Abstract
HIV-1 particles contain RNA species other than the unspliced viral RNA genome. For instance, viral spliced RNAs and host 7SL and U6 RNAs are natural components that are non-randomly incorporated. To understand the mechanism of packaging selectivity, we analyzed the content of a large panel of HIV-1 variants mutated either in the 5′UTR structures of the viral RNA or in the Gag-nucleocapsid protein (GagNC). In parallel, we determined whether the selection of host 7SL and U6 RNAs is dependent or not on viral RNA and/or GagNC. Our results reveal that the polyA hairpin in the 5′UTR is a major packaging determinant for both spliced and unspliced viral RNAs. In contrast, 5′UTR RNA structures have little influence on the U6 and 7SL RNAs, indicating that packaging of these host RNAs is independent of viral RNA packaging. Experiments with GagNC mutants indicated that the two zinc-fingers and N-terminal basic residues restrict the incorporation of the spliced RNAs, while favoring unspliced RNA packaging. GagNC through the zinc-finger motifs also restricts the packaging of 7SL and U6 RNAs. Thus, GagNC is a major contributor to the packaging selectivity. Altogether our results provide new molecular insight on how HIV selects distinct RNA species for incorporation into particles.
INTRODUCTION
Retroviruses are RNA viruses that replicate through a DNA proviral intermediate which is integrated in the host genome. Transcription of this proviral DNA gives rise to a primary full-length transcript (FL RNA) that plays several essential roles in viral replication. In addition to the classical function as a precursor for spliced mRNA production, the FL RNA also acts as mRNA template for protein synthesis and as genomic RNA for packaging into progeny virions. Therefore, a proportion of FL RNA must be diverted from translation to the packaging pathway for virus assembly. Released viruses contain a genome composed of a dimer of two identical FL RNA molecules. Packaging the genome entails the problem of specificity of RNA selection among a substantial excess of non-viral and spliced viral RNAs. Packaging of FL RNA is mediated predominantly by its 5′UTR. The 5′UTR of HIV-1 forms a series of secondary structures, including the transactivation response element forming the TAR hairpin, the polyadenylation signal hairpin (polyA), a large folded structure including the primer binding site (called PBS domain) and three stem loops (SL1, SL2 and SL3) which form the Psi region (1–3) (Figure 1). Most attention has focused on the Psi region with identification of SL1 and SL3 as the major packaging determinants (4–6). RNA mapping studies indicated the presence of additional determinants throughout the entire 5′-terminal region of the HIV-1 RNA. Indeed, TAR, polyA, and PBS structures also influence the packaging efficiency, although their direct role remains controversial (7–14). Packageable FL RNA is recognized by the nucleocapsid (NC) domain of the Gag polyproteins (GagNC) during virus assembly. HIV-1 NC binds SL2 and SL3 motifs with high affinity (15,16), whereas about 1500 NC molecules coat the FL RNA in viral particles (17). Mutation of the highly conserved CCHC residues of the NC zinc fingers (ZFs) impairs FL RNA packaging and results in the production of non-infectious particles (18–22). Interestingly, it has been recently discovered that these non-infectious particles contain high level of viral DNA (22).
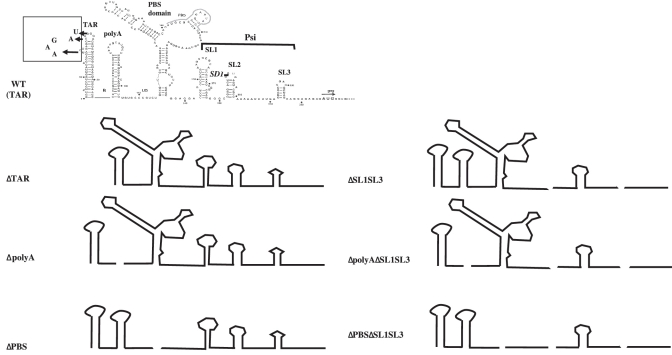
Figure 1.
Schematic representation of 5′UTR mutants of HIV-1 used in the study. The main structures are presented: TAR, polyA, PBS domain including the primer binding site for initiation of reverse transcription, as well as SL1, SL2 and SL3 hairpins forming Psi. SL2 includes the major splice donor site (SD1) that generates all spliced RNAs. TAR is transcribed from HIV-rtTA which is based on the HIV-1 molecular clone LAI in which TAR was inactivated by nucleotide substitutions in both the bulge and loop motifs (boxed).
The process of packaging selectivity remains unresolved. It is known that HIV-1 selectively incorporates RNAs other than the FL HIV-1 RNA genome. Notably, spliced viral mRNAs are present at measurable amounts in infectious wild-type (wt) HIV-1 particles and these transcripts can be reverse transcribed (23,24). Plasma analyses of AIDS patients revealed the accumulation of HIV particles with an abundance of spliced viral RNAs (25). The FL HIV-1 transcript undergoes complex alternative splicing to produce >46 spliced RNAs that can be divided in two classes: Multi Spliced (MS) mRNAs and singly spliced mRNAs with env mRNA (env) as the major representative (26). Like FL RNA, singly spliced RNAs are intron-containing transcripts and they contain the Rev Responsive Element (RRE) conferring nuclear export via the Rev/CRM1 pathway (27). Nevertheless, all spliced RNAs are equally encapsidated regardless of their nuclear export pathway (28). Determinants and mechanism involved in spliced RNA encapsidation remain unidentified. Since the major 5’ splice site (SD1) is contained within the SL2 hairpin, only unspliced RNA contains both the SL1 and SL3 determinants, which could explain, at least in part, why unspliced RNA is preferentially packaged (Figure 1). The main packaging signal SL1 is maintained in all spliced RNAs but was found to be dispensable for packaging (28). We examine the influence of 5′UTR structures such as TAR, polyA and PBS in the RNA selection process and we test the role of the GagNC protein.
In addition to viral RNAs, HIV-1 also packages cellular RNAs. Notably, several polymerase III (Pol III) transcripts are found selectively enriched in HIV-1 particles over their intracellular concentration (28–31). For instance, 7SL RNA, the common ancestor of Alu RNAs, and U6 snRNA were reported to be abundantly present in virions, but with different packaging efficiencies (28). It is unclear whether packaging of U6 and 7SL RNAs are governed by the same process. The only feature that these host RNAs have in common is Pol III transcription, which may suggest that the intracellular trafficking pathway contributes to packaging. The viral determinants for packaging of these small RNAs are still poorly defined and there is no consensus regarding their role in virus replication (31–34). Recent evidence supports a role for 7SL RNA in virion packaging of the antiviral cytosine deaminase APOBEC3G (A3G), although this function remains controversial (35).
The goal of the present study is to identify cis and trans viral determinants that drive the selection of these multiple viral and host RNA species for incorporation into HIV-1 particles. The present study explores the 5′UTR structures of viral RNAs and the GagNC protein requirements for packaging of unspliced and spliced viral RNAs and the consequences for incorporation of the cellular U6 and 7SL RNAs.
