Abstract
The interaction between single-stranded RNAs and liposomes was studied using UV, Fourier Transform Infrared spectroscopy (FTIR) and Circular Dichroism spectroscopy (CD). The effect of the surface characteristics of liposomes, which were composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and modified with cholesterol (Ch) or 1,2-dioleoyl-3-trimethylammonium propane (DOTAP), on the liposome–RNA interaction was investigated. The fluorescence of 6-(p-toluidino)naphthalene-2-sulfonate (TNS) embedded in the liposome surface (ε = 30–40) was decreased in the presence of tRNA, suggesting that single-stranded tRNA could bind onto the liposome. The dehydration of –PO2− –, guanine (G) and cytosine (C) of tRNA molecules in the presence of liposomes suggested both an electrostatic interaction (phosphate backbone of tRNA and trimethylammonium group of POPC, DOTAP) and a hydrophobic interaction (guanine or cytosine of tRNA and aliphatic tail of lipid). The tRNA conformation on the liposome was determined by CD spectroscopy. POPC/Ch (70/30) maintained tRNA conformation without any denaturation, while POPC/DOTAP(70/30) drastically denatured it. The mRNA translation was evaluated in an Escherichia coli cell-free translation system. POPC/Ch(70/30) enhanced expression of green fluorescent protein (GFP) (116%) while POPC/DOTAP(70/30) inhibited (37%), suggesting that the conformation of RNAs was closely related to the translation efficiency. Therefore, single-stranded RNAs could bind to liposomal membranes through electrostatic and hydrophobic attraction, after which conformational changes were induced depending on the liposome characteristics.
INTRODUCTION
Biomembranes play crucial roles in biological systems. Under normal conditions, the biomembrane acts as a physical boundary between the outside and inside of the cell, and regulates the transport of molecules. Under stress conditions, the biomembrane often induces biological functions in order to protect biomolecules from denaturation. For example, it has previously been reported that a damaged protein or a fragmented peptide could be reactivated on the liposome membrane (1). Furthermore, the biomembrane can transfer signals of environmental stress (heat, ROS, etc.) into the cell, resulting in the activation of anti-stress genes to induce a stress response. Kunimoto et al. (2,3) have reported that steryl glucoside could be a lipid mediator in stress–responsive signal transduction. Biomembranes clearly play important roles under various stress conditions; however, the interaction between biomembranes and biomolecules is not fully understood. In order to clarify this complicated and vital mechanism in the biological system, innovative approaches such as ‘Membranomics’ or ‘Membrane Stress Biotechnology’ that focus on the potential functions of biomembrane, are worth pursuing (4).
One of the essential roles of biomembranes is the localization of function through the binding of biomolecules (5). Artificial closed phospholipid-bilayer membranes, called as liposomes, have been well studied since the liposomes can mimic the biomembrane-like environment of the cell. Not only proteins or enzymes but also polynucleotides, such as DNA or RNA can be functionalized on the biomembrane or its mimics (6,7). Indeed, the biomembrane can (i) interact with biomolecules, (ii) induce minor conformational changes and then (iii) regulate their functions. This series of phenomena has been termed as ‘Biomembrane Interference’ (8), where the biomembrane has previously been shown to regulate in vitro gene expression (8–12). A zwitterionic liposome composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) enhanced expression of green florescent protein (GFP) in an Escherichia coli cell-free translation system; POPC modified with 30 mol% of cholesterol (Ch) most effectively enhanced GFP expression, while cationic liposome 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) or anionic liposome 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG) inhibited it. These regulations must be attributed to the interaction between liposomes and biomolecules, such as DNA, RNA, nascent peptides and transcriptional/translational enzymes. Details of the mechanisms were investigated, focusing on each elemental step (transcription, translation and folding), with the finding that the liposomes significantly affected the translation step; the POPC/Ch(70/30) liposome enhanced the polypeptide synthesis, while the cationic liposomes inhibited it (11). The anionic liposomes did not affect the translation; they rather inhibited the folding of the GFP peptide (9,13). Therefore, the heterogeneous liposomes or the cationic liposomes would have significant effects on the translation step. The key event was the functionalization of RNA on the liposomes, although the translation step was so complicated that the detailed mechanism of liposome-based interfering gene translation could not be clarified. Since there are many kinds of translation-related RNA molecules (mRNA, tRNA, rRNA, etc.), the interaction between liposomes and RNAs requires further investigation.
The final purpose of so-called ‘Biomembrane Interference’ research is to discover the mechanisms of the regulation of gene translation by liposomes and to understand the role of biomembranes in the cell system. In this study, we focused on the interaction between liposomes and single-stranded RNAs. Single-stranded RNAs, mRNA and tRNA, are the main components in translation and their biological functions are closely related to their conformations (14). Although the conformation of mRNA is very important for its translation efficiency (15), mRNA conformation is flexible and too complicated to be evaluated or predicted, such that special methods to evaluate RNA conformation are required. On the other hand, transfer RNA (tRNA) has a stereotypic conformation called ‘clover-leaf’, and its crystal structure has been investigated (16). It is thus rational and important to use tRNA, which has an A-form double helix conformation (8), as a model single-stranded RNA and to study the interaction between the liposome and tRNA as an initial step of ‘Biomembrane Interference’ research. There are several reports on liposome–RNA interaction, with findings that (i) the liquid-crystal phase liposome (ld) tends to be associated with RNA more than the liquid-gel phase one (lo) (17); (ii) heterogeneous lipid bilayers, such as phosphocholine/cholesterol or phosphocholine/sphingomyelin/cholesterol, can also interact with RNAs (18); (iii) size-specific interactions occur between tRNA and POPC/cetyl-trimethyl-ammonium bromide (CTAB) liposomes (19); (iv) these interactions induce minor conformational changes to RNA and liposome segregation (20) and (v) these interactions are driven through hydrophobic attraction, deriving from both the hydrophobic nucleobases of RNA and the exposed hydrophobic parts of the liposome surface (21).
