Abstract
Metazoan spliced leader (SL) trans-splicing generates mRNAs with an m2,2,7G-cap and a common downstream SL RNA sequence. The mechanism for eIF4E binding an m2,2,7G-cap is unknown. Here, we describe the first structure of an eIF4E with an m2,2,7G-cap and compare it to the cognate m7G-eIF4E complex. These structures and Nuclear Magnetic Resonance (NMR) data indicate that the nematode Ascaris suum eIF4E binds the two different caps in a similar manner except for the loss of a single hydrogen bond on binding the m2,2,7G-cap. Nematode and mammalian eIF4E both have a low affinity for m2,2,7G-cap compared with the m7G-cap. Nematode eIF4E binding to the m7G-cap, m2,2,7G-cap and the m2,2,7G-SL 22-nt RNA leads to distinct eIF4E conformational changes. Additional interactions occur between Ascaris eIF4E and the SL on binding the m2,2,7G-SL. We propose interactions between Ascaris eIF4E and the SL impact eIF4G and contribute to translation initiation, whereas these interactions do not occur when only the m2,2,7G-cap is present. These data have implications for the contribution of 5′-UTRs in mRNA translation and the function of different eIF4E isoforms.
INTRODUCTION
Eukaryotic translation initiation is the main regulatory step for translation and is controlled at multiple levels (1–3). The initiation complex eIF4F mediates ribosome recruitment to mRNA and is composed of three subunits: eIF4E, eIF4G and eIF4A (4). The binding of eIF4E to the cap is an essential step for cap-dependent translation (4–6). Efficient mRNA translation initiation also requires the scaffold protein, eIF4G, which interacts with eIF4E, eIF4A, the mRNA, and recruits the 40 S ribosome complex to generate the translation initiation complex (2–5).
The binding affinity of eIF4E for the mRNA cap is considered to play a key role in translation efficiency (7,8). In nematodes, two different types of capped mRNA are present, m7G- and m2,2,7G-cap. A large percentage of nematode mRNAs are matured by spliced leader RNA trans-splicing which generates the mature 5′-ends of mRNAs through the addition of a short leader sequence, the 22-nt spliced leader (SL), and a new cap (an m2,2,7G-cap in metazoa) (9–12). Previous studies suggest that some nematode eIF4E isoforms (Ascaris suum eIF4E-3 and Caenorhabditis elegans eIF4E-1, -2, -5) are able to recognize the two different nematode mRNA caps (10,13–15). Current data suggest that most eIF4E proteins have a strong preference for the m7G-cap compared with an m2,2,7G-cap evident from (i) their higher binding affinity for the m7G-cap (16) and (ii) inefficient translation of m2,2,7G-capped mRNAs in wheat germ and rabbit reticulocyte lysates (17,18). In contrast, a single nematode eIF4E isoform can translate both m7G- and m2,2,7G-capped mRNAs in a nematode in vitro translation system (17). Efficient translation of nematode m2,2,7G-capped mRNAs requires the downstream trans-spliced leader (10,12,17).
Trans-splicing is an essential process in nematodes (19). Importantly, many parasitic nematodes and flatworms have trans-spliced mRNAs with m2,2,7G-caps. These parasites remain a significant public health problem infecting over 2 billion people (20). Consequently, knowledge of the fundamental differences in nematode mRNA translation and the associated proteins may lead to the directed development of compounds that can specifically block parasite, but not human, gene expression.
Structural studies on the eIF4E-m7G-cap complex have revealed the details of eIF4E interactions with the m7G-cap (16,21–28). However, the mechanism of eIF4E binding to the atypical m2,2,7G-cap has not been determined. It has been proposed that the observed lower affinity of mammalian eIF4E for the m2,2,7G-cap is the consequence of a narrower binding pocket (14) or steric hindrance caused by the addition of two methyl groups at the N2 position (29). Alternatively, intrinsic protein conformational flexibility may be a major determinant that enables the eIF4E from a trans-splicing flatworm to bind both caps (30).
Here, we have used a combination of X-ray crystallography, Nuclear Magnetic Resonance (NMR) spectroscopy and other biophysical techniques to examine the interaction of nematode eIF4E with the m2,2,7G-cap. Our structural analyses represent a first step in understanding the structural specificity of eIF4E proteins for different caps and rational drug design against a parasitic nematode eIF4E. We present the first structure of an eIF4E-m2,2,7G complex and demonstrate how the addition of two methyl groups at the N2 position disrupts a critical hydrogen bond that is required for higher affinity binding of the m7G-cap. NMR analyses with nematode eIF4E and its interaction with m7G-cap, m2,2,7G-cap and m2,2,7G-SL demonstrate distinct conformational changes on binding these ligands. Notably, conformational changes associated with binding the m2,2,7G-cap are significantly reduced compared with binding the m7G-cap. Compensatory conformational changes on binding m2,2,7G-SL provide one explanation for the requirement of the 22-nt SL RNA sequence in nematode translation of m2,2,7G-capped, trans-spliced mRNAs. Our studies suggest that eIF4E proteins may undergo distinct conformational changes on binding a cap, and eIF4E interactions with the mRNA 5′-UTR could be important for the translation of some mRNAs.
MATERIAL AND METHODS
Synthesis of cap analogs and capped spliced leader sequence
Cap analogs and the m2,2,7GTP-Sepharose affinity resin were synthesized as previously described (31–33). The m2,2,7G-capped spliced leader sequence was prepared from a chemically synthesized 5′ phosphorylated RNA oligonucleotide, p-GGUUUAAUUACCCAAGUUUGAG (TriLink BioTechnologies, Inc.) by coupling with an imidazolide derivative of m2,2,7GDP under aqueous conditions. Final products were isolated from the reaction mixture and purified using High Performance Liquid Chromatography (HPLC) (a detailed protocol will be published elsewhere). bn2m7GMP was synthesized from N2-fluoro-2′,3′,5′-O-triacetyl-O6-[2-(4-nitrophenyl)ethyl]) guanosine (34) by nucleophilic substitution with benzyl iodide, subsequent 5′-phosphorylation using the standard method of Yoshikawa (35) and the methylation carried out as previously described (31). A detailed method for the synthesis of the compound will be described elsewhere.
Protein purification
Ascaris eIF4E-3 protein was prepared from bacterial cultures induced with 0.2 mM IPTG at A600 = 0.7 and incubated overnight at 25°C. The cells were collected by centrifugation at 4000 g, suspended in HEPES binding buffer (20 mM HEPES, pH 7.4, 100 mM KCl, 1 mM dithiothreitol, 1 mM EDTA), sonicated and clarified by centrifugation at 16 000 g. The supernatant was loaded onto m7GTP-Sepharose resin (GE Healthcare) and rotated at 4°C for 2 h. The resin was washed with the binding buffer, and eIF4E was eluted with 2 M KCl in binding buffer. The eluted protein was applied to a Superdex 75 (GE Healthcare) column equilibrated in binding buffer, and monomeric protein was collected, analyzed by Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS–PAGE) and concentrated using an Amicon 10 000 MW Ultra Centrifugal Filter Device (Millipore, Billerica, MA, USA).
Full-length human eIF4E was subcloned into pGEX6P-1 (GE Healthcare, Parsipanny, NJ, USA) and then transformed into Escherichia coli strain XA90. Protein expression was induced, the cells were collected and sonicated, and the supernatant was recovered as described above except that phosphate-buffered saline buffer was used. The cell lysate was incubated with Glutathione-Sepharose 4B (GE Healthcare) for 4 h at 4°C with rotation. The resin was washed with phosphate-buffered saline buffer, and eIF4E was cleaved from the glutathione S-transferase fusion protein by incubation with PreScission Protease at 4°C overnight. Eluted protein was further purified as described above on a Superdex 75 column.