MATERIALS AND METHODS
Plasmids
For the construction of pNL4-3 ΔpolyA, and ΔPBS, the plasmid pLTR5’-NL4-3 containing the AatII-Sph1 fragment of the molecular clone HIV-1 pNL4-3 (positions 13 269–1443) in a pUC119 vector was mutated with the QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene) which was used according to the manufacturer’s instructions. The mutated oligonucleotides used were sense-ΔpolyA (5′-GGCTAACTAGGGAACCTGTGCCCGTCTGTTGTG) and antisense-ΔpolyA (5′-CACAACAGACGGGCACAGGTTCCCTAGTTAGCC) for the pLTR5′-ΔpolyA mutant, s-ΔPBS (5′-GTGTGTGCCCGTCTGCAGGACTCGGC) and a-ΔPBS (5′-GCCGAGTCCTGCAGACGGGCACACAC) for the pLTR5′-ΔPBS mutant. The pLTR5′ plasmids were restricted with AatII and SphI enzymes and the obtained fragments were replaced in the homologous region of pNL4-3 (positions 13 269–1443) to obtain the pNL4-3 ΔpolyA, and ΔPBS plasmids, respectively. For the construction of pNL4-3 ΔpolyAΔSL1SL3, and ΔPBSΔSL1SL3, the plasmid pLTR5′-NL4-3ΔSL1SL3 containing the AatII-Sph1 fragment of the molecular clone HIV-1 pNL4-3 with the SL1 and SL3 deletions was mutated as mentioned above. The oligonucleotides are the same as before for the ΔpolyA deletion but for the ΔPBS deletion two other oligonucleotides were used taking into account the SL1 deletion: s-ΔPBS.1 (5′-GTGTGTGCCCGTCTGCAGGAGGGC) and a-ΔPBS.1 (5′-GCCCTCCTGCAGACGGGCACACAC). The presence of the deletions was confirmed by restriction and DNA sequencing. The rtTA and rtTA-E plasmids mutant of TAR are molecular clones of the LAI HIV-1 strain. They are described in detail in (11,36). The pNL4-3 plasmids with NC mutations are described elsewhere: ZF deletion ΔZF1 and ΔZF2 or mutation H23C and H44C (37,38), and mutations of the flanking regions: R7R10K11S and K14D (39), P31L (40) and K59L (41).
Cell culture, transfection and infection
HEK293T cells were grown in DMEM medium (Dulbecco’s modified Eagle’s medium) supplemented with glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 µg/ml) and heat-inactivated fetal calf serum (10% v/v) at 37°C. For the expression of the rtTA and rtTA-E vectors, the cells were grown with doxycyclin (1 µg/ml).
Transfections were performed as previously described (28). In a standard experiment, 3 × 106 cells were grown in 10 cm dishes. The next day, 2 µg of plasmid DNA and 6 µg of pSP72 carrier (Promega) were transfected by phosphate calcium precipitation. For cotransfections, 2 µg of each plasmid were used along with 4 µg of pSP72 carrier. In all cases, in order to eliminate the plasmid in excess in the medium, 6 h after the transfection, cells were trypsinized, centrifuged and then transferred in a new dish. The supernatant was harvested 24 h after the transfection, centrifuged at 1500 rpm during 10 min and filtered at 0.45 µm. Cells were collected by pipetting with PBS and centrifuged 5 min at 1500 rpm.
RNA extractions
RNA extractions from cells and virions were realized as described previously (42). Briefly, cell pellets were treated with 500 µl of TriReagent (MRC) according to the manufacturer's instructions and then RNA was dissolved in 50 µl of ultra-pure water.
Virions were purified from 10 ml of supernatant by ultracentrifugation through a 1 ml sucrose cushion (20% in PBS) at 30 000 r.p.m. during 90 min at 4°C. Virions in the pellet were dissolved in 80 µl of TES (50 mM Tris pH 7.5; 5 mM EDTA; 0.1% SDS), 5 µl were collected to evaluate the p24 quantity in the pellet with the ELISA Innotest HIV kit (Innogenetics), then 180 µl of TES containing 20 µg of tRNA carrier were added to the remaining virions. RNA was then extracted with phenol–chloroform, chloroform and precipitated with ethanol, washed with 70% ethanol and dissolved in ultra-pure water.
All RNA samples were treated with RQ1 DNase (Promega) in presence of RNaseOUT (Invitrogen) during 25 min at 37°C. RNA was extracted with phenol–chloroform then chloroform and finally precipitated with ethanol 100% and washed with ethanol 70%. RNA pellets were dissolved with 15 µl of ultra-pure water. RNAs were quantified by measuring optical absorption at 260 nm.
RT–PCR
Reverse transcription was performed with the expand RT (Roche) with 2 µg of RNA sample for the quantification of viral RNAs and GAPDH RNA and 0.5 µg for the U6 and 7SL RNAs. Oligo(dT) primer (4 µM) was used with viral RNAs and GAPDH cell RNA. Specific internal primers (4 µM) were used for U6 (aU6-103 5′-TATGGAACGCTTCACGAATTTGCG) and 7SL (a7SL148 5′-CCCGGGAGGTCACCATATT) RNAs. For each assay, a control experiment was systematically performed without RT to check the absence of DNA contamination.
After RT, the samples were diluted with ultra-pure water at 1/5 except for U6 and 7SL samples which were diluted at 1/200. A total of 2.5 µl of sample were used for each quantitative PCR reaction performed with the LightCycler® FastStart DNA MasterPLUS SYBR Green I kit (Roche) and the RotorGene apparatus (Labgene). The RT products were amplified by 35 cycles of PCR: 95°C for 15 s; 58°C for 12 s and 72°C for 20 s. 0.5 µM of each of the following oligonucleotides pairs were used: for FL, sHIV-1306 5′-TCAGCATTATCAGAAGGAGCCACC sense and aHIV-1541 5′-TCATCCATCCTATTTGTTCCTGAAG antisense; for MS, sHIV-5967 5′-CTATGGCAGGAAGAAGCGGAG sense and aHIV-8527 5′-CAAGCGGTGGTAGCTGAAGAG antisense; for env, sHIV-729SD1A5 5′-GAGGGGCGGCGACTGGAAGAA sense and aHIV-6134 5′-ACTATGGACCACACAACTATTGC antisense; for GAPDH, GA-721 5′-GCTCACTGGCATGGCCTTCCGTGT sense and GA-931 5′-TGGAGGAGTGGGTGTCGCTGTTGA antisense; for 7SL, s7S-22 5′-CTGTAGTCCCAGCTACTCG sense and a7S-148 5′-CCCGGGAGGTCACCATATT antisense; and for U6, sU6-3 5′-GCTCGCTTCGGCAGCACATATACT sense and aU6-103 5′-TATGGAACGCTTCACGAATTTGCG antisense.
A standard curve was generated from 102 to 106 copies of pNL4-3 plasmid. For analysis of the different RNAs produced in transfected cells, RNA samples were normalized to the GAPDH mRNA level. As negative control, GAPDH RNA was measured in viruses and showed no significant level of this RNA compared to negative control with H2O.
Standard PCR were performed with Taq DNA polymerase (Biolabs) and primers specific to singly spliced RNAs: sHIV490 5′-CTCTCTGGCTAACTAGGGAAC sense and aHIV-6134 antisense (see above). Each 32 PCR cycles consisted of denaturation for 15 s at 94°C, annealing for 1 min at 60°C and extension for 1 min at 72°C. PCR products were run on 2% agarose gel and bands corresponding to env RNA were visualized by GelRed (Biotium) staining and their intensities measured by ImageQuant.