Judging from these previous findings, the surface properties of liposomes play an important role in RNA regulation. We have previously reported circular dichroism (CD) spectroscopic findings that the heterogeneous POPC/Ch(70/30) liposome (ld + lo) destabilized tRNA under heat stress conditions (8). Although it is difficult to predict or evaluate the mRNA conformation owing to its larger molecular weight, CD spectroscopy analysis demonstrated that the conformation of mRNA could be regarded as an A-form, as was similarly observed in the case of tRNA. In addition, the liposomes were shown to affect the translational activity of GFP mRNA, a key fact needed to assess the relationship between mRNA conformation and its activity. The dependence of the liposome–RNA interaction on liposome characteristics is discussed based on the obtained results, such as those summarized in Table 1.
Table 1.
Summary of liposome surface characteristics and the melting temperature (Tm) of tRNA or mRNA on liposomal membrane
| Liposome | Phase | Surface charge densitya (C/nm2) | Membrane fluidity, 1/P (−) |
Tmb (°C) |
|
|---|---|---|---|---|---|
| tRNA | mRNA | ||||
| tRNA/mRNA | – | – | – | 48 | 38 |
| (+)POPC | ldc | ±0 | 5.58 ± 0.42 | 46 | – |
| (+)POPC/Ch (90/10) | ldc | ±0 | 5.28 ± 0.59 | 50 | – |
| (+)POPC/Ch (70/30) | ld+loc | ±0 | 3.54 ± 0.05 | 38 | 33 |
| (+)POPC/Ch (50/50) | loc | ±0 | 2.08 ± 0.09 | 44 | – |
| (+)POPC/DOTAP (70/30) | ldd | + 0.42 | 5.91 ± 0.27 | 53 | – |
| (+)POPC/DOTAP (50/50) | ldd | + 0.70 | 6.17 ± 0.43 | ND | – |
| (+)DOTAP | ldd | + 1.4 | 6.44 ± 0.54 | 62 | 48 |
aSurface charge density of liposome was calculated assuming unilamellarity, spherical shape, a bilayer thickness of 3.7 nm and a mean head group area of 0.72 nm2 (22). bTm was caluculated according to previou report (8). The methods were also shown in Supplementary Figure S3.
cPhase-state of liposome was shown according to De Almedia’s report (23).
dPhase state of POPC/DOTAP liposome was determined by membrane fluidity, 1/P.
MATERIALS AND METHODS
Materials
POPC and DOTAP were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Transfer RNA originating from E. coli and Ch were purchased from Sigma-Aldrich (St Louis, MO, USA). The lipid structure is shown in Supplementary Figure S1. A Rapid Translation System (RTS) 100 E. coli HY Kit (RTS-Kit) was purchased from Roche Diagnostics (Indianapolis, IN, USA). T7 RiboMAXTM Expression Large-scale RNA Production System and SV Total RNA Isolation System were purchased from Promega (Madison, WI, USA). Other chemicals were purchased from Wako Pure Chemical (Osaka, Japan) and were used without further purification.
Liposome preparation
A solution of POPC containing 0–50 mol% of Ch and 0–100 mol% of DOTAP in chloroform was dried in a round-bottom flask by rotary evaporation under a vacuum. The lipid films obtained were dissolved in chloroform twice, and the solvent was evaporated. The lipid thin film was kept under a high vacuum for at least 3 h, and was then hydrated at room temperature with 100 mM Tris–HCl buffer (pH 8.0) and water, for POPC/Ch and DOTAP, respectively. The vesicle suspension was frozen at −80°C and thawed at 50°C to enhance the transformation of small vesicles into larger multi lamellar vesicles (MLVs). This freeze–thaw cycle was performed five times. MLVs were used to prepare the small unilamellar vesicles (SUVs) by extruding the MLV suspension 11 times through two layers of polycarbonate membranes with mean pore diameters of 100 nm using an extruding device (Liposofast; Avestin Inc., Ottawa, ON, Canada).
Transcription and purification of GFP mRNA
As the plasmid DNA, pIVEX Control Vector GFP (Roche) was used. The plasmid DNA was treated once with restriction enzyme ApaL I for 1 h incubation at 37°C in order to cleave the AmpR gene and to obtain line DNA fragments harboring the GFP gene before transcription. The transcription of mRNA encoding the GFP gene (861 bp) was carried out using T7 RiboMAXTM Expression Large Scale RNA Production System (Promega, Madison, WI, USA), which includes T7 RNA polymerase as a transcriptional enzyme. Transcription was achieved for 30 min at 37°C. The obtained mRNA was recovered and purified with the SV total RNA Isolation Kit (Promega, Madison, WI, USA), as described in our previous study (9). The mRNA products were quantified from the absorbance at 258 nm and the electrophoresis on 1% agarose gel.
Evaluation of UV spectra
The turbidity of liposome suspension in the presence or absence of tRNA was evaluated using a UV-1800 Spectrophotometer (SHIMAZU, Kyoto) and an Ultramark microplate reader (Bio-Rad Japan, Tokyo). The turbidity at 405 nm (OD405) was measured using a quartz cell (1 cm path length) for a spectrophotometer and with 96-well plate for the microplate reader at 30°C (Table 2). The increase of OD405 was defined as ΔOD405; ΔOD405 = OD405, (+) RNA – OD405, (−) RNA.
Table 2.
Turbidity increase of liposomes in the presence of tRNA
| Liposomea | ΔOD405 (a.u.)b |
|---|---|
| POPC | 0.003 ± 0.015 |
| POPC/Ch(90/10) | −0.001 ± 0.001 |
| POPC/Ch(70/30) | 0.074 ± 0.026 |
| POPC/Ch(50/50) | 0.029 ± 0.045 |
| POPC/DOTAP(70/30) | 0.357 ± 0.003 |
| POPC/DOTAP(50/50) | 0.142 ± 0.002 |
| DOTAP | 0.162 ± 0.003 |
The final concentration of tRNA was 2.2 µM.a The lipid concentration was 1 mM for POPC/Ch and 1.17 mM for POPC/DOTAP.
bΔOD405 = OD405, (+) RNA − OD405, (−) RNA
It was confirmed that no absorbance (at 405 nm) was observed in the case of tRNA only. The final concentration of tRNA was 2.2 µM, where the lipid concentration was 1 mM for the zwitterionic liposomes (POPC/Ch) and 1.17 mM for the cationic liposomes (POPC/DOTAP).