Crystallization conditions, data collection and refinement
Purified A. suum eIF4E-3(49–236) was concentrated to 10 mg/ml (Amicon, 10 000 MW Ultra Centrifugal Filter Device, Millipore, Billerica, MA, USA) and then incubated with 1 mM 4E-BP peptide (RIIYDRKFLMECRNSPV, corresponding to the residues 51–67 of the human 4E-BP1 sequence) and 0.5 mM m7G or m2,2,7G-cap at 4°C for 1 h before crystallization setup using the hanging drop vapor diffusion method. Needle-like crystals developed in 20% MME PEG 2000, 0.2 M (NH4)2SO4 and 100 mM Na C3H5O(COO)3 (pH 5.6) at 4°C within 1 week using 0.5 ml 1 M NaCl as the well solution (30). Before data collection, the crystal was transferred into the equivalent mother solution containing 30% MMG PEG 2000 for 15 min, and then flash cooled in liquid nitrogen. X-ray diffraction data were collected using the Advanced Light Source beamline 4.2.2 at the Lawrence Berkley National Laboratory and 19-ID of the Structural Biology Center, Advanced Photon Source (APS) at Chicago. The Ascaris eIF4E-3 complex structures were solved by molecular replacement using PDB:2V8W as the initial model and the program PHASER in the CCP4 suite (36). Model building and manual refinement were performed in COOT (37) and REFMAC from the CCP4 suite (36), respectively. Figures were generated using Pymol (38). At the final stage of refinement, the TLS refinement with REFMCA was used using the TLS parameter generated by the TLSMD server (39).
NMR spectroscopy
All NMR spectra were acquired on a 900 or 800 MHz Varian spectrometer at a probe temperature of 25°C. For the backbone assignment, standard 3D resonance NMR experiments were conducted including HNCACB, HNCOCA, HNCOCACB, CCONH-TOCSY using 0.5 mM triple-labeled Ascaris eIF4E-3 in complex with m7GTP at a 1:1.2 molar ratio. The HNCACB and CBCACONH were recorded to facilitate resonance assignments for the m2,2,7GTP complex using 0.6 mM 13C/15N Ascaris eIF4E-3. Titration experiments with m2,2,7GTP were used to facilitate apo-form backbone assignments. Titration experiments were performed at the protein concentration of 50 µM in the presence of different molar ratios of ligands. All the NMR data were processed using NMRPipe (40) and analyzed using CcpNmr (41).
In vitro translation
In vitro translation and depletion/reconstitution assays were carried out as previously described (17). eIF4F complex consisting of full-length eIF4G and wild-type or mutant eIF4E were co-expressed and the complex purified using His-tagged eIF4G (17). eIF4F complex addition was normalized to both eIF4E and eIF4G using antibodies directed to the Ascaris proteins. Preparation and use of an optimized cap-dependent rabbit reticulocyte lysate translation system was done as described (42).
Isothermal titration calorimetry
The calorimetric titration was carried out at 20°C using a VP-ITC calorimeter (MicroCal Inc., Northampton, MA, USA). eIF4E proteins were purified as described above and used for ITC in 20 mM HEPES (pH 7.4), 1 mM EDTA and 100 mM KCl buffer. Each titration experiment consisted of a 5 µl injection followed by 29 injections of 10 µl of cap analogs. Data were processed using the single binding site model in Origin (Version 7.0 MicroCal). Control titration experiments demonstrated that the dilution heat was trivial compared with the real binding heat (e.g. buffer titrated into protein and cap titrated into buffer; data not shown).
Fluorescence titration
Titrations of eIF4E-3 with the cap analogs were performed as described previously (16). Absorption and emission spectra were recorded on Lambda 20 UV/VIS and LS-55 (Perkin-Elmer, CT, USA) spectrometers, respectively. The protein solution was spun down at 18 000 g for 10 min at 4°C before the experiments to remove any possible precipitants. The measurements were carried out in 50 mM HEPES/KOH, pH 7.20 ± 0.02, 200 mM KCl, 1 mM DTT and 0.5 mM EDTA, at a temperature of 20°C and repeated multiple times at eIF4E-3 concentrations of 0.4–0.6 µM. The samples of m2,2,7GpppG used for determination of the binding constant were synthesized in seven independent batches. The temperature was controlled to ±0.2 K with a thermocouple inside the thermostated cuvette, and the sample was stirred magnetically. The fluorescence, excited at 280 nm (slit 2.5 nm, auto cut-off filter), was monitored at 335 nm (slit 2.5–4 nm, 290 nm cut-off filter), with an integration time of 30 s and a gap of 30 s for adding the cap analogue. Dilution during the titration did not exceed 2.5%. Fluorescence intensity was corrected for the inner filter effect. The emission of the free cap analogues was explicitly included in the numerical analysis. A theoretical curve for the fluorescence intensity as a function of the total ligand concentration was fitted to the experimental data points by means of non-linear, least-squares method, using Prism 3.02 (GraphPad Software, USA).
Thermofluor assays
Thermofluor assays were carried out using an RT–PCR instrument (Bio-Rad iCycler 5). The protein concentration used in this assay is 1 µM in the 20 mM HEPES buffer (pH 7.4) containing 100 mM KCl and 1 mM EDTA. The various caps were incubated with the protein for 10 min before the addition of SYPRO orange dye (1:1000 dilution) (Invitrogen). The fluorescence was measured with the increase of temperature at the rate of 1°C/min (excitation wavelength at 492 nm and emission wavelength at 610 nm). The data were analyzed using the Boltzmann equation in Prism 3.02 (GraphPad Software).
Accession numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank for Ascaris eIF4E-3-m7GTP-4E-BP complex (PDB code 3M93) and Ascaris eIF4E-3- m2,2,7GTP-4E-BP complex (PDB code 3M94).
RESULTS
Structure of Ascaris eIF4E-3 in complex with m7G-/m2,2,7G-cap
Previous qualitative studies suggested that Ascaris eIF4E-3 recognizes both the m7G and m2,2,7G-cap (10), whereas studies on mammalian eIF4E proteins indicated they had very low affinity for m2,2,7GTP (∼100-fold less) compared with m7GTP (16). We carried out quantitative fluorescent cap-binding studies with Ascaris eIF4E-3. These studies suggested that Ascaris eIF4E-3 had equal affinity for m7GpppG and m2,2,7GpppG-cap (KD = ∼1 µM) (Table 1 and Supplementary Figure S1A). Sequence alignments comparing eIF4E proteins did not provide any obvious explanation for why the nematode proteins would have higher affinity for the m2,2,7GpppG-cap compared with mammalian eIF4E (Supplementary Figure S1B).
Table 1.
Fluorescent titration analysis of Ascaris eIF4E-3 and human eIF4E cap-binding affinity
| Cap analog |
KD (µM) |
ΔG° (kcal/mol) |
||
|---|---|---|---|---|
| Ascaris eIF4E-3 | Human eIF4E (28–217) | Ascaris eIF4E-3 | Human eIF4E (28–217) | |
| m7GTP | 0.161 ± 0.005 | 0.0274 ± 0.0002 | −9.11 ± 0.02 | −10.138 ± 0.003 |
| m7GpppGa | 0.787 ± 0.025 | 0.335 ± 0.036 | −8.19 ± 0.03 | −8.69 ± 0.06 |
| m2,2,7GpppGa | 0.662 ± 0.057 | 3.1 ± 1.1 | −8.28 ± 0.05 | −7.38 ± 0.20 |
| GTP | 83 ± 14 | 140 ± 6b | −5.47 ± 0.10 | −5.17 ± 0.03b |
| GpppG | 116 ± 8 | 250 ± 19b | −4.87 ± 0.09 | −4.43 ± 0.06b |
| GpppGc | 232 ± 36 | 500 ± 38b | ||
aResults similar to these were obtained for m7GpppA and m2,2,7GpppA.
bCalculated from the values at 100 mM KCl and the slope of ionic-strength dependence of the association constants for the triphosphate mono- and dinucleotide cap analogs (16).
cMicroscopic association constant derived from the measured one by dividing by a factor of two due to entropic effects for the symmetric cap analog.