RESULTS
Rationale for HIV-1 mutagenesis
Models for the conformation of the 5′UTR indicate the presence of at least six secondary structure elements (TAR, polyA, PBS, SL1, SL2 and SL3) (1–3) (Figure 1). The U5 junction sequence between the polyA and PBS domain (nt 105–115) is shown unpaired, but can participate in a long-range interaction with nucleotides surrounding the Gag initiation codon (8,43–45) (Figure 1). Based on these models, deletion of complete structural motifs was undertaken in order to minimize structural changes in adjacent domains that could affect packaging in an indirect manner through gross folding defects (46). To determine whether the 5′UTR motifs contribute to packaging selectivity, we singly deleted each of the aforementioned motifs (TAR, polyA and PBS). To study the possible packaging contribution of the TAR hairpin in the absence of its essential function in Tat-mediated activation of transcription, the effect of TAR deletion was examined with an HIV-1 variant (HIV-rtTA) that does not need TAR for transcription (Supplementary Figure S1). This HIV-rtTA vector carries substitutions in TAR that abolish transactivation of the viral promoter by Tat and was used as parental plasmid to construct the TAR-deleted mutant (rtTA-E) (11). For convenience, these rtTA and rtTA-E vectors were renamed TAR and ΔTAR in the present study (Figure 1 and Supplementary Figure S1).
When SL1SL3-driven packaging of FL RNA is abrogated (mutant ΔSL1SL3), the FL and spliced RNAs were packaged with approximate similar efficiency, with an optimal packaging efficiency for the spliced RNAs (28). This context appears to be useful to determine packaging determinants of spliced RNAs. Thus, the polyA and PBS motifs were deleted together with the SL1 and SL3 hairpins (ΔpolyAΔSL1SL3 and ΔPBSΔSL1SL3 in Figure 1).
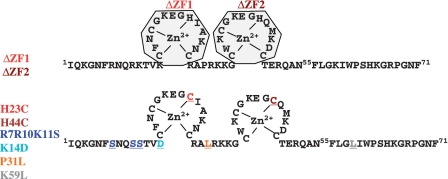
It was reported that Gag binds to the first 261 nt of the 5′UTR, precisely corresponding to the sequence that is shared by all spliced and FL RNAs (47). Thus, we hypothesize that Gag could interact with the spliced RNAs through its nucleocapsid domain (GagNC) known to drive FL RNA packaging (4,5,48). Since the two NC ZFs are major functional elements, we analyzed mutants with deletion of the first (ΔZF1) or the second ZF (ΔZF2) or with substitution, namely His23 and His44 to Cys, referred as the H23C and H44C mutants (Figure 2). These mutations maintain the Zn2+-binding residues (CCCC motifs), but trigger partial misfolding of the NC central globular domain. Basic residues of the N-terminal and the linker sequences stabilize the nucleoprotein complex through electrostatic interactions (49). Thus, the basic residues Arg7, Arg10 and Lys11 were changed to neutral Ser residues (R7R10K11S) and the basic Lys was substituted by the acidic Asp (K14D) (Figure 2). We analyzed the effect of changing Pro31 to Leu31 (mutant P31L) which is located in the highly conserved 29RAPRKKG35 linker involved in spatial orientation of the ZF motifs (15,50–52) (Figure 2). To study the influence of the SP2 domain of Gag, we used the K59L mutant in which Lys59 was changed to Leucine (Figure 2). We undertook a systematic survey of the GagNC requirements for packaging selectivity by monitoring the uptake of viral (FL and spliced) and cellular (7SL and U6) RNAs into HIV-1 particles.
Figure 2.
Sequence of HIV-1 NC-SP2 domain of Gag protein. Deletions (top) and substitutions (bottom) targeting the N-terminus, the zinc fingers and the linker domains of NC and the SP2 domain of Gag are shown by colored residues.
Multiple RNA domains (polyA, PBS, SL1 and SL3) are determinants for FL RNA packaging
Packaging of the FL HIV-1 RNA requires the two hairpins, SL1 and SL3, forming the Psi region (Figure 1) [for review see (4,5,48) and references herein]. In the absence of these two hairpins the packaging of FL RNA (ΔSL1SL3) was strongly decreased (3-fold reduction, Figure 3A) but not abolished and it remained selective despite the reduced efficiency (28). These findings suggest the presence of residual packaging signal(s) within the RNA genome of the ΔSL1SL3 mutant. We hypothesize the presence of additional cis-packaging signals upstream of the splice donor site (SD1) that includes three well-characterized structures, the TAR and polyA hairpins and the PBS domain (Figure 1). The contribution of these motifs was examined by individual deletion in the presence or absence of the major SL1 and SL3 packaging signals. A previous study showed that the level of HIV-1 expression in 293 T cells did not impact on the selectivity of RNA packaging (28). Thus, moderate-level expression of HIV-1 as previously determined (28) was chosen to prevent cell lysis, which could disturb the intracellular RNA traffic. The content of wt and mutant HIV-1 particles was rigorously assessed with highly quantitative techniques such as real-time RT–PCR and ELISA assays for quantitation of RNA and virion capsids (CA-p24), respectively. For RNA analysis, two control RT–PCR reactions were systematically performed. One control was used to check for DNA contamination by running RT reactions without reverse transcriptase, followed by quantitative PCR amplification. The other control experiment evaluated the background level by running a real-time PCR with RNA samples extracted from supernatant of mock-transfected cells. These background signals were removed from the RNA copy numbers measured in HIV-1 positive samples and the RNA data were normalized for viral particle production as quantitated by CA-p24 ELISA.
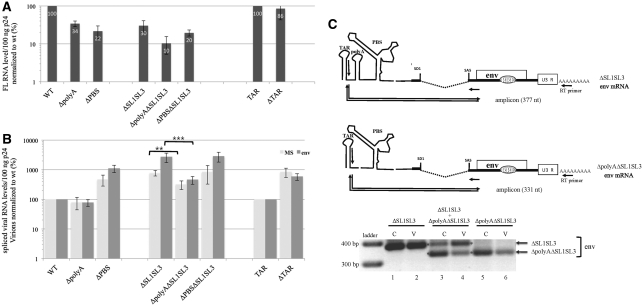
Figure 3.
Effects of the 5′UTR mutations on packaging of viral RNAs. (A) Quantitation of FL RNA in wt and mutant particles. Levels of released virions and FL RNA content were determined by p24 ELISA and quantitative RT–PCR, respectively. Copy numbers of FL RNA were given for 100 ng p24 and normalized to wt. Error bars indicate SD from five independent experiments. (B) Effects of mutations on spliced RNA levels in virions. MS for overall multi-spliced RNA class and env (for mRNA encoding envelope) is one major representative of the singly spliced RNA class. Spliced RNA were measured from the same RNA samples as used for FL RNA quantitation. Error bars indicate SD from five independent experiments. (C) Comparison of spliced RNA encapsidation when polyA+ and polyA− viruses are co-expressed. 293T cells were transfected with ΔSL1SL3 and ΔpolyAΔSL1SL3 either together or individually as control. RNA was extracted from cells (C) and viruses (V). Spliced env-RNAs were detected by standard RT–PCR and the amplified products were run on agarose gel. The mutant mRNA templates, the RT and PCR primers and the amplified cDNA products were drawn in the top part. Since polyA deletion shortened the amplified product by 46 nt, the different mutant forms of env-RNAs were distinct.