Fluorescence measurement of TNS
The fluorescent emission of 6-(p-toluidino)naphthalene-2-sulfonate (TNS) has been reported to be affected by environmental polarity; the fluorescence emission is weak in a polar environment (in water, the relative permittivity ε = 78.5) but strong in an organic solvent (ε = 2.2) (24) (see Supplementary Figure S2). TNS was directly added to a liposome suspension and was then incubated for 1 h at room temperature in order to complete TNS insertion into lipid membrane. After the addition of tRNA, samples were incubated in the dark for 30 min at 30°C. The fluorescence spectra of TNS (Ex: 340 nm) were measured from 380 to 500 nm using a fluorescence spectrophotometer (FP-6500; JASCO, Tokyo, Japan) with 5 nm light path. The final concentrations of TNS and lipid were 20 µM and 0.5 mM, respectively. TNS fluorescence was not observed with tRNA only.
Infrared spectroscopy of tRNA
A 30 µl tRNA sample was applied in 50 μm-thick cell with a CaF2 window. The infrared (IR) spectra were measured using an Fourier Transform Infrared spectroscopy (FTIR) 4100 spectrometer (JASCO, Japan) equipped with an Hg–Cd–Te detector. The resolution was set up at 2 cm−1; the frequency range from 1750 to 1150 cm−1 was collected for each sample. The IR spectrum of water was subtracted from those of the samples. The accuracy of the frequency reading was better than ±0.1 cm−1. One hundred scans excluding buffer and liposome background signals were accumulated. The spectra were smoothed with the Savitzky-Golay procedure (41,42). The concentration of tRNA and the lipid was adjusted at 3.94 mM and 0–98.5 µM, respectively, and the lipid/tRNA molar ratios were 0/100, 1/100 and 1/40. Measured wavenumbers, relative intensity and assignments are shown in Table 3.
Table 3.
Peak shift and peak assignment of tRNA measured by FTIR
| Wavenumber (cm−1) |
Δν (cm−1) |
Assignment (21,27,28)a | ||||||
|---|---|---|---|---|---|---|---|---|
| POPC | POPC/ Ch (90/10) | POPC/ Ch (70/30) | POPC/ Ch (50/50) | POPC/DOTAP (70/30) | POPC/DOTAP (50/50) | DOTAP | ||
| 1706 | +0.96 | +0.48 | +0.96 | +1.45 | +2.84 | +2.41 | +1.45 | Guanine (C=O stretching, vs) |
| 1656 | +1.93 | 0 | +0.96 | −2.41 | – | +0.96 | +3.86 | Uracil (C=O stretching, vs) |
| 1603 | +1.45 | 0 | 0 | +0.48 | +1.45 | +2.89 | +4.38 | Adenine (C=N stretching, s) |
| 1492 | 0 | −1.93 | +3.38 | +0.96 | +2.41 | +0.96 | +3.38 | Cytosine (plane ring vibration, m) |
| 1236 | +4.34 | +2.89 | +4.82 | −0.44 | +2.89 | +3.38 | +6.27 | -PO2−- (asymmetric stretching, vs) |
aPeak intensities are shown using the following abbreviations: vs, very strong; s, strong and m, medium.
Evaluation of the conformation of RNA by CD spectra
The conformation of RNA was evaluated using a JASCO J-820 SFU spectropolarimeter (JASCO, Tokyo) (8). The CD spectrum from 300 to 200 nm was measured with a quartz cell (0.1 cm path length) at a scan speed of 50 nm min−1 and a width of 2 nm. Five scans excluding buffer and liposome background signals were accumulated, and the data were calculated as molar ellipticity. The sample was prepared with 2.2 µM of tRNA or 0.77 µM of mRNA, together with 10 mM Tris–HCl at pH 7.8 in the presence or absence of 1.17 mM lipids. The stability of RNAs under the heat stress conditions was defined according to the previous report (8). The negative peak at 208 nm (θ208) was normalized and the half-point of normalized θ208 was defined as the melting temperature Tm. Details are also shown in Supplementary Figure S3.
Evaluation of mRNA translational activity using E. coli cell-free translation system
GFP was expressed in the cell-free translation system, RTS-Kit. The liposomes and mRNA were pre-incubated for 15 min at 30°C, and then added to RTS-Kit. GFP expression was achieved for 6 h at 30°C, and the obtained GFP was kept at 4°C for 24 h. The amount of GFP synthesized using RTS-Kit was evaluated by SDS–PAGE analysis and the fluorescence of GFP (Ex = 395 nm, Em = 509 nm), based on the methods published previously (11).
Evaluation of liposome characteristics
The characteristics of liposomes were evaluated based on previous reports (9,29). To measure the inner membrane fluidity, a fluorescent probe 1,3,5-hexatriene (DPH), was added to the liposome suspension (molar ratio; lipid/DPH 250/1). The fluorescence polarization of DPH (Ex = 360 nm, Em = 430 nm) was measured after 1 h incubation at 30°C using a fluorescence spectrophotometer (FP-6500; JASCO, Tokyo, Japan). The sample was excited with vertically polarized light (360 nm), and emission intensities (430 nm) both parallel (I∥) and perpendicular (I⊥) to the excited light were recorded. Then, the polarization (P) of DPH was calculated based on the following equation:
The membrane fluidity was evaluated based on the reciprocal of the polarization (1/P). The surface charge density of liposome was calculated according to our previous work (9).
RESULTS AND DISCUSSION
The liposomes used in this study were composed of POPC, and the liposomes were modified with Ch or the cationic lipid DOTAP. POPC/Ch (0–50 mol% Ch) and POPC/DOTAP (0–100 mol% DOTAP) possess different phase states (membrane fluidity) and surface charge densities, respectively (Table 1) (23). The effect of liposome characteristics on the interaction with RNAs was studied as follows.