In order to understand the binding mechanism of Ascaris eIF4E-3 with the m2,2,7G-cap, we tried to determine the co-crystal structure of full-length Ascaris eIF4E-3 with the m2,2,7G-cap. Full-length Ascaris eIF4E-3 with the m2,2,7G-cap failed to produce diffraction quality crystals. Sequence alignment, trypsin digestion and isothermal titration calorimetry (ITC) cap-binding analyses indicated that N-terminal truncation of the protein (49–236) retained cap-binding ability (Table 2). Using this shorter construct in the presence of the human 4E-BP1 peptide, we obtained diffraction quality crystals and determined the co-crystal structures of Ascaris eIF4E-3 with m2,2,7GTP and m7GTP (Table 3). The overall structures of the Ascaris m2,2,7GTP and m7GTP complexes are very similar to each other (RMSD for Cα = 0.31 Å) and to the human eIF4E-m7GTP complex structures (PDB:2V8W, RMSD for Cα = 0.79 Å) (25) (Figure 1A–C and Supplementary Figure S1C). The human 4E-BP1 peptide binds to the opposite surface of the protein and forms interactions with Ascaris eIF4E-3 (Figure 1A and B) almost identical to those previously observed in the human eIF4E/4E-BP complex structure (43).
Table 2.
Isothermal titration calorimetry analysis of Ascaris eIF4E-3 and human eIF4E cap-binding affinity
| Protein | Ligand | KD (μM) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| Ascaris eIF4E-3 | m7GTP | 0.24 ± 0.04 | −14.5 ± 1.7 | 5.5 ± 1.4 | −9.0 |
| Ascaris eIF4E-3 | m2,2,7GTP | 8.4 ± 1.0 | −10.4 ± 0.3 | 3.54 ± 0.42 | −6.86 |
| Ascaris eIF4E-3 (49–236) | m7GTP | 0.17 ± 0.05 | −12.38 ± 0.02 | 3.3 ± 0.56 | −9.08 |
| Ascaris eIF4E-3 (49–236) | m2,2,7GTP | 12.2 ± 3.9 | −11.58 ± 0.52 | 5.02 ± 0.72 | −6.56 |
| Ascaris eIF4E-3 (E116D) | m7GTP | 1.98 ± 0.2 | −11.04 ± 0.84 | 3.38 ± 0.78 | −7.66 |
| Ascaris eIF4E-3 (E116D) | m2,2,7GTP | 7.0 ± 0.1 | −10.65 ± 0.05 | 3.69 ± 0.1 | −6.96 |
| Ascaris eIF4E-3 (49–236) | m7GpppG | 0.75 ± 0.19 | −11.5 ± 0.8 | 3.3 ± 0.7 | −8.2 |
| Ascaris eIF4E-3 (49–236) | m2,2,7GpppGa | 17.2 ± 2.9 | −4.52 ± 0.38 | 1.3 ± 0.1 | −3.22 |
| Human eIF4E | m7GTP | 0.032 ± 0.008 | −12.54 ± 0.45 | 2.5 ± 0.3 | −10.4 |
| Human eIF4E | m2,2,7GTP | 4.56 ± 2.1 | −12.1 ± 0.36 | 4.76 ± 1.01 | −7.34 |
aData were analyzed using the fixed stoichiometry (N = 1) due to the weak binding affinity (Supplementary Figure S1A).
Table 3.
Crystal data and refinement statistics for Ascaris eIF4E-3
| Data collection | m7GTP-cap | m2,2,7GTP-cap |
|---|---|---|
| Space group | I4 (1) | I4 (1) |
| Cell dimensions | ||
| a (Å) | 104.62 | 104.98 |
| b (Å) | 104.62 | 104.98 |
| c (Å) | 47.22 | 46.91 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90 |
| γ (°) | 90 | 90 |
| Measured reflections | 22 966 | 104 640 |
| Resolutiona | 39.99–2.9 (3.0–2.9) | 41.73–2.00 (2.07–2.00) |
| Rsym or Rmerge | 0.113 (0.299) | 0.113 (0.275) |
| I/σ | 6.7 (2.2) | 6.6 (3.2) |
| Completeness (%) | 98.5 (98.8) | 98.3 (98.1) |
| Redundancy | 3.4 (3.3) | 3.68 (3.7) |
| Refinement | ||
| Resolution (Å) | 2.9 | 2.05 |
| Number of reflections | 5713 | 15 993 |
| Rwork/Rfree | 23.6/29.8 | 20.4/24.4 |
| Number of atoms | ||
| Protein | 1429 | 1441 |
| Peptide | 113 | 117 |
| Ligand | 29 | 31 |
| Water | 2 | 22 |
| B factors | ||
| Protein | 45.6 | 21.4 |
| Peptide | 46.2 | 91.3 |
| Ligand | 77.5 | 41.6 |
| Water | 17 | 19.9 |
| Rms deviations | ||
| Bond length (Å) | 0.011 | 0.01 |
| Bond angles (°) | 1.35 | 1.27 |
| Ramachandran plot | ||
| Residues in most favorable regions (%) | 95.7 | 98.9 |
| Residues in disallowed regions (%) | 0 | 0 |
aValues in parentheses are for the highest resolution shell.
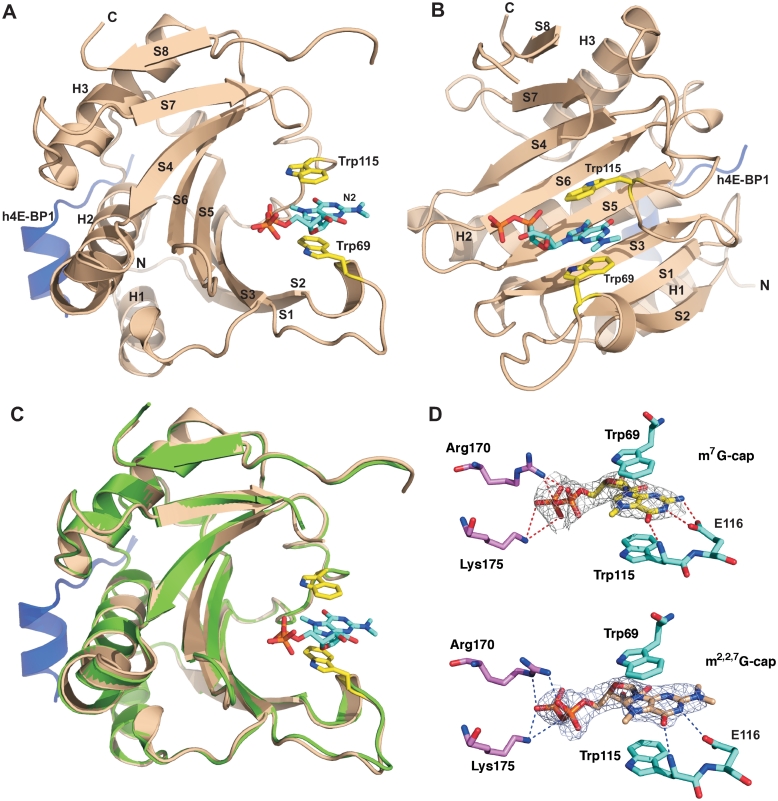
Figure 1.