The role of the TAR motif was investigated with a Tat-independent vector (TAR) that allows TAR hairpin deletion without abolishing RNA production (11) (Supplementary Figure S1). We found that deletion of the TAR hairpin (ΔTAR) had little effect on the intravirion FL RNA level, indicating that TAR hairpin is not crucial for packaging of FL RNA. In contrast, deletion of the polyA hairpin (ΔpolyA) or the PBS domain (ΔPBS) reduces the packaging by about 70% when compared to that of the corresponding wt control virus. Interestingly, such decreases in RNA packaging efficiency are similar to that observed for ΔSL1SL3 RNA (Figure 3A). The absence of cumulative effect of ΔPBS and ΔSL1SL3 in the double mutant (ΔPBSΔSL1SL3) suggests that the PBS effect might result from SL1SL3 disturbance. A more prominent decrease was observed when the polyA deletion was combined with SL1SL3 removal in ΔpolyAΔSL1SL3 leading to FL RNA incorporation levels of 10% when compared to the wt control. Consistent with previous reports (9,13,14,53), the polyA and the PBS motifs appeared important for packaging of FL HIV-1 RNA. Additionally, our results reveal similar contributions of these two structures and the major SL1SL3 determinants.
The polyA hairpin contributes to packaging of spliced viral RNAs
In addition to the FL viral genome, the spliced viral RNAs are also specifically incorporated in HIV-1 virions. A previous quantitative and comparative analysis showed that multi-spliced (RRE-) and singly-spliced (RRE+) mRNAs display similar encapsidation capabilities that do not required the presence of SL1 (28). Up to now, there is no data available on the packaging determinants of viral spliced mRNAs. Since the TAR, polyA and PBS structures are included in all spliced RNA molecules, we measured the effects of their deletion on spliced mRNAs packaging.
These 5′UTR deletions could affect the usage of the adjacent SD1 site that is used for production of all spliced mRNA species (2) (Figure 1) and could complicate the RNA packaging analysis. Thus, we first examined the splicing efficiencies of the 5′UTR mutated transcripts. RNA was extracted from transfected cells and the intracellular levels of spliced and unspliced RNAs were determined. Quantitative analysis of spliced and unspliced viral RNAs was performed by using the same initial RT reaction, followed by specific qPCR amplifications. We used a primer pair specific for the SD4/SA7 exon–exon junction that allowed quantification of the MS RNA class. Intracellular ratios of MS to FL RNA were determined. Results for ΔSL1SL3 (Table 1) correlate with a previous study showing that the ΔSL1SL3 mutation did not influence the splicing (28). Except for the ΔTAR mutant that exhibited a modest 2-fold decrease of splicing, the ΔpolyA, ΔPBS and ΔpolyAΔSL1SL3 mutations do not seem to significantly modify the splicing (P > 0.1) (Table 1). However, the ΔPBSΔSL1SL3 shows a 1.6-fold splicing increase (P = 0.005) and a possible effect on packaging cannot be excluded (Table 1).
Table 1.
Effect of 5′UTR deletions on splicing
| Wt | ΔpolyA | ΔPBS | ΔSL1SL3 | ΔpolyA ΔSL1SL3 | ΔPBS ΔSL1SL3 | TAR | ΔTAR |
|---|---|---|---|---|---|---|---|
| MS/FL (%) | |||||||
| 100 | 93 ± 34 | 119 ± 27 | 142 ± 57 | 101 ± 25 | 163 ± 20 | 100 | 52 ± 22 |
The ratio of MS to FL RNAs were normalized to ratio determined in cells transfected with wt HIV-1. The values represent mean ± SD of at least three independent experiments.
Despite the 2-fold decreased splicing efficiency (Table 1), the TAR mutant (ΔTAR) displayed about a 7-to 9-fold increase of spliced mRNA level in virions compared to the corresponding wt construct (TAR) (Figure 3B). These results indicate that the TAR hairpin is not required for packaging of the spliced mRNAs and rather acts as a negative determinant. Similarly, deletion of the PBS domain (ΔPBS) increased the amount of MS and env mRNAs in virion by 4- and 11-fold, respectively, compared to wt (Figure 3B). As expected, ΔSL1SL3 mutant particles exhibit a strong increase of the MS and env mRNA levels, yielding the highest ratios of RNA to CA-p24 (increases about 700 and 2700% of wt, respectively, Figure 3B). The ΔPBSΔSL1SL3 mutant contains levels of spliced RNAs close to those of the ΔSL1SL3 mutant (Figure 3B). Since the mutant mRNAs are not packaged less efficiently than the wild-type mRNAs, the PBS domain appears dispensable for packaging of these mRNAs.
In contrast, deletion of the polyA hairpin (ΔpolyA) leads to a 20% packaging decrease compared to wt (Figure 3B). To confirm the importance of the polyA signal, packaging was monitored in the ΔSL1SL3 context (ΔpolyAΔSL1SL3) where packaging of FL RNA, possibly a competitor of spliced RNAs, was impaired (trans-effect). In the ΔSL1SL3 context, the packaging of spliced RNAs was not affected by a direct cis-effect, since the SL1 motif was dispensable and SL3 is absent from the wt spliced sequences. A previous analysis of the ΔSL1, ΔSL3 and ΔSL1SL3 mutants revealed that the highest increase of spliced RNA packaging was obtained with the co-deletion (ΔSL1SL3) (28). Therefore, the ΔSL1SL3 context appears the most appropriate for the detection of packaging reduction when deleting a putative cis-determinant. Interestingly, the ΔpolyAΔSL1SL3 virions contained 2.5- and 6-fold less MS and env mRNAs, respectively, compared to ΔSL1SL3 particles. These results combined with with the absence of splicing reduction (Table 1) suggest that the polyA motif contributes to the spliced RNA packaging. To directly address the role of the polyA hairpin, we undertook competition experiments. The ΔSL1SL3 and ΔpolyAΔSL1SL3 plasmids were cotransfected in 293 T cells. RNA was extracted from cells and virions, and analyzed by standard PCR, followed by agarose gel electrophoresis and visualization of the band corresponding to env RNAs. This approach allowed discrimination of the ΔSL1SL3 and ΔpolyAΔSL1SL3 RT–PCR products, due to the ΔpolyA deletion (Figure 3C). Although ΔpolyAΔSL1SL3 env-RNAs were more abundant than ΔSL1SL3 env-RNAs in cells (lane 3), the ΔpolyAΔSL1SL3 env-RNAs were less efficiently (∼2-fold) incorporated than ΔSL1SL3 in virions (lane 4). Thus, when the two mutant env-RNAs were present together, HIV-1 preferentially packaged the RNA with the polyA motif, demonstrating that the polyA structure confers an advantage to the spliced RNA for packaging. Altogether, these results reveal that the polyA hairpin is a packaging determinant of the viral spliced RNAs and of FL RNA.
Influence of 5′UTR mutations on packaging of host 7SL and U6 RNAs
Several host cell derived RNAs, such as the 7SL and U6 transcripts, are natural components of HIV-1 particles that are packaged in a non-random manner. Previous study of their abundance in virions reported only 10-times less copies of 7SL and U6 RNA than that of FL RNA in the same virion sample (28). To examine whether 7SL/U6 incorporation is influenced by viral RNA packaging, we measured their amounts in the different 5′UTR-mutated viruses. As a control for non-specific secretion of cellular RNAs, the culture supernatant from mock-transfected 293 T cells was always analyzed in parallel. A low level of spontaneous host RNA secretion was measured for both RNAs (0.004%) and this value was subtracted from the quantitated RNA levels. Host RNA copy numbers were also normalized for viral particle production (CA-p24) (Figure 4A).