Binding of tRNA onto liposomes
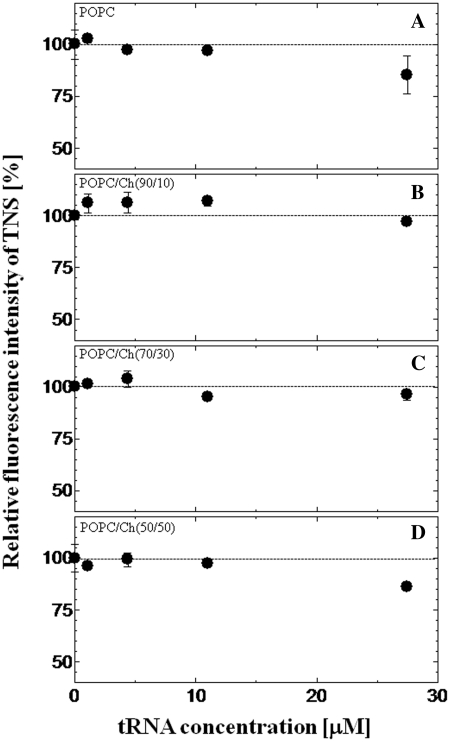
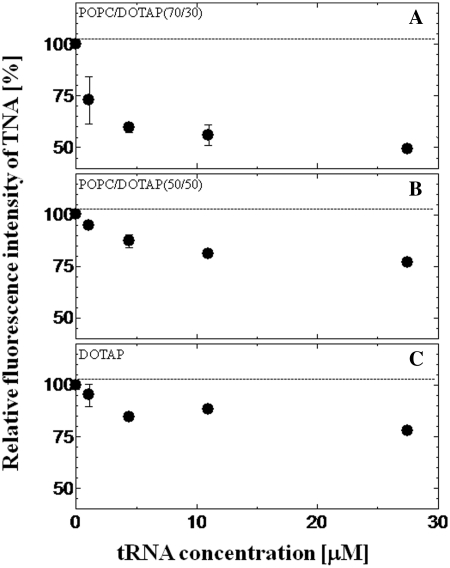
The interaction between liposomes and tRNA, which is a model single-stranded RNA, was investigated using a fluorescent probe TNS. It has been reported that the emission peak and fluorescence intensity of TNS are sensitive to the polarity of the environment (30) (see Supplementary Figure S2). TNS can also be applied to the evaluation of the binding of tRNA onto the liposome membrane. Although TNS fluorescence in bulk water solution was very weak, TNS inserted into the vesicle was found to emit strong fluorescence (31). The fluorescence properties of TNS inserted into the lipid membrane are therefore expected to depend on the micropolarity of the lipid vesicles or liposomes (32,33). The relative fluorescence intensity of TNS proved to be a function of tRNA concentration is shown in Figures 1 and 2. The emission peaks of TNS embedded in the membrane of various liposomes were 441–446 nm, indicating that TNS was inserted into the water/lipid interface with the dielectric constant of ∼30–40, and that was ∼1 nm below the liposome surface (24). The value of the relative fluorescence intensity of TNS became lower as the tRNA concentration increased: 85 ± 9% for POPC, 96 ± 2% for POPC/Ch(70/30) and 49 ± 1% for POPC/DOTAP(70/30), where the fluorescence intensity of TNS without tRNA was defined as 100%. Since no peak shift was observed at higher tRNA concentrations, the fluorescence intensity decrease was possibly due to the replacement of TNS by tRNA, and to the dissipated TNS losing its fluorescence in the bulk water phase, which could be evidence for the binding of tRNA to the liposome surface. In contrast to POPC or POPC/DOTAP (30–100 mol%), POPC modified with 10 or 30 mol% Ch showed slight decrease of the TNS fluorescence. As possible reasons, it was considered that either the tRNA did not bind to the liposomes or the binding was so weak that bound tRNA was not inserted into the liposome (TNS was not altered). To confirm this point, the turbidity of the liposome suspension (OD405) in the presence or absence of tRNA was measured (Table 2). The turbidity of POPC/Ch(70/30) was markedly increased in the presence of tRNA although no increase was observed with POPC/Ch(90/10), suggesting that the liposome–RNA interaction was dependent on the phase state of the liposome membrane (20). This turbidity increase suggested the formation of a liposome–RNA complex (19). Cationic liposomes were shown to have a stronger interaction with negatively charged tRNA than the other liposomes tested here. These results indicated that the binding mechanism differs depending on the liposome type. The lipid packing density may be closely related with the surface charge density of the membrane. Therefore, the surface charge density and phase state may be key parameters of the lipid membrane–oligonucleotide interaction (34).
Figure 1.
Effect of tRNA on the relative fluorescence intensity of TNS with zwitterionic liposomes. TNS was excited at 340 nm, and the TNS emission peak was 446 nm for POPC (A), 444 nm for POPC/Ch(90/10) (B), 442 nm for POPC/Ch(70/30) (C) and 440 nm for POPC/Ch(50/50) (D). The dotted line shows the relative fluorescence intensity without tRNA. The final concentrations of lipids and TNS were 0.5 mM and 20 µM, respectively.
Figure 2.
Effect of tRNA on the relative fluorescence intensity of TNS with cationic liposomes. TNS was excited at 340 nm, and the TNS emission peak was 445 nm for POPC/DOTAP(70/30) (A), 444 nm for POPC/DOTAP(50/50) (B) and 441–444 nm for DOTAP (C). The dotted line shows the relative fluorescence intensity without tRNA. The final concentrations of lipids and TNS were 0.5 mM and 20 µM, respectively.