Structure comparison of Ascaris eIF4E-3 m7GTP and m2,2,7GTP complexes. (A and B) Ascaris eIF4E-3 structure in complex with m2,2,7GTP and human 4E-BP1 peptide (blue) shown in two orientations. The three α helices and eight β strands in the structure are labeled. m2,2,7GTP and the two stacking aromatic residues of eIF4E (Trp69 and Trp115—yellow) are shown as colored sticks. (C) Superimposition of Ascaris eIF4E-m2,2,7GTP (Wheat) and m7GTP-cap complexes (Green). The m7GTP-cap is omitted from the figure. (D) m7GTP and m2,2,7GTP interactions in the Ascaris eIF4E-3 cap-binding pocket. The colored dashed lines illustrate hydrogen bonding. Note the additional hydrogen bond present with the m7GTP-cap from E116. 2Fo-Fc Electron density map is contoured at a level of 1σ.
The electron density of both caps in the binding pocket is well defined except for the third phosphate (Figure 1D). Three main interactions occur between the m2,2,7GTP-cap and the protein that include: (i) cation-π stacking between the guanine base and Trp69 and Trp115; (ii) electrostatic and hydrogen bonding interactions between Arg170 and Lys175 and the α and β phosphates of the m2,2,7GTP-cap; and (iii) two H-bonds between the guanine N1 and O6 with the carboxyl group of Glu116 and the backbone amide of Trp115, respectively. Comparison of the Ascaris eIF4E-3 m2,2,7GTP and m7GTP complexes and other eukaryotic eIF4E complex structures (22,30) demonstrates that the only major difference on binding m2,2,7G-cap is the loss of a single H-bond between the N2 of the guanine base and the carboxyl group of Glu116 (Figure 1D).
To further evaluate the role of Glu116 in m2,2,7GTP-cap binding, two mutants were generated (E116A and E116D). We predicted that the substitution of Glu by Asp or Ala would disrupt one H-bond between Glu carboxyl group and guanine N2 of the m7G-cap. However, the loss of the H-bond would not affect binding to the m2,2,7GTP-cap since this H-bond is already absent, as shown in Figure 1D. The E116A substitution resulted in an insoluble protein precluding its use in cap-binding analysis. In contrast, the E116D substituted protein was readily purified. Binding studies using ITC demonstrated that the E116D substitution led to a decreased binding affinity for the m7GTP-cap (10-fold), with little effect on m2,2,7GTP-cap binding (Table 2). To examine the role of these substitutions on translation, we were able to functionally co-express both E116A and E116D with full-length Ascaris eIF4G and use them in translation assays. The substitutions in E116A or E116D led to a loss in translation of m7G-capped mRNAs compared with m2,2,7G-capped mRNAs (Figure 2A and B). Overall, these data are consistent with our structural observations indicating the importance of the hydrogen bonds at the N2 position for binding of the m7G-cap (16,22).
Figure 2.
Mutations in Ascaris eIF4E-3 E116 have a greater affect on the translation of m7G-capped compared with m2,2,7-capped mRNAs. (A) Comparison of translation of m7G- to m2,2,7G-capped mRNAs by wild-type and mutant E116 eIF4E. W69A or W115A represent mutations in aromatic tryptophan residues required for cap binding and abolish translation. (B) Ratio of m2,2,7G- to m7G-capped mRNA translation as function of the wild-type and mutant E116 eIF4E. Ascaris embryo extracts were depleted of the eIF4F complex. eIF4F complex consisting of wild-type eIF4G and wild-type or mutant eIF4E complex were then added back to the extract to reconstitute translation as described (17).
Characterization of Ascaris eIF4E-3 binding to m7G- and m2,2,7G-caps
A comparison of the Ascaris eIF4E-m2,2,7G and Ascaris eIF4E-m7G complexes presented here with the human eIF4E-m7G complexes did not show any other major differences in the cap-binding pocket or the protein's interactions with the ligands (Supplemental Figure S1C). The two methyl groups at the N2 position of the m2,2,7G-cap are solvent exposed and have no apparent direct interaction with the protein. These observations suggest that mammalian eIF4E might be able to bind m2,2,7G-cap with a similar binding affinity as that observed for the Ascaris eIF4E-3. In support of this prediction, both human eIF4E and Ascaris eIF4E-3 were retained on m2,2,7GTP-sepharose and could be eluted by the m2,2,7GTP-cap analog (Supplementary Figure S2A and S2B). We next carried out ITC binding studies on Ascaris and human eIF4E. These analyses indicated that Ascaris eIF4E-3 has a lower binding affinity for either m2,2,7GTP or the m2,2,7GpppG-cap compared with m7GTP- and m7GpppG caps (Table 2 and Supplementary Figure S2C–F). NMR studies of Ascaris eIF4E-3 with dinucleotide caps (m2,2,7GpppG and m7GpppG) further support that the affinity of Ascaris eIF4E-3 for m2,2,7GpppG is lower than for m7GpppG (Supplementary Figure S2G) as do SPR studies (data not shown). We note that while similar Ascaris eIF4E-3 binding affinities for the two caps were observed using fluorescence titration methods described above (Table 1), overall our interpretation of the structural, NMR and translation data (see below) are most consistent with the view that the affinity of Ascaris eIF4E-3 for the m2,2,7G-cap is lower than for the m7G-cap (see Discussion in Supplementary Data). Furthermore, the ITC data indicate that Ascaris eIF4E-3 and human eIF4E have similar and lower binding affinities for the m2,2,7G-cap. Similar observations were made for eIF4E protein from the parasitic flatworm Schistosoma mansoni that also has m2,2,7G-capped mRNAs (30). Thus, the major difference between the human, nematode and flatworm eIF4E is that the human protein has a higher binding affinity (6–9 fold) for m7G-cap (Table 2) (14,30,44).
The lower binding affinity of mammalian eIF4E for the m2,2,7GTP-cap was previously proposed to be a result of both a narrower binding pocket and steric hindrance caused by the two additional methyl groups at N2 (14,15,29). Our structures of the Ascaris eIF4E-m2,2,7G and eIF4E-m7G suggest there is no direct interaction between the protein and the two methyl groups at the N2 position. Furthermore, we have shown here that human and Ascaris eIF4E-3 have a similar affinity for the m2,2,7GTP (Table 2 and Supplementary Figure S2). To examine if steric hindrance is responsible for the reduced affinity for the m2,2,7G-cap, we tested the binding of a cap analog, bn2m7GMP (Supplementary Figure S3A), which has a large benzyl substituent at N2 and find that it binds to both human and Ascaris eIF4E-3 with a 10-fold higher affinity compared with m7GMP (Supplementary Table S1). These data suggest that additional N2 methyl groups are not likely to preclude m2,2,7GTP from entering the cap-binding pocket and that the pocket may be able to adapt its conformation to accommodate different caps. This is consistent with recent data indicating that the eIF4E cap binding pocket is relatively flexible (25). Further, as mammalian eIF4E undergoes a major conformational change on binding the cap (24,29,45), it is apparent that the N2 substituent does not impede any conformational change required to form the cap-binding pocket or access of the cap to the pocket. Consistent with the increased affinity of bn2m7GMP for Ascaris eIF4E-3, it is also a strong competitive inhibitor of either m7G- or m2,2,7G-capped mRNA translation in vitro (Supplementary Figure S3B and S3C) and is a stronger inhibitor than m7GTP. Furthermore, in an optimized cap-dependent rabbit reticulocyte lysate translation system (Supplementary Figure S3D), addition of the benzyl group to m7GMP significantly increases its efficacy as a translational inhibitor. Enhanced binding of eIF4E for bn2m7GMP may be due to a change in the methylated ring character (46) or an increased favorable entropy contribution from the N2 benzyl group (47). The benzyl group of bn2m7GMP may form additional interactions with eIF4E to enhance cap binding.
