletion duplex is significantly stabilized (by ∼4 kcal/mol) according to thermal melting studies (52). While the deletion duplexes share the feature of repair resistance in human HeLa cell assays, our results here show that unique topological elements can play an important part in determining susceptibility to NER.
Recognition and verification steps in NER
The efficiencies of dual incisions catalyzed by the complex, multi-step mammalian global genomic NER system is often described in terms of a bipartite recognition mechanism (53,54). The first step involves the recognition of the distorted/destabilized lesion site by XPC/Rad23B; the second is believed to be the verification that a lesion exists, by a mechanism involving helicases in the TFIIH multi-protein complex (55). Following recognition by XPC/Rad23B, the DNA duplex containing a cis-B[a]P-dG adduct is opened within a 6 bp stretch containing the lesion (4), and the adducted duplex is no longer double-stranded. In the case of our cis-B[a]P-dG (or C8-dG-PhIP) full and deletion duplexes, the verification step is unlikely to contribute to the observed striking differences in NER efficiencies, since the lesions are identical in each pair of full and deletion duplexes but the NER efficiencies are markedly different. However, the recruitment of subsequent NER factors might be influenced by the structural features of the initial XPC/Rad23B complexes formed, and these features may exert an effect on the efficiencies of dual incisions (5).
Local thermodynamic stabilization may hinder β-hairpin insertion
A common structural feature of both prokaryotic and eukaryotic NER mechanisms revealed in crystal structures is the insertion of a β-hairpin of UvrB (12), or the yeast Rad4/Rad23 (11) (an ortholog of the mammalian XPC/Rad23B recognition factors) between the two strands of the duplex at or close to the lesion site. The insertion of the β-hairpin, with the concomitant extrusion of the bases opposite the lesion in the complementary strand and their interactions with the protein, stabilize the DNA–protein complex. These structural properties suggest the hypothesis that the local thermodynamic stability of the damaged site within the modified DNA duplex determines the ease or difficulty of DNA strand separation and concerted base flipping (11,56); the insertion of the β-hairpin is facilitated or hindered by the local thermodynamic stability. Our results reveal that the severe distortions/destabilizations produced by complete rupturing of Watson–Crick base pairing, observed in our full duplexes, favor β-hairpin insertion and thus the subsequent cascade of NER steps that lead to the dual incisions of the damaged strand; this is because stacking of the adduct aromatic ring system with adjacent base pairs is insufficient to compensate energetically for the distortions produced by two unstacked, solvent-exposed bases. Our studies of modified deletion duplexes show that sufficient stabilizing stacking interactions between the bulky aromatic lesions with the DNA bases can overcome the effects of the local distortions/destabilizations caused by bulky lesions; these stabilizing stacking interactions may hinder the insertion of the β-hairpin and the subsequent NER steps.
A pre-flipped base from the complementary strand is not absolutely required for lesion recognition
The full duplexes with cis-B[a]P-dG lesions have a flipped out cytosine partner base in the complementary strand (34). It has been argued that this flipped out base plays a key role (13) in the efficient removal of this lesion by the NER apparatus in human cell extracts, since the absence of this cytidine residue in deletion duplexes abolishes NER (13,15). Yet, we show here that, even with the partner nucleotide absent, the topological features of a lesion can provide sufficient destabilization to, presumably, permit β-hairpin insertion and NER. This is demonstrated with the C8-dG-PhIP-modified deletion duplex, where stacking is more modest than for the cis-B[a]P-dG adduct, and steric hindrance imposed by the dynamic non-planar phenyl ring causes neighboring base pair perturbations. While the NER of the C8-dG-PhIP-modified deletion duplex is significantly smaller than that of the C8-dG-PhIP-modified full duplex, it still exhibits significant NER (Figure 4) even though the partner dCp is missing entirely in the complementary strand. We suggest that the ease of flipping the partner base out of the duplex, which is determined by the local thermodynamic destabilization, rather than the existence of a pre-flipped base, governs β-hairpin intrusion. Strong base stacking interactions between lesions with aromatic ring systems and neighboring base pairs can prevent not only the insertion of the β-hairpin between the two strands, but also impede the flippability of the bases in the complementary strand, an area we are currently investigating (57). Therefore, the recognition of a bulky lesion is a function of a combination of destabilizing effects and stabilizing vdW interactions that depend on the structural features of the lesions; these together determine the local thermodynamic properties of the modified duplexes. Hence, the presence of a pre-flipped partner nucleotide in the complementary strand is not an absolute requirement for NER.