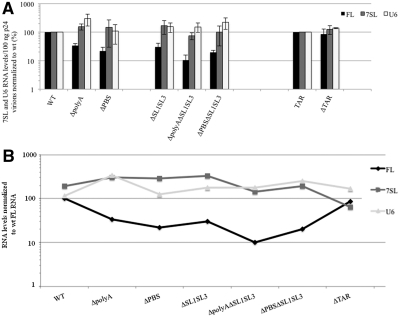
Figure 4.
Consequences of 5′UTR mutations on packaging of host U6 and 7SL RNAs. (A) For each RNA species, specific primers were used for both RT and qPCR reactions. As in Figure 3, RNA levels are expressed as copy numbers for 100 ng p24 and normalized to wt. Error bars indicate SD from five independent experiments. (B) To compare level variations between FL and host RNAs among the 5′UTR mutants, RNA levels/100 ng p24 were expressed as percentage of the wt FL RNA and reported in a log-scale graph.
All 5′UTR mutant virions harbor modest increases of both cellular 7SL and U6 RNAs compared to the wt construct (Figure 4A). To know whether the variation of host cell RNA packaging depends on FL RNA, the 7SL and U6 RNA amounts were plotted as percentage of wt FL RNA (Figure 4B). This analysis showed no correlation, suggesting that host RNA incorporation is not influenced directly by that of FL RNA.
Role of different GagNC domains in FL RNA packaging
GagNC protein plays a key role in virus assembly and the RNA packaging process, which require its nucleic acid-binding and chaperone activities (20). To study the control exerted by GagNC on the packaging selectivity, we studied the effects of mutating the two highly conserved CCHC ZFs, the basic RAPRKKG linker sequence and the flanked N-terminal and C-terminal domains (52) (Figure 2). After transfection of cells with the wt and mutant HIV-1 plasmids, the level of intravirion RNA was determined as described above for the 5’UTR mutants and as described in ‘Materials and Methods’ section.
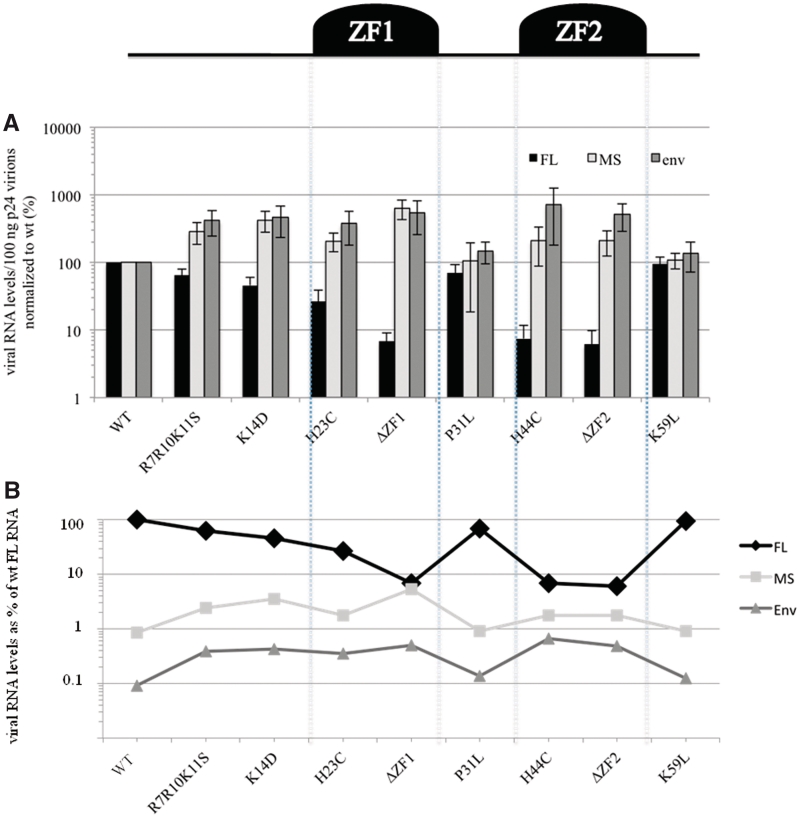
As expected, deletion or mutation of either ZF1 or ZF2 decreased the FL RNA level in virions, with the strongest decrease scored for ΔZF1 and ΔZF2 (6% of wt) (Table 2). These severe effects confirm the importance of the two ZFs for FL RNA packaging (37,38,54–56,60). The basic residues interact with viral RNA in vitro (57), especially with the SL3 hairpin (15). We therefore investigated the impact of the N-terminal mutants R7R10K11S and K14D and the linker mutant P31L. These substitutions caused moderate decreases of the FL RNA levels in virions (∼30–55% of wt) (Table 2), consistent with previous studies (39,58–62). In contrast, the K59L substitution in the Sp2 domain of Gag had no effect on RNA packaging. Altogether, these results support the notion that the ZFs are intimately involved in the interaction with viral RNA, and the basic residues may neutralize the phosphates of the RNA backbone, leading to specific packaging of the genome.
Table 2.
Effect of GagNC mutations on FL RNA level in HIV-1 particles
| Wt | R7R10K11S | K14D | H23C | ΔZF1 | H44C | ΔZF2 | P31L | K59L |
|---|---|---|---|---|---|---|---|---|
| FL RNA (%) | ||||||||
| 100 | 64 ± 15 | 46 ± 15 | 27 ± 12 | 7±2 | 7 ± 4 | 6 ± 4 | 70 ± 24 | 96 ± 25 |
For comparison purposes, averaged values were normalized to wt. The values represent mean ± SD of at least three independent experiments.
N-terminal basic residues and ZFs of GagNC negatively regulate packaging of viral spliced mRNAs
As for the 5′UTR RNA mutants, the effects of the GagNC mutations on splicing were first investigated. We measured the amounts of the viral RNAs produced in transfected cells and determined the intracellular ratio of MS to FL RNA for each mutant (Table 3). These results indicated that splicing is not affected. To investigate the role of GagNC in spliced RNA packaging, we determined the levels of MS and env mRNAs in wt and mutant viruses. Ratios of RNA to CA-p24 were calculated and normalized to that of wt. All mutations increased the incorporation of spliced mRNAs in virions, with exception of the P31L and K59L virions that exhibit about wt level (Figure 5A). The most prominent effect was obtained for the ΔZF1 mutant with a 5-to 6-fold increase of the env and MS mRNAs, respectively. It appears that spliced mRNA packaging increases when the intravirion level of FL RNA decreases and vice versa. These inverse co-variations can be illustrated when the spliced mRNA levels are plotted as percentage of wt FL RNA (Figure 5B). These results indicate that the two ZFs and the basic residues of the N-term restrict the incorporation of spliced RNA, while favoring FL RNA incorporation into virions.
Table 3.
Effect of GagNC mutations on splicing
| wt | R7R10K11S | K14D | H23C | ΔZF1 | H44C | ΔZF2 | P31L | K59L |
|---|---|---|---|---|---|---|---|---|
| MS/FL (%) | ||||||||
| 100 | 81 ± 28 | 83 ± 34 | 112 ± 7 | 94 ± 25 | 108 ± 4 | 99 ± 21 | 76 ± 21 | 129 ± 28 |
The ratio of MS to FL RNAs were normalized to ratio determined in cells transfected with wt HIV-1. The values represent mean ± SD of at least three independent experiments.
Figure 5.