eIF4E conformational differences on binding m2,2,7G- versus m7G-caps
To further explore the mechanistic basis for m2,2,7G-cap binding, we used NMR chemical shift perturbation analyses to examine the changes that occur in Ascaris eIF4E-3 upon binding m2,2,7G- and m7G-caps (Supplementary Figure S4A). Approximately 25 residues from Ascaris eIF4E-3 exhibit significant chemical shift changes upon addition of m7G- or m2,2,7G-cap (Supplementary Figure S4B and S4D), indicating that these residues interact with the cap and/or undergo significant conformational rearrangements on binding the cap. These residues fall into three categories based on their location in the structure (Figure 3B): (i) residues involved in the formation of the cap-binding pocket or around the binding pocket; (ii) residues associated with the eIF4E/eIF4G interface (compare H1 and S2 in Figure 3B and D); and (iii) residues at a distance from these two regions. These data are consistent with chemical shift perturbations observed for the human eIF4E protein on binding m7GTP-cap (24). The majority of residues with NMR chemical shift perturbations are similar on binding both the m7G- and m2,2,7G-cap (Supplementary Figure S4B–D), consistent with the minimal differences observed between the crystal structures of the eIF4E-m2,2,7G and eIF4E-m7G complexes (Figure 1C).
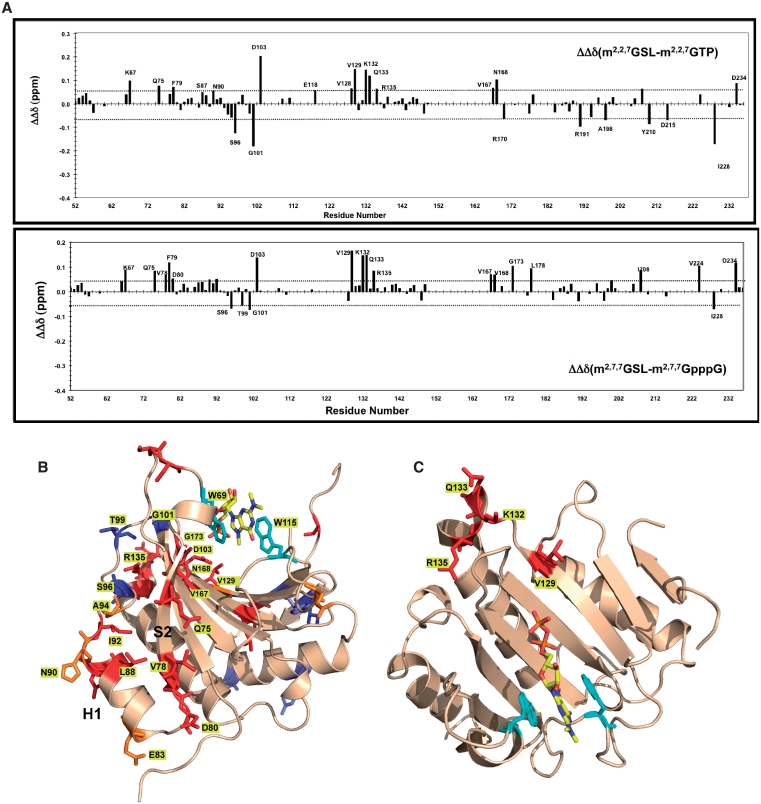
Figure 3.
Ascaris eIF4E-3 chemical shift perturbations are less on binding the m2,2,7G- compared to m7G-cap. (A) Differences in eIF4E-3 chemical shift perturbation upon binding m2,2,7G- compared to m7G-cap. The reported difference in the chemical shift changes (ΔΔδ) is the difference in the weighted chemical shift changes, calculated as Δδ = {Δδ(1H)2 + 0.2 × Δδ(15N)2}1/2, where Δδ(1H) and Δδ(15N) are the chemical-shift differences of the amide proton and nitrogen, relative to chemical shifts observed in the apo-protein, respectively (24,54). The dashed horizontal lines represent ±1 SD from the mean chemical shift difference. (B) Location of eIF4E-3 residues that show significant chemical shift perturbations on binding m7GTP compared to the apo protein (Supplementary Figure S4A and S4B). Residues with major chemical shifts are colored as follows: red = >2.5 SD; orange = >2.0 SD; yellow = >1.5 SD above the mean. (C) Location of eIF4E-3 residues with significantly different chemical shift perturbations on binding m2,2,7G- compared with the m7G-cap. Data are derived from (A). Residues with major chemical shifts are colored as follows: red = chemical shifts >1 SD above the mean and blue = residues with chemical shift changes <1 SD below the mean. Orange (increase) and slate (decrease) residues are those that show less significant changes for one of the two comparisons (m2,2,7GTP versus m7GTP or m2,2,7GpppG versus m7GpppG).(D) Location of eIF4E-3 residues predicted to be involved in the interaction with eIF4G. eIF4E residues impacted by yeast eIF4G (393–490) (55) were mapped onto Ascaris eIF4E-3 and colored in cyan.
However, there are clear differences in the magnitude of chemical shift changes on binding m7G- versus m2,2,7G-cap (Figure 3A–C). Most of the changes are significant reductions in the chemical shifts associated with residues located in and around the cap-binding pocket and near or at the eIF4E/eIF4G interface (see Figure 3D for cyan residues at the eIF4E/G interface). Reduced changes are observed in Lys112 and Asp117 (Figure 3A–B and Supplementary Figure S5) that may be a direct consequence of the loss of the H-bond between the N2 of the base and carboxyl group of Glu116 on binding the m2,2,7G-cap. Interactions with eIF4G may be affected by perturbations of residues Lys74, Gln75, Val76, Ala77 and Phe79 of S2 as well as Tyr104 (Figure 3B and C). These residues pack against alpha helix 1 and are connected to the key Trp69 that stacks with the methylated cap base (Figure 3C and D). Notably, both of these regions are part of the eIF4E interface with eIF4G. In addition, there are a number of changes at some distance from the binding pocket, including Asp157 that is also part of the eIF4E/G interface. Data examining mammalian eIF4E cap-binding (m7GTP versus m2,2,7GTP) conformation changes by increased susceptibility to pepsin cleavage showed conformational differences in regions of H1, H2 and S2 that support the changes we observe in the eIF4E/eIF4G interface on binding these two caps (29). While we envision potential networks of interactions that would lead to these changes emanating either from the alterations in Lys112 and Asp117 or residues adjacent to Trp69, peaks from a number of residues in the NMR spectra are missing or not assignable due to the effects of intermediate conformational exchange. Thus, the exact mechanism of how these perturbations are transmitted from the cap-binding pocket to these regions of Ascaris eIF4E-3 interface with eIF4G is unclear.