CONCLUSION
Thermodynamic stabilization of a damaged DNA duplex through effective carcinogen–base stacking interactions can overcome the impact of destabilizing distortions imposed by the lesion, and thus convey resistance to NER. The presence of the normal dCp in the complementary strand opposite the lesion exerts a destabilizing effect, hindering the development of optimal aromatic lesion–base stacking interactions seen in the deletion duplexes and thus favoring efficient NER. However, the existence of the pre-flipped dCp partner nucleotide is not an essential requisite for the initiation of NER.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant CA-099194 to N.E.G., CA-75449 and CA-28038 to S.B.); (CA-046533 to D.J.P.). Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement (grant C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the MALDI-TOF mass spectrometer used in this work was supported by the National Science Foundation (CHE-0958457). Funding for open access charge: Grants from the National Institutes of Health CA-099194 to N.E.G., CA-75449 and CA-28038 to S.B.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge TeraGrid resources provided by the Texas Advanced Computing Center supported by the National Science Foundation. D.A.R., M.K. and K.K. carried out the NER experiments. D.A.R., A.K., M.K. and Y.C. collaborated on the thermal melting experiments. D.A.R., A.K. and M.K. synthesized the adducts utilizing diolepoxide and the N-acetoxy-PhIP reactive intermediates generated by J.K. and S.A. The computer modeling was carried out by H.M. with the help of Y.C. and S.D. The B[a]P-DNA adduct NMR studies were performed in the laboratory of D.J.P, while S.B. supervised the modeling studies, and N.E.G. supervised the experimental work. N.E.G., S.B. and H.M. analyzed the results. N.E.G., H.M., S.B. and D.J.P. wrote the manuscript.
REFERENCES
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Tornaletti S. DNA repair in mammalian cells: Transcription-coupled DNA repair: directing your effort where it's most needed. Cell Mol. Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 4.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scharer OD. Mechanisms of Base Excision Repair and Nucleotide Excision Repair. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Germany: Wiley-VCH, Weinheim; 2010. pp. 239–260. [Google Scholar]
- 7.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair. 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J. Mol. Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/174252. Article ID 174252, 9 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 12.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 13.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 15.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu P, Oum L, Geacintov NE, Broyde S. Nucleotide selectivity opposite a benzo[a]pyrene-derived N2-dG adduct in a Y-family DNA polymerase: a 5′-slippage mechanism. Biochemistry. 2008;47:2701–2709. doi: 10.1021/bi701839q. [DOI] [PubMed] [Google Scholar]
- 17.Xu P, Oum L, Lee YC, Geacintov NE, Broyde S. Visualizing sequence-governed nucleotide selectivities and mutagenic consequences through a replicative cycle: processing of a bulky carcinogen N2-dG lesion in a Y-family DNA polymerase. Biochemistry. 2009;48:4677–4690. doi: 10.1021/bi802363f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel TA. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 19.Felton JS, Knize MG. Occurrence, identification, and bacterial mutagenicity of heterocyclic amines in cooked food. Mutat. Res. 1991;259:205–217. doi: 10.1016/0165-1218(91)90118-6. [DOI] [PubMed] [Google Scholar]
- 20.Wakabayashi K, Nagao M, Esumi H, Sugimura T. Food-derived mutagens and carcinogens. Cancer Res. 1992;52:2092s–2098s. [PubMed] [Google Scholar]
- 21.Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 22.Sugimura T, Nagao M, Wakabayashi K. Carcinogenicity of food mutagens. Environ. Health Perspect. 1996;104(Suppl. 3):429–433. doi: 10.1289/ehp.96104s3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knize MG, Felton JS. Formation and human risk of carcinogenic heterocyclic amines formed from natural precursors in meat. Nutr. Rev. 2005;63:158–165. doi: 10.1111/j.1753-4887.2005.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 24.Turesky RJ. Aromatic Amines and Heterocyclic Aromatic Amines: From Tobacco Smoke to Food Mutagens. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Germany: Wiley-VCH, Weinheim; 2010. pp. 157–184. [Google Scholar]
- 25.Boobis AR, Lynch AM, Murray S, de la Torre R, Solans A, Farre M, Segura J, Gooderham NJ, Davies DS. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 26.Buonarati MH, Felton JS. Activation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) to mutagenic metabolites. Carcinogenesis. 1990;11:1133–1138. doi: 10.1093/carcin/11.7.1133. [DOI] [PubMed] [Google Scholar]
- 27.Buonarati MH, Turteltaub KW, Shen NH, Felton JS. Role of sulfation and acetylation in the activation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine to intermediates which bind DNA. Mutat. Res. 1990;245:185–190. doi: 10.1016/0165-7992(90)90048-o. [DOI] [PubMed] [Google Scholar]