Role of the GagNC in selective uptake of HIV-1 RNAs. A schematic representation of NC is reported in the upper part. (A) Quantitation of spliced and unspliced RNAs in wt and mutant particles. Copy numbers of each RNA species were given for 100 ng p24 and normalized to wt. Error bars indicate SD from at least three independent experiments. (B) Comparative analysis of the effects of mutations on the different RNA species. RNA levels/100 ng p24 were expressed as percentage of the wt FL RNA.
Role of GagNC in packaging of 7SL and U6 RNAs
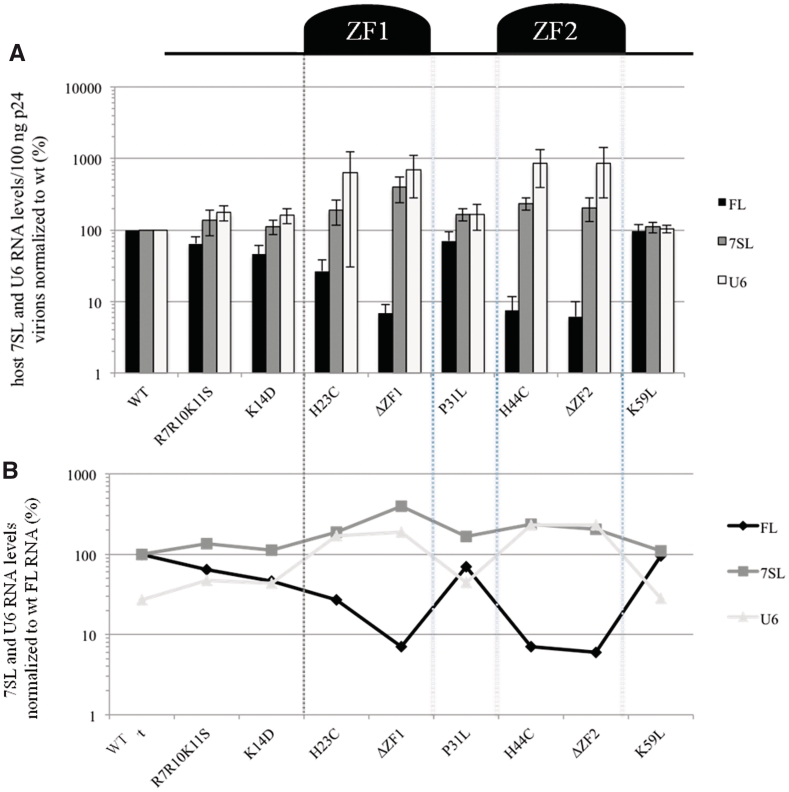
Since GagNC is involved in the selective uptake of viral RNAs in virions, it could also play a role in the selection of the host RNAs. Thus, we examine whether deletion or mutation of GagNC changes the virion levels of 7SL and U6 RNA. The intravirion amounts of 7SL and U6 RNAs were determined for each GagNC mutant by RT-qPCR with a specific set of primers, as described in the ‘Materials and Methods’ section. The results show that the ZF mutants H23C, ΔZF1, H44C and ΔZF2 contain an increased amount of the two cellular RNAs. This up-regulation is more pronounced for U6 (up to 8-fold) than for 7SL (up to 4-fold) (Figure 6A). In contrast, U6 and 7SL RNAs respond similarly to mutation of the other NC domains (R7R10K11S, K14D, P31L and K59L), which cause an average increase of 1.4-fold compared to wt (Figure 6A). These results are expressed as percentage of the wt FL RNA level in the overview graph in Figure 6B. This graph suggests an inverse co-variation between the FL and the cellular RNAs. Indeed, when the level of FL RNA decreased in particular mutants, the level of 7SL and U6 increased. Altogether, these results indicate that GagNC contributes, mostly through its zinc-fingers, to discrimination between the viral genome and cellular U6 and 7SL RNAs.
Figure 6.
GagNC restricts the incorporation of the host 7SL and U6 RNAs in virions. (A) Quantitation of 7SL, U6 and FL in wt and mutant particles. Copy numbers of each RNA species were given for 100 ng p24 and normalized to wt. Error bars indicate SD from at least three independent experiments. (B) Comparative analysis of the effects of mutations on the different RNA species. RNA levels/100 ng p24 were expressed as percentage of the wt FL RNA.
DISCUSSION
This study indicates that non-genomic RNAs, such as spliced viral RNAs and the cellular 7SL and U6 RNAs, are incorporated into HIV-1 particles through a selective mechanism. We studied packaging of the host U6 and 7SL RNAs in virions by mutating two key components of HIV-1 particles: the 5′UTR of the viral RNA genome and the GagNC protein. The variation in 7SL encapsidation efficiency among the mutants was similar to the effects on U6 RNA. There is no obvious correlation between incorporation of viral and host RNAs for the 5′UTR mutants (Figure 4B). Thus, the virion recruitment of 7SL and U6 is not dependent on FL RNA, a result that is consistent with previous studies (31,34). The GagNC mutations suggest an opposite correlation for incorporation of host and FL RNAs (Figure 6B). Thus, the level of 7SL and U6 RNAs seems affected differently by disturbing viral RNA packaging through mutation of cis (RNA) versus trans (NC protein) elements. For instance, the ΔpolyAΔSL1SL3 (cis) and ΔZF1 (trans) mutants incorporate the same low amount of FL RNA (∼10% of wt) (Figures 3A and 6A), but an increased level of U6 RNA ranging from 154% to 630% of wt, respectively (Figures 4A and 6A). Taken together, these findings exclude the hypothesis of co-packaging that is based on RNA–RNA interactions between cellular and viral RNAs. The results also indicate that incorporation of the cellular RNA is not triggered by the space left by the HIV-1 RNA genome, but rather that it involves a specific RNA packaging process. This process seems negatively regulated, either directly or indirectly, by the GagNC protein that favors packaging of FL RNA. Thus, GagNC is a major contributor to the discrimination between viral and cellular RNAs.
Several recent studies focused on the incorporation of 7SL RNA in HIV-1 particles because 7SL is thought to account for the incorporation of the antiviral cytidine deaminase A3G (35). However, the role of NC in 7SL packaging remains controversial. Some studies reported that 7SL incorporation depends on the NC protein (31,63,64). Other studies did not score reduced 7SL packaging with mutant HIV-1 constructs expressing either a Gag protein deleted for the NC domain or NC replaced by a leucine zipper (Zwt-p6), such that the authors concluded that 7SL is packaged in an NC-independent manner (30,34). Keene et al. explain this discrepancy by the different approaches used to monitor 7SL RNA, because the 7SL molecule is truncated in minimal virus-like particles (VLPs) lacking the NC protein, thus generating undetectable molecules (65). Our PCR measurements specifically detect intact 7SL molecules. We think that the use of HIV particles instead of VLPs provides a more rigorous assessment. In agreement with some studies (30,34), we found that GagNC mutation does not reduce 7SL incorporation. We measured an increased level of 7SL RNA packaging for the ZFs mutants (2- to 4-fold), suggesting that GagNC negatively controls 7SL incorporation, either directly or indirectly, through co-factor(s) present in HIV particles such as the viral Vif protein (66,67) or cellular proteins like Staufen1 (68), ABCE1 (69), Bro1 (70) or others (71).