Binding of nucleobases of tRNA onto liposomes
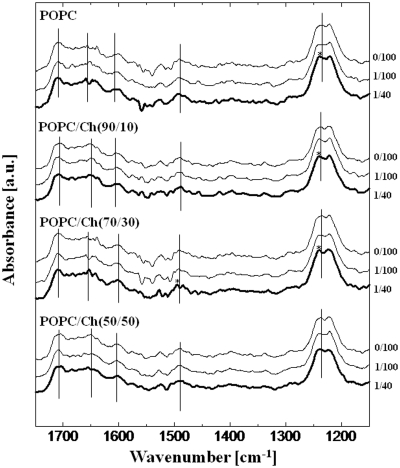
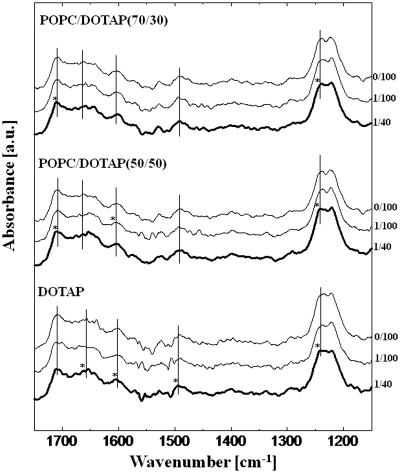
The liposome–tRNA interaction was investigated in detail using FTIR (Figures 3 and 4). Marty et al. (21) reported the lipid–tRNA interaction by FTIR, identifying a hydrophobic interaction between the lipid acyl-chain and the hydrophobic parts of tRNA. In the presence of liposomes, increases in the peak intensity derived from nucleobases were observed (data not shown): 1706 cm−1 (guanine C=O stretching), 1656 cm−1 (uracil C=O stretching), 1603 cm−1 (adenine C=N stretching) and 1492 cm−1 (cytosine plane ring vibration). This result was confirmed by the previous report (21). Shifts of peaks not only of the nucleobases but also of the phosphate backbone were observed as summarized in Table 3. In common with all kinds of liposomes, the peak of the –PO2− – asymmetric stretching at 1236 cm−1 was shifted to a higher frequency (1240–1242 cm−1). The phosphate backbone of tRNA has a negative charge and the lipid head group of POPC or DOTAP (trimethylammonium group) has a positive charge, suggesting that the head group of the lipid and tRNA backbone could interact through electrostatic attraction. No peak shift was observed at 1118 cm−1 (ribose C–C stretching) or at 1087 cm−1 (–PO2− – symmetric stretching) in the presence of liposomes (data not shown). Giel-Pietraszuk and Barciszewski have previously reported that the blue shift of the IR peak is due to an increase of bond energy as a result of the shortening of the hydrogen bonds followed by dehydration of tRNA (35). It is, therefore, considered that the higher frequency peak shift was caused by liposome binding, and that the tRNA phosphate backbone was dehydrated. Focusing on the nucleobases, the peak shifts were shown to depend on the liposome characteristics. In the case of POPC, the peaks of guanine (1707 cm−1), uracil (1658 cm−1) and adenine (1604 cm−1) were shifted to a higher frequency by ∼2 cm−1, suggesting that nucleobases were inserted into the phospholipid bilayer of the POPC liposome and their surfaces were then dehydrated. A hydrophobic interaction between the lipid aliphatic tail and tRNA has been reported to occur in a specific nucleobases, such as guanine, adenine and uracil (21). However, no significant peak shifts were observed in the presence of other Ch-modified POPC liposomes, although a specific peak shift at cytosine (1495 cm−1) was observed in POPC/Ch(70/30). Among the Ch-modified liposomes, the membrane of the POPC/Ch(70/30) liposome exists at a heterogeneous phase state (ld + lo), whereas the POPC/Ch(90/10) and the POPC/Ch(50/50) exist at homogeneous phase state, (ld) and (lo), respectively (23). In fact, the POPC/Ch(70/30) liposome was most effective in enhancing the in vitro gene expression (10). One possible reason for this enhancement could be that the heterogeneous membrane interacted with cytosine residue of RNAs in order to accumulate them on the membrane, and then the in vitro gene expression was enhanced on the liposome membrane. The water hydration on Ch-incorporating liposome membranes was increased above 33 mol% Ch and, as a result, the expressed peptide could be recruited on the liposome surface through the hydrogen bonds (36). Such a specific interaction of the nucleobases with lipid membranes would be important from the viewpoint of the functionalization of biomolecules on the membrane surface (37). On the contrary, peak shifts of all four nucleobases were observed in DOTAP, showing the strong hydrophobic interaction among them.
Figure 3.
FTIR spectra of tRNA in the presence of zwitterionic liposomes. The molar ratios of lipid/tRNA were 0/100, 1/100 and 1/40. Axial bars indicating peak assignments of tRNA are shown in Table 3. Stars indicate significant peak shifts (>2.5 nm). The final concentration of tRNA and the lipid was 3.94 mM and 0–98.5 µM, respectively.
Figure 4.
FTIR spectra of tRNA in the presence of cationic liposomes. The molar ratios of lipd/tRNA were 0/100, 1/100 and 1/40. Axial bars indicating peak assignments of tRNA are shown in Table 3. Stars indicate significant peak shifts (>2.5 nm). The final concentration of tRNA and the lipid was 3.94 mM and 0–98.5 µM, respectively.
Although the above results were just obtained in the case of tRNA with ∼80 nt, the interaction between liposomes and tRNA was investigated much more in details. The interaction at phosphate backbone or at nucleobases should hold true for other single-stranded RNAs. As can be seen in the obtained IR spectra (Figures 3 and 4), the peak shifts of the phosphate backbone and nucleobases towards higher frequencies indicated the dehydration of tRNA (35) induced by the interaction with liposomes. This dehydration was due to the binding of tRNA onto the liposome membranes, and the tRNA binding was found to be driven through electrostatic interactions (the lipid head group and tRNA phosphate backbone), hydrophobic interactions (the lipid aliphatic tail and the nucleobases) and the formation of hydrogen bonds. Increases of peak intensity at G, U, A and C were also observed (data not shown), indicating that the base stacking of nucleobases was decreased in the presence of liposomes. In previous studies, double-stranded DNA was found to bind onto the phospholipid vesicles, including the conformational transition on the membrane (6,34). It is expected that not only DNAs, but also single-stranded RNAs change their conformation on the phospholipid membrane.
Conformational change of single-stranded RNAs induced by liposome binding
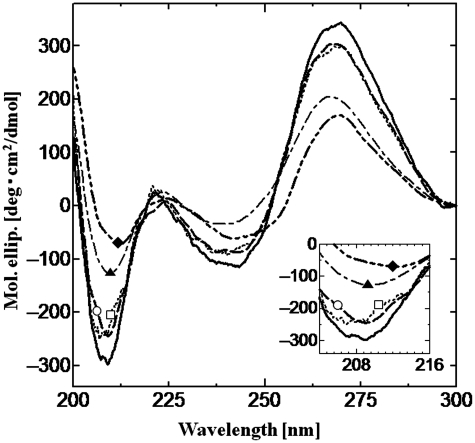
The conformation of tRNA is an A-form double helix, which has a negative CD peak at 208 nm and a positive peak at 265 nm (38,39); these peak intensities were decreased with its denaturation under the heat stress conditions (8). Thus, the decrease of peaks at 208 and 265 nm indicates the denaturation of tRNA. CD spectra of tRNA were measured in the presence of liposomes (Figure 5). Under the experimental condition, tRNA conformation in the presence of POPC or POPC/Ch(70/30) was maintained although there was no significant difference between the spectra obtained with POPC or POPC/Ch(70/30) at 30°C. On the contrary, the tRNA conformation was drastically denatured in the presence of the cationic liposomes [POPC/DOTAP(70/30) and POPC/DOTAP(50/50)], and the CD peak intensity was decreased in proportion to the surface charge density of liposomes. The anionic liposome [POPC/POPG(70/30)] had little effect on the tRNA conformation due to the electrostatic repulsion (data not shown).