- 28.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu D, McNaughton L, Lemaster D, Lake BG, Gooderham NJ, Kadlubar FF, Turesky RJ. A comprehensive approach to the profiling of the cooked meat carcinogens 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and their metabolites in human urine. Chem. Res. Toxicol. 2010;23:788–801. doi: 10.1021/tx900436m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis AJ, Walle UK, King RS, Kadlubar FF, Falany CN, Walle T. Bioactivation of the cooked food mutagen N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by estrogen sulfotransferase in cultured human mammary epithelial cells. Carcinogenesis. 1998;19:2049–2053. doi: 10.1093/carcin/19.11.2049. [DOI] [PubMed] [Google Scholar]
- 31.Brown K, Guenther EA, Dingley KH, Cosman M, Harvey CA, Shields SJ, Turteltaub KW. Synthesis and spectroscopic characterization of site-specific 2-amino-1-methyl-6-phenylimidazo. Nucleic Acids Res. 2001;29:1951–1959. doi: 10.1093/nar/29.9.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr, Kadlubar FF. Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-a]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem. Res. Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 33.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl Acad. Sci. USA. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 35.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove. Biochemistry. 1994;33:11518–11527. doi: 10.1021/bi00204a014. [DOI] [PubMed] [Google Scholar]
- 36.Pirogov N, Shafirovich V, Kolbanovskiy A, Solntsev K, Courtney SA, Amin S, Geacintov NE. Role of hydrophobic effects in the reaction of a polynuclear aromatic diol epoxide with oligodeoxynucleotides in aqueous solutions. Chem. Res. Toxicol. 1998;11:381–388. doi: 10.1021/tx980006q. [DOI] [PubMed] [Google Scholar]
- 37.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, et al. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolbanovskiy A, Kuzmin V, Shastry A, Kolbanovskaya M, Chen D, Chang M, Bolton JL, Geacintov NE. Base selectivity and effects of sequence and DNA secondary structure on the formation of covalent adducts derived from the equine estrogen metabolite 4-hydroxyequilenin. Chem. Res. Toxicol. 2005;18:1737–1747. doi: 10.1021/tx050190x. [DOI] [PubMed] [Google Scholar]
- 39.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 40.Arghavani MB, SantaLucia J, Jr, Romano LJ. Effect of mismatched complementary strands and 5′-change in sequence context on the thermodynamics and structure of benzo[a]pyrene-modified oligonucleotides. Biochemistry. 1998;37:8575–8583. doi: 10.1021/bi980237w. [DOI] [PubMed] [Google Scholar]
- 41.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, et al. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 42.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J. Mol. Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hingerty BE, Figueroa S, Hayden TL, Broyde S. Prediction of DNA structure from sequence: a build-up technique. Biopolymers. 1989;28:1195–1222. doi: 10.1002/bip.360280703. [DOI] [PubMed] [Google Scholar]
- 44.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 46.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 47.Cosman M, Hingerty BE, Luneva N, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (-)-cis-anti-benzo[a]pyrenyl-dG adduct opposite dC in a DNA duplex: intercalation of the covalently attached BP ring into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1996;35:9850–9863. doi: 10.1021/bi9605346. [DOI] [PubMed] [Google Scholar]
- 48.Gillet LC, Alzeer J, Scharer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified ‘ultra-mild’ DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoare S, Zou Y, Purohit V, Krishnasamy R, Skorvaga M, Van Houten B, Geacintov NE, Basu AK. Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures. Biochemistry. 2000;39:12252–12261. doi: 10.1021/bi0013187. [DOI] [PubMed] [Google Scholar]
- 51.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1994;33:11507–11517. doi: 10.1021/bi00204a013. [DOI] [PubMed] [Google Scholar]
- 52.Nolan SJ, McNulty JM, Krishnasamy R, McGregor WG, Basu AK. C8-guanine adduct-induced stabilization of a -1 frame shift intermediate in a nonrepetitive DNA sequence. Biochemistry. 1999;38:14056–14062. doi: 10.1021/bi991342o. [DOI] [PubMed] [Google Scholar]
- 53.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl Acad. Sci. USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maillard O, Camenisch U, Blagoev KB, Naegeli H. Versatile protection from mutagenic DNA lesions conferred by bipartite recognition in nucleotide excision repair. Mutat. Res. 2008;658:271–286. doi: 10.1016/j.mrrev.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Clement FC, Camenisch U, Fei J, Kaczmarek N, Mathieu N, Naegeli H. Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat. Res. 2010;685:21–28. doi: 10.1016/j.mrfmmm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Scharer OD. Achieving broad substrate specificity in damage recognition by binding accessible nondamaged DNA. Mol. Cell. 2007;28:184–186. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Zheng H, Cai Y, Ding S, Tang Y, Kropachev K, Zhou Y, Wang L, Wang S, Geacintov NE, Zhang Y, et al. Base Flipping Free Energy Profiles for Damaged and Undamaged DNA. Chem. Res. Toxicol. 2010;23:1868–1870. doi: 10.1021/tx1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]