There are several literature reports on the virion packaging of spliced HIV-1 RNAs that are natural components of infectious HIV-1 particles (23,24). Their presence has been reported mainly in particles with reduced levels of FL RNA, e.g. due to Gag or Psi mutation (72–76). But the packaging mechanism remains undefined, mostly because the spliced RNAs are encapsidated only as trace amount in wt particles, which requires sensitive analysis methods. Recently, we showed that their presence in HIV-1 particles results from an active and selective process, and not from random incorporation (28). In addition, this previous packaging study concluded that neither SL1 nor SL3 (absent in spliced sequences) acts as cis-packaging determinants of the spliced RNAs. Their deletions (ΔSL1SL3) induced increased packaging of spliced RNAs that resulted from a competitive compensation of the defect in FL RNA packaging, rather than from a direct cis-effect on spliced RNAs (28). Here we show for the first time a decrease of spliced RNA incorporation by deletion of the polyA hairpin structure of the ΔSL1SL3 mutant, revealing the polyA hairpin as a new cis-packaging determinant for spliced mRNAs.
Our screen for a trans-packaging determinant within the GagNC domain indicates that ZF and the N-terminal basic residues restrict packaging of the spliced RNAs. Consistent with these results, increased packaging of spliced RNA was previously reported for the R10A and K11A NC mutants (75). This phenotype was suppressed by the compensatory T12I mutation in the SP1 domain of Gag (75,76). The matrix (MA) domain of Gag could also play a redundant role with NC in Gag/RNA association (77). A recent study selected compensatory mutations for the ΔSL1 deletion that exhibits increased spliced RNA packaging. It was shown that the MA and SP1 domains of Gag restore the exclusion of spliced RNAs from virions, indicating a role of MA and SP1 in spliced RNA packaging. Further immunoprecipitation experiments indicated that MA and SP1 are responsible for the association between Gag and spliced RNAs (78).
We show that viral spliced and FL RNAs share the polyA hairpin as packaging determinant, but the packaging of FL RNA relies on several additional structures such as PBS, SL1 and SL3 that are either inactive (PBS, SL1) or simply absent (SL3) in spliced RNAs (Figure 3B; 28). Although the TAR hairpin seems to restrict spliced RNA packaging, TAR was found dispensable for HIV-1 RNA packaging, which is in agreement with a previous study (11), but not with others (9,10,79). This discrepancy could be explained by adverse 5′UTR folding effects induced by mutational destabilization of the TAR hairpin. Specifically, its destabilization has been reported to influence the neighbouring polyA hairpin, which could affect this packaging determinant (46).
The present study does not only confirm the importance of NC domains for FL RNA packaging, that is the N-terminal basic residues and especially the ZFs, but these NC domains also control the packaging specificity by favoring FL RNA over spliced RNA (Figure 5B). This inverse correlation between FL and spliced RNA incorporation suggests a competitive process. Preferential packaging of FL RNA is attributable to the high affinity of GagNC for the 5′UTR. The 5′UTR likely adopts different conformations in spliced and unspliced molecules. For instance, spliced RNAs could not form the long-range U5–AUG interaction that is important for genome dimerization and that is probably induced by Gag association (8,43–45,80,81). This could reduce the affinity of NC for spliced RNAs and explain their reduced packaging efficiency.
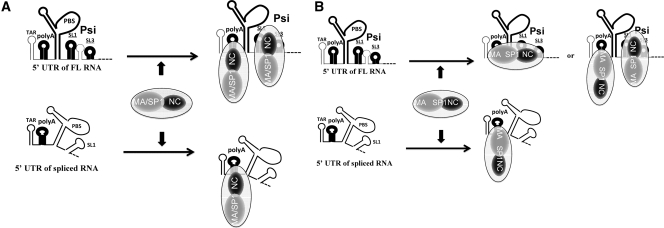
A model that explains the packaging specificity can be proposed (Figure 7A). The Gag protein, through its NC domain, binds with similar affinity to the polyA hairpin in spliced and FL RNAs. This Gag/RNA association is reinforced by the strong affinity of the GagNC for properly folded Psi region (notably SL1 and SL3) of FL RNA. With wild-type Gag, mainly FL RNA is packaged due to its tight interactions with multiple Gag molecules at the expense of spliced RNAs which lack active Psi elements. In contrast, in the presence of mutated NC, the RNA/Gag association is only ensured by the polyA site. In this case, spliced and FL RNAs equally compete for packaging, leading to increased levels of spliced RNAs in virions. Based on studies showing that the MA/SP1 domains of Gag can bind the spliced RNAs and participate into their packaging process (75–78), a more complex model could be proposed. In this alternative model, Gag binds loosely to the polyA site of the FL and spliced RNAs through its MA/SP1 domains (instead of GagNC in the first model), while GagNC binds tighly to the Psi region of the FL RNA (Figure 7B). This model predicts that GagNC mutation do not affect packaging of the ΔpolyA spliced RNA that remains to be determined.
Figure 7.
Models for selective encapsidation of viral unspliced and spliced RNAs. (A) The FL RNA includes the Psi site formed by SL1 and SL3 hairpins that is properly folded and specifically recognized by Gag. GagNC domain is indicated in black and Gag MA/SP1 domains in grey. (A) The polyA hairpin harbored by FL and spliced RNAs, could also be recognized by the GagNC, conferring to the FL RNA a second Gag binding site. Thus, FL RNA is more competitive than spliced RNAs, since the FL RNA through its polyA and Psi sites can bind more Gag molecules than spliced RNAs. This common cis-packaging determinant (polyA) may explain how FL and spliced RNA species are packaged in a competitive manner and why they harbored similar (but low) packaging efficiencies in ΔSL1SL3 context (28). (B) In this alternative model, Gag MA/SP1 domains bind loosely to the polyA, while GagNC, as in (A), tightly binds to the Psi region. These two types of Gag/RNA interaction illustrate how FL RNA is more competitive that spliced RNAs for packaging.
Furthermore, the viral spliced RNAs are not the only transcripts to increase their virion content when GagNC is mutated. Indeed, HIV-1 mutated in the ZFs or the basic residues also packaged 100 times more viral cDNA than wt particles (54,82,83). We demonstrated that this DNA packaging resulted from the deregulation of viral reverse transcription activity during late steps of replication, called late RT (22). Taken together, these data and the present results showed a correlation between levels of DNA and FL RNA among the NC-mutant particles (Supplementary Figure S2). This is consistent with the notion that viral reverse transcriptase copies the FL RNA to generate the double-stranded viral DNA while degrading the template RNA via its associated RNase H activity. Consequently, decreased incorporation of FL RNA in NC-mutant particles could result from decreased affinity of NC for FL RNA unable to recruit RNA but also from degradation by RNAse H activity. This new role of GagNC in the spatio-temporal control of reverse transcription makes the RNA packaging process appear more complex than initially thought. RNA dimerization as prerequisite for packaging could also be mentioned as complicating factor. Indeed, spliced HIV-1 RNA can dimerize in vitro (84). However, the hypothesis of spliced RNA homodimerization or heterodimerization with FL RNA as positive signal for packaging is not supported by the fact that SL1, the dimerization initiation site, is dispensable for spliced RNA packaging (28) and by the absence of systematic inverse co-variation between FL and spliced RNA packaging (Figure 3). To better understand the packaging process and its selectivity, it will be interesting to know where and when viral RNA is first recruited by Gag to target the assembly sites in the cell. Several recent studies based on live cell microscopy of single RNA molecule have begun to reveal the initial events of HIV-1 RNA packaging, bringing evidences that Gag/RNA interact at a perinuclear/centrosomal site (85) and in the cytosol (86–88).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS and ANRS; Fellowship from ANRS (to P.J.R.); NWO-CW (TOP grant to B.B.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the team, including S. Lainé, L. Pessel-Vivares and B. Yu for helpful discussions and constant support.