Figure 5.
CD spectra of tRNA in the presence of liposomes at 30°C. Black bold line only, tRNA without liposomes; dotted line (opened square), with POPC; dashed bold line (opened circle), with POPC/Ch(70/30); dot-dashed line (closed triangle), with POPC/DOTAP(70/30) and two dot-dashed bold line (closed diamond), with POPC/DOTAP(50/50). Final tRNA and lipid concentrations were 2.2 µM and 1.17 mM, respectively. Insert shows close-up spectra at around 208 nm.
In order to discuss the conformation of tRNA on the liposomes, the melting temperature (Tm) of tRNA was calculated under heat stress conditions (see Supplementary Figure S3). Herein, the Tm value is a parameter to describe the stability of RNA. The coexistence of Mg2+, that stabilizes the RNA conformation (40,41), resulted in the increase of the Tm value (48–60°C), whereas POPC/Ch(70/30) destabilized the tRNA together with the decrease of Tm (48–38°C) (8). Although zwitterionic liposomes did not significantly affect the tRNA conformation at 30°C, its stability was varied, depending on the state of the liposomes (Table 1), where interaction with POPC/Ch(70/30) (ld + lo) and POPC/Ch(50/50) (lo) resulted in tRNA destabilization (Tm: 38 and 44°C, respectively). Since lipid molecules in the (lo) phase liposomes are tightly packed, tRNA insertion would be difficult, such that the tRNA binding sites would be limited to the surface. In fact, the hydrophobic interaction between liposomes and tRNA was rather decreased in the case of the Ch-modified liposomes, implying that the possible binding site of the liposome is the phosphate backbone at the liposome-bulk interface. In the presence of the cationic liposomes, the Tm value was increased in proportion to the surface charge density of liposomes. This result indicated that the cationic liposomes denatured tRNA and then the denatured tRNA was stabilized on the liposomes. In other words, the cationic liposomes-induced tRNA denaturation and then the denatured tRNA conformation was maintained on the cationic liposomes. It is therefore suggested that conformational changes of tRNA were induced by liposome binding at the tRNA phosphate backbone, whereas the electrostatic interaction was dominant rather than the hydrophobic one.
Relationship between mRNA conformation and translational activity
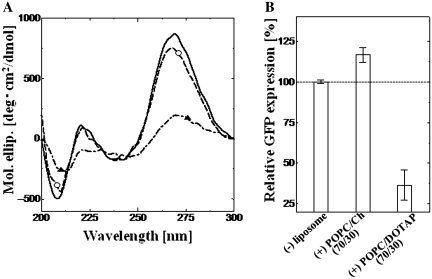
Finally, the conformation of mRNA on liposomes and its resulting translational activities were determined. Although it is difficult to predict the conformation of mRNA, the CD data indicated that the mRNA was in the A-form double helix conformation, similarly to the case of tRNA (see Supplementary Figure S4). Since the binding of single-stranded tRNA onto liposomes was shown to induce a conformational change in the above section, similar conformational changes were expected for the case of single-stranded mRNA. The conformational changes of mRNA in the presence of POPC/Ch(70/30) or POPC/DOTAP(70/30) were analyzed (Figure 6A). POPC/Ch(70/30) was found to maintain the mRNA conformation while POPC/DOTAP(70/30) drastically denatured it; this tendency was exactly the same as the result for tRNA. As for the heat stress stability of mRNA (Tm = 38°C), POPC/Ch(70/30) also destabilized the mRNA conformation (33°C) under heat stress conditions, while DOTAP stabilized it (48°C).
Figure 6.
Effect of liposomes on mRNA conformation and mRNA translational activity. (A) CD spectra are shown: black line only, mRNA without liposome; dashed line (opened circle), with POPC/Ch(70/30) and dot-dashed line (closed triangle), with POPC/DOTAP(70/30). Final mRNA and lipid concentrations were 0.77 µM and 1.17 mM, respectively. (B) GFP expression with and without liposomes. Final lipid concentration was 1.17 mM; GFP was expressed for 6 h at 30°C. The excitation and emission peaks of GFP were 395 and 509 nm, respectively.
The translational activity of mRNA was investigated using a cell-free translation system (Figure 6B). The POPC/Ch(70/30) liposomes enhanced GFP expression to 116% compared to the control (without liposomes). On the other hand, the POPC/DOTAP(70/30) liposome drastically inhibited it to 37%. Our previous studies investigated the effect of liposomes on in vitro GFP expression using a cell-free system (9–11,13), where the POPC/Ch(70/30) liposomes could recognize tRNA or mRNA, then induce their conformational change (destabilization of RNAs towards heat stress). Since the folding of mRNA affected its translation (15), a suitable destabilization of mRNA was found to enhance mRNA translational activity. One possible reason for the enhancement of the mRNA translation is the condensation effect. Liposomes recognized not only mRNA, but also other enzymes such as tRNA, ribosomes, etc. On the contrary, POPC/DOTAP(70/30) liposomes formed complexes with RNAs through electrostatic attraction and then induced their denaturation. The anionic liposomes did not inhibit the translation step because they were difficult to interact with negatively charged RNAs. Previous studies have demonstrated that liposomes or lipid vesicles could assist in protein folding like molecular chaperones (42), indicating that liposomes added to the cell-free translation system would affect the folding step of the nascent peptide of GFP (9,10). In this study, it has been shown that the liposomes could induce conformational changes in polynucleotides. DNA or RNA must take an active form in order to perform its crucial role. Tsuji et al. (6) have recently reported how the ON–OFF switching of DNA transcriptional activity occurs through conformational transitions in relation to phospholipid membranes. Taken together, these results imply that biomembranes and liposomes can functionalize biomolecules on their surfaces (5). Therefore, lipid membranes have the important role of inducing such conformational changes in single-stranded RNAs.