m2,2,7GpppG-SL and eIF4E interaction reveals a possible explanation for the ‘SL effect’
Nematode trans-splicing generates mRNAs with an m2,2,7GpppG-cap followed by a conserved 22-nt RNA spliced leader. Translation of mRNA with only the m2,2,7GpppG-cap is poor in nematodes (10,12). Efficient nematode translation of the m2,2,7GpppG-capped mRNAs requires specific nucleotides and structural elements within the downstream spliced leader (SL) sequence including a stem loop that contains a 3-bp stem immediately following the cap (17). We have described this phenomenon as the ‘SL effect’. The requirement for the SL in the translation of m2,2,7GpppG-capped RNA appears consistent with the low affinity of Ascaris eIF4E-3 and the reduced conformational changes on binding this cap. We hypothesize that the SL is required to compensate for these deficiencies and facilitate translation. To examine where the SL might interact with Ascaris eIF4E-3, we used NMR spectroscopy to compare the binding of m2,2,7GpppG-SL, m2,2,7GpppG and m2,2,7GTP-caps to Ascaris eIF4E-3. Comparison of the chemical shift perturbations in eIF4E on association with these three ligands (Figure 4A and B) demonstrated that the interaction with m2,2,7G-SL leads to additional perturbation of residues around the cap-binding pocket (Gly101, Asp103, Val 128, Val 129, Val 167 and Asn168) and at the eIF4E/G interface including: (i) residues in the S2 strand (Lys75-Asp80) and (ii) residues in the H1 helix area (Glu83-Ala94). Unique chemical shift changes observed only in the presence of the SL occur in the region of Val128-Arg135 (Figure 4C). These perturbations may reflect the position of some portion of the SL RNA sequence on Ascaris eIF4E-3. Overall, these observations suggest that binding of m2,2,7GpppG-SL may contribute to the structure or interactions within the cap-binding pocket and may influence Ascaris eIF4E-3 interactions with eIF4G and other translation factors through protein conformational changes (Figure 4B).
Figure 4.
Chemical shift changes on Ascaris eIF4E-3 binding m2,2,7G-spliced leader RNA increase changes at the eIF4E/G interface. (A) Chemical shift perturbations in eIF4E-3 on binding the m2,2,7G-spliced leader RNA compared to either m2,2,7GTP or m2,2,7GpppG. Determined as described in Figure 3A. The dashed horizontal lines represent ±1 SD from the mean chemical shift difference. Residues with significant chemical shifts are colored as follows: red = chemical shifts >1 SD above the mean and blue = residues with chemical shift changes <1 SD below the mean. Orange residues represent residues that show smaller differences when comparing m2,2,7GTP or m2,2,7GpppG. (B) Location of eIF4E-3 residues with different significant chemical shift perturbations on binding m2,2,7G-SL RNA compared to the m2,2,7G-cap. Residues with major chemical shifts are colored as follows: red = chemical shifts >1 SD above the mean and blue = residues with chemical shift changes <1 SD below the mean. Orange residues represent residues that show less increased shifts or differences when comparing m2,2,7GTP or m2,2,7GpppG. (C) Location of residues that exhibit increased chemical shift perturbations only in the presence of the 22-nt SL-RNA. These regions do not show chemical shift changes in the presence of triphosphate or dinucleotide cap analogs and most likely represents a region that interacts with the SL-RNA.
DISCUSSION
Spliced leader trans-splicing in metazoa generates mRNAs with an atypical mRNA-cap, m2,2,7G-cap, followed by a spliced leader (9–12). In nematodes, two populations of mRNA are always present: (i) non-trans-spliced RNAs with a typical m7GpppN-cap and variable 5′ sequence and (ii) trans-spliced RNAs with an m2,2,7GpppG-cap and a common 22-nt 5′ spliced leader sequence. Ascaris eIF4E-3 can initiate the translation of both types of nematode mRNAs (17). Here, we describe the first structure of an eIF4E with an m2,2,7G-cap and compare it to the cognate m7G-eIF4E complex. These structures and NMR data indicate that the nematode A. suum eIF4E binds the two different caps in a similar manner except for the loss of a single hydrogen bond on binding the m2,2,7G-cap. Consistent with the structures, both mammalian and nematode eIF4E have a similar low affinity for m2,2,7G-cap compared with the m7G-cap. The low affinity of eIF4E for the m2,2,7G-cap is not a result of a narrower cap-binding pocket or steric hindrance as previously postulated. Nematode eIF4E binding to the m7G-cap, m2,2,7G-cap and m2,2,7G-SL leads to distinct eIF4E conformational changes. Overall, these data lead us to propose a model for how the nematode spliced leader facilitates translation of m2,2,7G-capped mRNAs.
Nematode eIF4E structure and conformation changes on cap binding
The overall structures of Ascaris eIF4E-3 bound to both caps are similar and resemble those determined for other eukaryotic eIF4E-cap complexes. (16,21–28). The major difference in Ascaris eIF4E-3 on binding m2,2,7G-cap compared with the m7G-cap is the loss of a single H-bond between the N2 of the guanine base and the carboxyl group of Glu116 (Figure 1D), which leads to a significant drop in m2,2,7G-cap binding affinity (Table 2). From ITC analysis, the reduced binding affinity for the m2,2,7G-cap results from a reduced enthalpy (ΔH) presumably from the loss of the H-bond to Glu116, and also an increase in the unfavorable entropy. The crystal structures with the m2,2,7G-cap and m7G-cap reveal no significant differences in the crystallographic B-factors or atomic packing density in the vicinity of the ligand. However, formation of the H-bond between Glu116 and the N2 in the m7G-cap would lead to release of water back into bulk solvent and would be entropically favorable. Therefore, the presence of the two methyl groups prevents this release, requires solvation of the exposed E116 side chain carboxyl, and additionally leads to increased solvent ordering around the two methyl groups leading to less favorable enthalpy.
Several lines of data suggest binding to the m7G-cap compared with the m2,2,7G-cap results in different protein conformations. First, although NMR spectroscopy reveals that most of the residues that change on binding each cap are the same, the magnitude of the chemical shift differences for a subset of these residues (20 of 25) is smaller on binding the m2,2,7G-cap (Figure 3A and C). Second, the different entropy change on binding m7GTP compared with m2,2,7GTP (Table 2) further suggests that different protein conformational changes occur upon binding the different caps. Third, the thermafluor assays demonstrate that cap-binding stabilizes Ascaris eIF4E-3 against thermal denaturation and the stabilization is much greater with the m7GTP compared with the m2,2,7GTP-cap (Supplementary Figure S6). Overall, these data suggest that conformational changes are different on binding these two caps and these could have important ramifications in translation (see below).
Nematode eIF4E and the trans-spliced spliced leader
Efficient translation of m2,2,7G-capped mRNAs in Ascaris requires specific sequence and structure within the downstream trans-spliced 22-nt SL sequence, the ‘SL effect’ (10,12,17). Notably, these SL elements do not facilitate translation of m7G-capped RNAs in nematodes or m2,2,7G-capped mRNAs in mammalian or plant translation systems (17). Furthermore, adaptations in the Ascaris eukaryotic translation initiation factor eIF4E/G complex enable efficient translation of the m2,2,7G -SL RNAs in diverse in vitro translation systems (17).
One possible explanation for the requirement of the SL in the translation of m2,2,7G-capped mRNAs is that the SL sequence facilitates or increases eIF4E-3 binding to the m2,2,7G-cap. However, studies on Ascaris eIF4E-3 and eIF4E/G indicate that the SL sequence does not increase either Ascaris eIF4E-3 or eIF4E/G affinity for the m2,2,7G-cap (17).