REFERENCES
- 1.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mougel M, Smagulova F, Ramirez J, Houzet L. RNA Packaging in MuLV and HIV-1 Retroviral Particles. Curr. Topics Virol. 2004;4:201–218. [Google Scholar]
- 5.D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 6.Jouvenet N, Lainé S, Pessel-Vivares L, Mougel M. Cell biology of retroviral RNA packaging. RNA Biol. 2011;8:1–9. doi: 10.4161/rna.8.4.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrich D, Hooker CW, Parry E. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 2000;74:5639–5646. doi: 10.1128/jvi.74.12.5639-5646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song R, Kafaie J, Laughrea M. Role of the 5′ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47:3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 9.McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helga-Maria C, Hammarskjold ML, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 2007;81:7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 13.Clever JL, Mirandar D, Jr, Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303:152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 15.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 17.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, Barsov E, II, Hood BL, Fisher RJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 19.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 20.Muriaux D, Darlix JL. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mougel M, Cimarelli A, Darlix J-L. Implications of the nucleocapsid and the microenvironment in retroviral reverse transcription. Viruses. 2010;2:939–960. doi: 10.3390/v2040939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mougel M, Houzet L, Darlix JL. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C, Hu J, Russell RS, Kameoka M, Wainberg MA. Spliced human immunodeficiency virus type 1 RNA is reverse transcribed into cDNA within infected cells. AIDS Res. Hum. Retroviruses. 2004;20:203–211. doi: 10.1089/088922204773004923. [DOI] [PubMed] [Google Scholar]
- 24.Houzet L, Morichaud Z, Mougel M. Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology. 2007;4:30. doi: 10.1186/1742-4690-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saurya S, Lichtenstein Z, Karpas A. Defective rev response element (RRE) and rev gene in HAART treated AIDS patients with discordance between viral load and CD4+ T-cell counts. J. Clin. Virol. 2005;33:324–327. doi: 10.1016/j.jcv.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi R, Bogerd HP, Cullen BR. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J. Virol. 2002;76:2036–2042. doi: 10.1128/jvi.76.5.2036-2042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007;35:2695–2704. doi: 10.1093/nar/gkm153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rulli SJ, Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA. 2006;12:542–546. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian C, Wang T, Zhang W, Yu XF. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007;35:7288–7302. doi: 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giles KE, Caputi M, Beemon KL. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA. 2004;10:299–307. doi: 10.1261/rna.2150604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 2001;75:979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix JL. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix JL. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ottmann M, Gabus C, Darlix JL. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J. Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Khorchid A, Wang J, Parniak MA, Darlix JL, Wainberg MA, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160(gag-pol) and tRNA(Lys) incorporation into human immunodeficiency virus type 1. J. Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smagulova F, Maurel S, Morichaud Z, Devaux C, Mougel M, Houzet L. The highly structured encapsidation signal of MuLV RNA is involved in the nuclear export of its unspliced RNA. J. Mol. Biol. 2005;354:1118–1128. doi: 10.1016/j.jmb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 44.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Spriggs S, Garyu L, Connor R, Summers MF. Potential intra- and intermolecular interactions involving the unique-5' region of the HIV-1 5′-UTR. Biochemistry. 2008;47:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vrolijk MM, Ooms M, Harwig A, Das AT, Berkhout B. Destabilization of the TAR hairpin affects the structure and function of the HIV-1 leader RNA. Nucleic Acids Res. 2008;36:4352–4363. doi: 10.1093/nar/gkn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geigenmuller U, Linial ML. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J. Virol. 1996;70:667–671. doi: 10.1128/jvi.70.1.667-671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lever AM. HIV-1 RNA packaging. Adv. Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 49.Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry. 2011;49:3525–3533. doi: 10.1021/bi901279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mely Y, Jullian N, Morellet N, De Rocquigny H, Dong CZ, Piemont E, Roques BP, Gerard D. Spatial proximity of the HIV-1 nucleocapsid protein zinc fingers investigated by time-resolved fluorescence and fluorescence resonance energy transfer. Biochemistry. 1994;33:12085–12091. doi: 10.1021/bi00206a011. [DOI] [PubMed] [Google Scholar]
- 51.Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 52.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berkhout B. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 2000;48:29–73. doi: 10.1016/s1054-3589(00)48003-8. [DOI] [PubMed] [Google Scholar]
- 54.Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008;36:2311–2319. doi: 10.1093/nar/gkn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demene H, Dong CZ, Ottmann M, Rouyez MC, Jullian N, Morellet N, Mely Y, Darlix JL, Fournie-Zaluski MC, Saragosti S, et al. 1H NMR structure and biological studies of the His23–>Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 56.Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 57.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poon DT, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cen S, Huang Y, Khorchid A, Darlix JL, Wainberg MA, Kleiman L. The role of Pr55(gag) in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology. 2008;375:592–610. doi: 10.1016/j.virol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Laughrea M, Shen N, Jette L, Darlix JL, Kleiman L, Wainberg MA. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin. Virology. 2001;281:109–116. doi: 10.1006/viro.2000.0778. [DOI] [PubMed] [Google Scholar]
- 62.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 65.Keene SE, King SR, Telesnitsky A. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J. Virol. 2010;84:9070–9077. doi: 10.1128/JVI.00714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan MA, Aberham C, Kao S, Akari H, Gorelick R, Bour S, Strebel K. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 2001;75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 2006;281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- 70.Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goff SP. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 72.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 73.Clever JL, Parslow TG. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill MK, Shehu-Xhilaga M, Campbell SM, Poumbourios P, Crowe SM, Mak J. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 2003;77:8329–8335. doi: 10.1128/JVI.77.15.8329-8335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roy BB, Russell RS, Turner D, Liang C. The T12I mutation within the SP1 region of Gag restricts packaging of spliced viral RNA into human immunodeficiency virus type 1 with mutated RNA packaging signals and mutated nucleocapsid sequence. Virology. 2006;344:304–314. doi: 10.1016/j.virol.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 76.Russell RS, Roldan A, Detorio M, Hu J, Wainberg MA, Liang C. Effects of a single amino acid substitution within the p2 region of HIV-1 on packaging of spliced viral RNA. J. Virol. 2003;77:12986–12995. doi: 10.1128/JVI.77.24.12986-12995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ott DE, Coren LV, Gagliardi TD. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 2005;79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ristic N, Chin MP. Mutations in matrix and SP1 repair the packaging specificity of a Human Immunodeficiency Virus Type 1 mutant by reducing the association of Gag with spliced viral RNA. Retrovirology. 2010;7:73. doi: 10.1186/1742-4690-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das AT, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 81.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Didierlaurent L, Houzet L, Morichaud Z, Darlix JL, Mougel M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008;36:4745–4753. doi: 10.1093/nar/gkn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 2008;82:9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sinck L, Richer D, Howard J, Alexander M, Purcell DF, Marquet R, Paillart JC. In vitro dimerization of human immunodeficiency virus type 1 (HIV-1) spliced RNAs. RNA. 2007;13:2141–2150. doi: 10.1261/rna.678307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 86.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl Acad. Sci. USA. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 2010;84:6352–6366. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5:e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.