CONCLUSION
It was investigated whether liposomes could bind to single-stranded RNA, such as tRNA and mRNA, through electrostatic and hydrophobic attraction, and by doing so, induce conformational changes in them. There was a significant difference in the tRNA localization between liposome treatments; tRNA was bound to the membrane surface of POPC/Ch(70/30) liposome, at its bulk-lipid head group interface, and into the inner membrane of the cationic liposomes POPC/DOTAP. According to FTIR studies, the phosphate group of tRNA and the trimethyl ammonium group of lipids interacted through electrostatic attraction. Although the nucleobases (A, U, C and G) and lipid aliphatic tails interacted through hydrophobic attraction, the interaction moieties of tRNA differed depending on the liposome phase state and surface charge density. Such differences in dielectric surroundings induced different RNA conformational changes for both tRNA and mRNA. POPC/Ch(70/30) induced only minor conformational changes in RNAs. On the other hand, POPC/DOTAP(70/30) induced drastic denaturation of RNAs. Such conformational change of RNA on liposomes is one possible mechanism of gene regulation. It has thus, been shown that liposomes and biomembranes can regulate RNA functions; that is, a liposome can recruit RNA to its membrane and then induce conformational changes in it, a process known as ‘Biomembrane Interference’.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Next Generation World-Leading Researchers (NEXT Program, GR066) from the Council for Science and Technology Policy, Cabinet Office and Japan Society for the Promotion of Science (JSPS). Ministry of Education, Science, Sports and Culture of Japan (MEXT), Grants-in-Aid for Scientific Research (Nos. 19656203, 19656220 and 20360350); Global COE program ‘Bio-Environmental Chemistry’ of JSPS. One of the authors (K.S.) acknowledges scholarships as JSPS and GCOE fellow. Funding for open access charge: NEXT, JSPS, MEXT.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The fundamental concept of this study was supported by the Research Group of ‘Membrane Stress Biotechnology’ and the Sigma Multidisciplinary Research Laboratory Group (Graduate School of Engineering Science, Osaka University) ‘Membranomics’. We acknowledge Prof. Dr Ryoichi Kuboi (Executive Director of San Francisco Center, Osaka University) for his fruitful discussion and advice on this research.
REFERENCES
- 1.Tuan LQ, Umakoshi H, Shimanouchi T, Kuboi R. Liposome-recruited activity of oxidized and fragmented superoxide dismutase. Langmuir. 2008;24:350–354. doi: 10.1021/la702690a. [DOI] [PubMed] [Google Scholar]
- 2.Kunimoto S, Murofushi W, Kai H, Ishida Y, Uchiyama A, Kobayashi T, Kobayashi S, Murofushi H, Murakami-Murofushi K. Steryl glucoside is a lipid mediator in stress-responsive signal transduction. Cell Struct. Funct. 2002;27:157–162. doi: 10.1247/csf.27.157. [DOI] [PubMed] [Google Scholar]
- 3.Kunimoto S, Murofushi W, Yamatsu I, Hasegawa Y, Sasaki N, Kobayashi S, Kobayashi T, Murofushi H, Murakami-Murofushi K. Cholesteryl glucoside-induced protection against gastric ulcer. Cell Struct. Funct. 2003;28:179–186. doi: 10.1247/csf.28.179. [DOI] [PubMed] [Google Scholar]
- 4.Umakoshi H, Shimanouchi T, Kuboi R. Development of LIPOzymes Based on Biomembrane Process Chemistry. In: Shioiri T, Izawa K, Konoike T, editors. Pharmaceutical Process Chemistry. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. [Google Scholar]
- 5.Walde P. Phospholipid membranes as regulators of localized activity. Chem. Biol. 2010;17:922–923. doi: 10.1016/j.chembiol.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Tsuji A, Yoshikawa K. ON–OFF switching of transcriptional activity of large DNA through a conformational transition in cooperation with phospholipid membrane. J. Am. Chem. Soc. 2010;132:12464–12471. doi: 10.1021/ja105154k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García JM, García V, Peña C, Domínguez G, Silva J, Diaz R, Espinosa P, Citores MJ, Collado M, Bonilla F. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008;14:1424–1432. doi: 10.1261/rna.755908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suga K, Umakoshi H, Tomita H, Tanabe T, Shimanouchi T, Kuboi R. Liposomes destabilize tRNA during heat stress. Biotechnol. J. 2010;5:526–529. doi: 10.1002/biot.200900289. [DOI] [PubMed] [Google Scholar]
- 9.Umakoshi H, Suga K, Bui HT, Nishida M, Shimanouchi T, Kuboi R. Charged liposome affects the translation and folding steps of in vitro expression of green fluorescent protein. J. Biosci. Bioeng. 2009;108:450–454. doi: 10.1016/j.jbiosc.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Bui HT, Umakoshi H, Ngo KX, Nishida M, Shimanouchi T, Kuboi R. Liposome membrane itself can affect gene expression in the Escherichia coli cell-free translation system. Langmuir. 2008;24:10537–10542. doi: 10.1021/la801962j. [DOI] [PubMed] [Google Scholar]
- 11.Bui HT, Umakoshi H, Suga K, Tanabe T, Ngo KX, Shimanouchi T, Kuboi R. Cationic liposome can interfere mRNA translation in an E. coli cell-free translation system. Biochem. Eng. J. 2010;52:38–43. [Google Scholar]
- 12.Bui HT, Umakoshi H, Suga K, Tanabe T, Shimanouchi T, Kuboi R. Cationic liposome inhibits gene expression in an E.coli cell-free translation system. Membrane. 2009;34:146–151. [Google Scholar]
- 13.Bui HT, Umakoshi H, Suga K, Nishida M, Shimanouchi T, Kuboi R. Negatively charged liposome as a potent inhibitor of post-translation during in vitro synthesis of green fluorescent protein. Biochem. Eng. J. 2009;46:154–160. [Google Scholar]