Our NMR data indicate that the SL leads to increased chemical shift changes in Ascaris eIF4E-3 S2 (Lys75-Asp80) and residues in the H1 helix region (Glu83-Ala94) (Figure 4A and B). Recent molecular dynamics data using C. elegans eIF4E isoforms also suggest that the flexible S1–S2 loop is affected by m2,2,7G-cap binding (48). Residues located in S2 exhibit different chemical shifts upon interaction with the two caps. These chemical shifts are significantly less on binding the m2,2,7G-cap (Figure 3A). We propose that binding of the m7G-cap leads to perturbation of residues at the eIF4E/G interface (S2 and H1) (Figure 3B) and this is a result of a hydrogen bond interaction with Glu116, and possibly other changes. On binding the m2,2,7G-cap, the loss of a hydrogen bond between the cap and Glu116 (and possibly other changes) leads to reduced changes in eIF4E at the interface of eIF4E with eIF4G. We propose that these interactions are required for recruitment of the 43S ribosome complex. When the m2,2,7G-cap is bound, the loss of these interactions leads to reduced recruitment of the 43S ribosome complex. When the m2,2,7G-cap is present with the SL, the SL leads to increased chemical shift changes including those in S2 (Lys75-Asp80) and residues in the H1 helix region (Glu83-Ala94) at the eIF4E/G interface. Therefore, we propose that the SL sequence in the m2,2,7G-capped RNA is required to induce the requisite conformational changes necessary for interactions at the Ascaris eIF4E/eIF4G interface. The reduced binding affinity of nematode eIF4E for m7G-cap compared with human eIF4E may allow additional degrees of conformational flexibility in Ascaris eIF4E-3 that enable alternate conformations of the eIF4E/G/cap ternary complex necessary for translation of the two populations of mRNAs (m7G- and m2,2,7G-capped) in nematodes. Additional adaptations in the Ascaris eIF4E/G are also required (17).
Nematode eIF4E cap binding specificity
Our initial interest in determining the structure of Ascaris eIF4E-3 was based on the premise that Ascaris eIF4E-3 and eIF4E from other metazoa with trans-splicing had adaptations in their eIF4Es to specifically recognize the m2,2,7G-cap (10,13–16,30,44). However, our new data demonstrate that both human and Ascaris eIF4E-3 recognize the m2,2,7G-cap with broadly similar, but low affinity, and that the major difference in cap-binding between the Ascaris and human proteins is the higher affinity of the human protein for the m7G-cap (Table 2 and Discussion in Supplementary Data).
The lower binding affinity of mammalian eIF4E for the m2,2,7GTP-cap was previously proposed to be a result of both a narrower binding pocket and steric hindrance due to the introduction of two additional methyl groups at N2, thus preventing this cap from entering the cap binding pocket (14,15,29). However, our structures of the Ascaris eIF4E-m2,2,7G and eIF4E-m7G complexes rule this out as there is no direct interaction between the protein and the two methyl groups at N2 in the base (Figure 1). We show that both human and Ascaris eIF4E-3 have a similar affinity for the m2,2,7GTP (Table 2 and Supplementary Figure S2). In addition, the cap analog, bn2m7GMP, with a large benzyl substituent at N2 position, has increased binding affinity to both Ascaris and human eIF4E further suggesting that the N2 substituent does not impede the known eIF4E conformational change required to form the cap binding pocket or prevent access of the cap to the pocket. Our observation that addition of an N2 benzyl group substituent on a monophosphate (this bn2m7GMP) is a better ligand for mammalian eIF4E and leads to significant translation inhibition may provide a novel avenue for cap analog inhibitor development (Supplementary Figure S3D).
Five isoforms of eIF4E have been described in C. elegans (13,15,49) with two isoforms, described as primarily specific for the m7G-cap, whereas the remaining three interact with both the m7G- and m2,2,7G-caps. Mutagenesis studies on the C. elegans dual cap-binding protein eIF4E-5 suggested that alteration of two specific residues (N64Y/V65L) led to a 2-fold decrease in binding specificity for m2,2,7GTP-cap (14). These substitutions were suggested to influence both the width and depth of the cap-binding pocket. However, native Ascaris eIF4E-3 has both of these substitutions (Tyr105 and Leu106) (10). The position of these two residues in the Ascaris eIF4E-3 complex structures (m7GTP or m2,2,7GTP-cap) and their lack of change on binding the two caps (Supplementary Figure S7) suggests they are not likely the key determinants of cap binding specificity.
The only other trimethylguanosine cap-binding protein whose structure is known is snurportin1 (PDB: 1XK5) (46). Snurportin specifically binds the trimethylguanosine cap of snRNAs. The m2,2,7GpppG-cap-binding mechanism for snurportin1 (PDB:1XK5) is different from that observed in Ascaris eIF4E-3 (Supplementary Figure S8). The hypermethylated base of the cap is stacked with an aromatic group (Trp107) of snurportin and the second base forms the third residue in a stacked sandwich. Consistent with this structure, snurportin binds a dinucleotide cap (m2,2,7GpppG) with a 10-fold higher affinity than a mononucleotide cap (46). While the methyl group at N7 introduces a net positive charge into the guanine ring and thus enhances the stacking energy between the base and eIF4E Trp (16), the two additional methyl groups at the exocyclic N2 have been proposed to have no effect on the stacking interaction (50). Thus, the two additional methyl groups at N2 of the cap appear to play very different roles in their interaction with eIF4E compared with snuportin1 where they form a hydrophobic interaction with Trp107 (Supplementary Figure S8).
CONCLUSIONS
Our data provide new insight into the mechanism of eIF4E binding to the atypical m2,2,7G mRNA cap, identify eIF4E conformational changes on binding m7G and m2,2,7G mRNA caps, and suggest a key interaction between the trans-spliced nematode 5′-UTR and eIF4E necessary for efficient translation of trans-spliced mRNAs in nematodes. Some mammalian mRNAs are more dependent on eIF4E levels or eIF4E cap affinity for efficient translation (51–53). Given our data, we suggest that eIF4E binding to the cap may be dependent on or influenced by the RNA 5′-UTR sequence and structure and that this may influence translation efficiency of some eukaryotic mRNAs.
Most eukaryotes have several isoforms of eIF4E (6,28). Alignment of eIF4E isoform sequences and residues associated with their cap-binding pockets indicates that most are very similar. However, based on our findings, inherent differences in eIF4E isoform protein flexibility, ternary complex formation and interaction with the mRNA 5′-UTR may have important consequences in translation initiation resulting in distinct physiological roles for eIF4E isoforms in translation of different mRNAs.
ACCESSION NUMBERS
3M93, 3M94.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants (AI049558 and AI080805 to R.E.D); Howard Hughes Medical Institute Grant (55005604 to E.D.); Polish Ministry of Science and Higher Education grant (N N301 267 137 to A.N.), (N N301 035 936 to R.S.), (N N301 096339 to E.D.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Katherine Borden for providing human eIF4E and NMR data, Shaun Bevers at the University of Colorado Denver biophysical core for help with ITC, and members of the structural biology community at the University of Colorado Denver School of Medicine, University of Colorado Denver X-ray and NMR facility (supported in part by the University of Colorado Cancer Center). We thank Katherine Borden, Laurent Volpon, and members of the Davis lab for their comments on the article.