- 14.Laso MRV, Zhu D, Sagliocco F, Brown AJP, Tuite MF, McCarthy JEG. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- 15.Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlassov A, Yarus M. Interaction of RNA with phospholipid membranes (Vzaimodeistvie RNK s fosfolipidnymi membranami.) Mol. Biol. 2002;36:496–502. [PubMed] [Google Scholar]
- 18.Janas T, Janas T, Yarus M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 2006;34:2128–2136. doi: 10.1093/nar/gkl220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas CF, Luisi PL. RNA selectively interacts with vesicles depending on their size. J. Phys. Chem. B. 2005;109:14544–14550. doi: 10.1021/jp0512210. [DOI] [PubMed] [Google Scholar]
- 20.Michanek A, Kristen N, Höök F, Nylander T, Sparr E. RNA and DNA interactions with zwitterionic and charged lipid membranes – a DSC and QCM-D study. Biochim. Biophys. Acta. 2010;1798:829–838. doi: 10.1016/j.bbamem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Marty R, N’Soukpoé-Kossi CN, Charbonneau DM, Kreplak L, Tajmir-Riahi H-A. Structural characterization of cationic lipid-tRNA complexes. Nucleic Acids Res. 2009;37:5197–5207. doi: 10.1093/nar/gkp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorovska-Taran V, Wick R, Walde P. A 1H nuclear magnetic resonance method for investigating the phospholipase D-catalyzed hydrolysis of phosphatidylcholine in liposomes. Analyt. Biochem. 1996;240:37–47. doi: 10.1006/abio.1996.0328. [DOI] [PubMed] [Google Scholar]
- 23.De Almeida RFM, Fedorov A, Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: Boundaries and composition of lipid rafts. Biophys. J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cevc G. Membrane electrostatics. Biochim. Biophys. Acta. 1990;1031:311–382. doi: 10.1016/0304-4157(90)90015-5. [DOI] [PubMed] [Google Scholar]
- 25.Savitzky A, Golay MJE. Smoothing and differentiation of data by simplified least squares procedures. Analyt. Chem. 1964;36:1627–1639. [Google Scholar]
- 26.Alex S, Dupuis P. FT-IR and Raman investigation of cadmium binding by DNA. Inorg. Chim. Acta. 1989;157:271–281. [Google Scholar]
- 27.Banyay M, Sarkar M, Gräslund A. A library of IR bands of nucleic acids in solution. Biophysical Chemistry. 2003;104:477–488. doi: 10.1016/s0301-4622(03)00035-8. [DOI] [PubMed] [Google Scholar]
- 28.Dovbeshko GI, Gridina NY, Kruglova EB, Pashchuk OP. FTIR spectroscopy studies of nucleic acid damage. Talanta. 2000;53:233–246. doi: 10.1016/s0039-9140(00)00462-8. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto M, Miyazaki Y, Umemoto A, Walde P, Kuboi R, Nakao K. Phosphatidylcholine vesicle-mediated decomposition of hydrogen peroxide. Langmuir. 2007;23:9416–9422. doi: 10.1021/la701277f. [DOI] [PubMed] [Google Scholar]
- 30.Edelman GM, Mcclure WO. Fluorescent probes and the conformation of proteins. Acc. Chem. Res. 1968;1:65–70. [Google Scholar]
- 31.Cócera M, López O, Estelrich J, Parra JL, De La Maza A. Influence of the temperature in the adsorption of sodium dodecyl sulfate on phosphatidylcholine liposomes. Chem. Phys. Lipids. 2003;124:15–22. doi: 10.1016/s0009-3084(03)00022-7. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg M, Gresalfi T, Riccio T, McLaughlin S. Adsorption of monovalent cations to bilayer membranes containing negative phospholipids. Biochemistry. 1979;18:5213–5223. doi: 10.1021/bi00590a028. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z, Rüegger H, Kissner R, Ishikawa T, Willeke M, Walde P. Vesicles as soft templates for the enzymatic polymerization of aniline. Langmuir. 2009;25:11390–11405. doi: 10.1021/la901510m. [DOI] [PubMed] [Google Scholar]
- 34.Kato A, Tsuji A, Yanagisawa M, Saeki D, Juni K, Morimoto Y, Yoshikawa K. Phase separation on a phospholipid membrane inducing a characteristic localization of DNA accompanied by its structural transition. J. Phys. Chem. Lett. 2010;1:3391–3395. [Google Scholar]
- 35.Giel-Pietraszuk M, Barciszewski J. A nature of conformational changes of yeast tRNAPhe: High hydrostatic pressure effects. Int. J. of Biol. Macromol. 2005;37:109–114. doi: 10.1016/j.ijbiomac.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Shimanouchi T, Tasaki M, Vu HT, Ishii H, Yoshimoto N, Umakoshi H, Kuboi R. Aβ/Cu-catalyzed oxidation of cholesterol in 1,2-dipalmitoyl phosphatidylcholine liposome membrane. J. Biosci. Bioeng. 2010;109:145–148. doi: 10.1016/j.jbiosc.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Guo X, Cui B, Li H, Gong Z, Guo R. Facilitation effect of oligonucleotide on vesicle formation from single-chained cationic surfactant—Dependences of oligonucleotide sequence and size and surfactant structure. J. Polym. Sci. Pol. Chem. 2009;47:434–449. [Google Scholar]
- 38.Gregoire CJ, Gautheret D, Loret EP. No tRNA3Lys Unwinding in a Complex with HIV NCp7. J. Biol. Chem. 1997;272:25143–25148. doi: 10.1074/jbc.272.40.25143. [DOI] [PubMed] [Google Scholar]
- 39.Carmona P, Rodriguez-Casado A, Molina M. Conformational structure and binding mode of glyceraldehydes-3-phosphate dehydrogenase to tRNA studied by Raman and CD spectroscopy. Biochim. Biophys. Acta. 1999;1432:222–233. doi: 10.1016/s0167-4838(99)00113-2. [DOI] [PubMed] [Google Scholar]
- 40.Madore E, Florentz C, Giege R, Lapointe J. Magunesium-dependent alternative foldings of active and inactive Escherichia coli tRNAGlu revealed by chemical probing. Nucleic Acids Res. 1999;27:3583–3588. doi: 10.1093/nar/27.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conn GL, Draper DE. RNA structure. Curr. Opinion Struct. Biol. 1998;8:278–285. doi: 10.1016/s0959-440x(98)80059-6. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimoto M, Kuboi R. Oxidative refolding of denatured/reduced lysozyme utilizing the chaperone-like function of liposomes and immobilized liposome chromatography. Biotechnol. Progr. 1999;15:480–487. doi: 10.1021/bp990050b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.