REFERENCES
- 1.Mathews MB, Sonenberg N, Hershey J. Origins and principles of translational control. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harbor: Cold Spring Harbor Press; 2007. pp. 1–40. [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mrna recruitment to ribosomes and regulators of translation. Ann. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 5.Von Der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 6.Rhoads RE. eIF4E - new family members, new binding partners, new roles. J. Biol. Chem. 2009;284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darzynkiewicz E, Stepinski J, Ekiel I, Goyer C, Sonenberg N, Temeriusz A, Jin Y, Sijuwade T, Haber D, Tahara SM. Inhibition of eukaryotic translation by nucleoside 5′-monophosphate analogues of mRNA 5′-cap: changes in N7 substituent affect analogue activity. Biochemistry. 1989;28:4771–4778. doi: 10.1021/bi00437a038. [DOI] [PubMed] [Google Scholar]
- 8.Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 9.Allen MA, Hillier LW, Waterston RH, Blumenthal T. A global analysis of C. elegans trans-splicing. Genome Res. 2011;21:255–264. doi: 10.1101/gr.113811.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lall S, Friedman CC, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE. Contribution of trans-splicing, 5′ -leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J. Biol. Chem. 2004;279:45573–45585. doi: 10.1074/jbc.M407475200. [DOI] [PubMed] [Google Scholar]
- 11.Liou RF, Blumenthal T. Trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maroney PA, Denker JA, Darzynkiewicz E, Laneve R, Nilsen TW. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 13.Jankowska-Anyszka M, Lamphear BJ, Aamodt EJ, Harrington T, Darzynkiewicz E, Stolarski R, Rhoads RE. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in C.elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 1998;273:10538–10542. doi: 10.1074/jbc.273.17.10538. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi H, Dwyer DS, Keiper BD, Jankowska-Anyszka M, Darzynkiewicz E, Rhoads RE. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 2002;21:4680–4690. doi: 10.1093/emboj/cdf473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 2000;275:10590–10596. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
- 16.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 17.Wallace A, Filbin ME, Veo B, McFarland C, Stepinski J, Jankowska-Anyszka M, Darzynkiewicz E, Davis RE. The nematode eukaryotic translation initiation factor 4E/G complex works with a trans-spliced leader stem-loop to enable efficient translation of trimethylguanosine-capped RNAs. Mol. Cell Biol. 2010;30:1958–1970. doi: 10.1128/MCB.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darzynkiewicz E, Stepinski J, Ekiel I, Jin Y, Haber D, Sijuwade T, Tahara SM. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson KC, Heid PJ, Rothman JH. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 1996;10:1543–1556. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- 20.Hotez PJ. Neglected infections of poverty in the United States of america. PLoS Negl. Trop. Dis. 2008;2:e256. doi: 10.1371/journal.pntd.0000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 22.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 23.Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS. The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 2007;143:1504–1518. doi: 10.1104/pp.106.093146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volpon L, Osborne MJ, Borden KL. NMR assignment of human eukaryotic translation initiation factor 4E (eIF4E) in its cap-free form. J. Biomol. NMR. 2006;36(Suppl. 1):65. doi: 10.1007/s10858-006-9051-8. [DOI] [PubMed] [Google Scholar]
- 25.Brown CJ, McNae I, Fischer PM, Walkinshaw MD. Crystallographic and mass spectrometric characterisation of eIF4E with N7-alkylated cap derivatives. J. Mol. Biol. 2007;372:7–15. doi: 10.1016/j.jmb.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 26.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Ishida T, Taniguchi T, Hasegawa H, Terashima A, et al. Crystal structures of 7-methylguanosine (m7GTP)- and P1-7-methylguanosine-P3-adenosine-5′, 5′-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem. J. 2002;362:539–544. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, et al. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 2003;328:365–383. doi: 10.1016/s0022-2836(03)00314-0. [DOI] [PubMed] [Google Scholar]
- 28.Rosettani P, Knapp S, Vismara MG, Rusconi L, Cameron AD. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J. Mol. Biol. 2007;368:691–705. doi: 10.1016/j.jmb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Rutkowska-Wlodarczyk I, Stepinski J, Dadlez M, Darzynkiewicz E, Stolarski R, Niedzwiecka A. Structural changes of eIF4E upon binding to the mRNA 5′ monomethylguanosine and trimethylguanosine cap. Biochemistry. 2008;47:2710–2720. doi: 10.1021/bi701168z. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Zhao R, McFarland C, Kieft J, Niedzwiecka A, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Jones DN, Davis RE. Structural insights into parasite eIF4E binding specificity for m7G and m2,2,7G mRNA caps. J. Biol. Chem. 2009;284:31336–31349. doi: 10.1074/jbc.M109.049858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darzynkiewicz E, Ekiel I, Tahara SM, Seliger LR, Shatkin AJ. Chemical synthesis and characterization of 7-methylguanosine cap analogs. Biochemistry. 1985;24:1701–1707. [Google Scholar]
- 32.Jankowska M, Temeriusz A, Stolarski R, Darzynkiewicz E. Synthesis of m2,7GTP- and m2,2,7GTP-sepharose 4B: new affinity resins for isolation of cap binding proteins. Collect. Czech. Chem. Commun. 1993;58:132–137. [Google Scholar]
- 33.Stepinski J, Bretner M, Jankowska M, Felczak K, Stolarski R, Wieczorek Z, Cai A-L, Rhoads RE, Temeriusz A, Haber D, et al. Synthesis and properties of P1,P2-, P1,P3- and P1,P4-dinucleoside di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides Nucleotides. 1995;14:717–721. [Google Scholar]
- 34.Avino AM, Espuny R, Bach M, Eritja R. A convenient method for the preparation of N2,N2-dimethylguanosine. Nucleosides Nucleotides. 1995;7:1613–1617. [Google Scholar]
- 35.Yoshikawa M, Kato T, Takenishi T. A novel method for phosphorylation of nucleosides to 5′-nucleotides. Tetrahedron Lett. 1967;8:5065–5068. doi: 10.1016/s0040-4039(01)89915-9. [DOI] [PubMed] [Google Scholar]
- 36.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Delano WL. The Pymol Molecular Graphics System. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- 39.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 42.Kowalska J, Lukaszewicz M, Zuberek J, Ziemniak M, Darzynkiewicz E, Jemielity J. Phosphorothioate analogs of m7GTP are enzymatically stable inhibitors of cap-dependent translation. Bioorg. Med. Chem. Lett. 2009;19:1921–1925. doi: 10.1016/j.bmcl.2009.02.053. [DOI] [PubMed] [Google Scholar]
- 43.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 44.Stachelska A, Wieczorek Z, Ruszczynska K, Stolarski R, Pietrzak M, Lamphear BJ, Rhoads RE, Darzynkiewicz E, Jankowska-Anyszka M. Interaction of three Caenorhabditis elegans isoforms of translation initiation factor eIF4E with mono- and trimethylated mRNA 5′ cap analogues. Acta Biochim. Pol. 2002;49:671–682. [PubMed] [Google Scholar]
- 45.Niedzwiecka A, Darzynkiewicz E, Stolarski R. Thermodynamics of mRNA 5′ cap binding by eukaryotic translation initiation factor eIF4E. Biochemistry. 2004;43:13305–13317. doi: 10.1021/bi0491651. [DOI] [PubMed] [Google Scholar]
- 46.Strasser A, Dickmanns A, Luhrmann R, Ficner R. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 2005;24:2235–2243. doi: 10.1038/sj.emboj.7600701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zidek L, Novotny MV, Stone MJ. Increased protein backbone conformational entropy upon hydrophobic ligand binding. Nat. Struct. Biol. 1999;6:1118–1121. doi: 10.1038/70057. [DOI] [PubMed] [Google Scholar]
- 48.Ruszczynska-Bartnik K, Maciejczyk M, Stolarski R. Dynamical insight into Caenorhabditis elegans eIF4E recognition specificity for mono-and trimethylated structures of mRNA 5′ cap. J. Mol. Mod. 2011;17:727–737. doi: 10.1007/s00894-010-0773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–3912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruszczynska K, Kamienska-Trela K, Wojcik J, Stepinski J, Darzynkiewicz E, Stolarski R. Charge distribution in 7-methylguanine regarding cation-pi interaction with protein factor eIF4E. Biophys. J. 2003;85:1450–1456. doi: 10.1016/S0006-3495(03)74577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 52.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 55.Gross JD, von Moerke NJ, der Haar T, Lugovsky AA, Sachs AB, McCarthy JEG, Wagner G. Ribosome Loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.