Abstract
In the course of analyzing 9 522 746 pyrosequencing reads from 23 stations in the Southwestern Pacific and equatorial Atlantic oceans, it came to our attention that misannotations of rRNA as proteins is now so widespread that false positive matching of rRNA pyrosequencing reads to the National Center for Biotechnology Information (NCBI) non-redundant protein database approaches 90%. One conserved portion of 23S rRNA was consistently misannotated often enough to prompt curators at Pfam to create a spurious protein family. Detailed examination of the annotation history of each seed sequence in the spurious Pfam protein family (PF10695, ‘Cw-hydrolase’) uncovered issues in the standard operating procedures and quality assurance programs of major sequencing centers, and other issues relating to the curation practices of those managing public databases such as GenBank and SwissProt. We offer recommendations for all these issues, and recommend as well that workers in the field of metatranscriptomics take extra care to avoid including false positive matches in their datasets.
INTRODUCTION
Ribosomes are the site of peptide bond formation in all living cells, from bacteria to humans (1). They are composed in part of highly conserved RNA sequences (2) usually coded on DNA in operons of three subunits (16S, 23S and 5S) in Bacteria and Archaea (3,4) and in tandem repeats of longer operons that ultimately mature to four subunits (18S, 25/28S, 5.8S and 5S) in Eukaryotes (5,6). The complete primary nucleotide sequences of representative rRNA subunits in the seven duplicated rRNA operons of Escherichia coli were published between 1967 and 1978 (7–10). The rRNA nucleotide sequences for Saccharomyces cerevisiae, which occur in ∼140 tandem repeats, were published between 1972 and 1981 (11–14).
While artificial overexpression of a pentapeptide sequence adjacent to a Shine–Dalgarno motif within E. coli 23S rRNA was found to impart drug resistance to erythromycin (15), rRNA operons in Bacteria and Archaea are not known to contain naturally expressed protein coding regions that also code for rRNA. Also, while antisense transcription was recently reported for Bacterial and Archaeal proteins, that study did not report antisense transcription from Bacteria and Archaea rRNA (16). To be sure, insertion elements can be found in rRNA operons of Bacteria and Archaea, but not sequences that code for rRNA and protein at the same time. Therefore, annotations of Bacteria and Archaea proteins embedded in rRNA operons and overlapping with rRNA coding regions within those operons have been rightly presumed to be misannotations (17) and should continue to be, until hard evidence to the contrary emerges. While these misannotations continue to exist, they have the potential to generate false positive matches of translated environmental rRNA sequences to proteins. To our knowledge, the potential for false positives in metatranscriptomic studies due to misannotations of rRNA operons has not been reported prior to this study.
Unlike Bacterial and Archaeal rRNA operons, the yeast rRNA operon has indeed been shown to contain an embedded protein coding domain sequence (CDS) called Tar1p that overlaps the 5′–end of the DNA sequence coding for the 25S rRNA subunit (18). Another substantive difference between rRNA operons in Bacteria, Archaea and Eukaryotes is that Eukaryotes are also known to contain rRNA sequences that have moved to other parts of the genome including expressed coding regions, putatively with regulatory functions (19). Other Eukaryotic proteins of unknown function having rRNA homology have been reported (20–22). All these Eukaryotic proteins with real rRNA homology are another source of potential false positives in metatranscriptomic studies, since translations of conserved rRNA sequences from other Eukaryotes will match to these protein sequences.
We observed both kinds of false positives in a metatranscriptome of 9 522 746 pyrosequencing reads from 23 stations in the Southwestern Pacific and equatorial Atlantic oceans. When we discovered that the misannotations of Bacterial and Archaeal rRNA sequences were so widespread that a spurious Pfam (23) protein family had been created, we paused in our ecological analysis to assess the extent of these misannotations and to make recommendations on how to address them.
MATERIALS AND METHODS
Analysis of known Candidatus Pelagibacter sp. HTCC7211 expressed intergenic regions
Fasta sequences of the 11 expressed intergenic regions (eIGRs) of Candidatus Pelagibacter sp. HTCC7211 (24) were compared to all RNA reads [CAMERA (25) project names CAM_PROJ_PacificOcean and CAM_PROJ_AmazonRiverPlume] using blastn and a bit score cut-off of 40. The blast results were parsed and loaded into a MySQL database containing sample metadata. Blast results and metadata were joined into Structured Query Language (SQL) logical views for analysis of eIGRs by sample. The data in the SQL logical views were summarized and visualized using Microsoft Access and Excel.
Analysis of gene contexts for PF10695 seed sequences
With the hypothesis that the seed sequences were all embedded in an rRNA operon, overlapping with the 3′-end of the 23S rRNA sequence on the opposite strand from the 23S rRNA sequence, we attempted to extract the full rRNA operon within which, we hypothesized each Pfam seed sequence to exist. Knowing the 3′-end of the 23S rRNA operon to be no >500 bp from the 3′-end of the entire rRNA operon in most Bacteria and Archaea, we chose 500 bp upstream (recall that we hypothesize the seed sequence to be on the opposite strand from the RNA sequence) of the seed sequence as the likely 3′-end of the rRNA operon in which we hypothesized the seed sequence to exist. Knowing that the 5′-end of the rRNA operon is usually no >5000 bp upstream from the 3′-end of the 23S rRNA sequence, we chose 5000 bp downstream of the seed sequence as the likely 5′-start of the rRNA operon in which we hypothesized the seed sequence to exist. For three seed sequences (GI 145845866, 47093546, 121729912), we could not extract the entire region of interest because the contig on which the seed sequence was found, ended prematurely. For two seed sequences (GI 149912432 and 90419149), 6000 and 5500 bp downstream of the seed sequence was required to reach the 5′-end of the rRNA operon.
The GenBank protein identifiers of the 10 seed sequences for PF10695 (GI 81390223, 122460149, 30316295, 122409581, 122668550, 149912438, 150010506, 121729919, 154505437, 154487654) were obtained from the Web site for the NCBI Conserved Domain (pfam10695) at http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=151191. The start and end coordinates for the seed sequences were found in two different ways, depending on the sequence. For SwissProt seed sequences, the Exon Information area at the bottom of the ExonView screen gave the strand (plus or minus), and the start and end coordinates for the seed sequence. For GenBank proteins, the ‘/coded_by’ entry in the CDS feature for the record gave the same information. The nucleotide accessions and coordinates for the gene contexts surrounding the PF10695 seed sequences can be found in an Excel file in Supplementary Data.
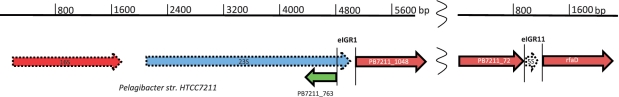
Using the nucleotide accession information for the seed sequences, we then calculated expected 5′- and 3′-ends of the rRNA operon containing the seed sequences, as described above, and extracted the context from GenBank in GenBank format. We used Artemis (26) to reannotate the rRNA sequences using either the RNAmmer 1.2 Server (http://www.cbs.dtu.dk/services/RNAmmer/) for complete rRNA operons, or blastn against GenBank's nucleotide database for incomplete rRNA operons. The reannotated Artemis screens were then exported to a graphics file and traced to scale in PowerPoint. The result is shown in Figure 1.
Figure 1.
Gene contexts of Candidatus Pelagibacter sp. HTCC7211 eIGR1 and eIGR11. The dotted lines indicate that the rRNA were not annotated originally, but were found in this study using the RNAmmer web site. The broken lines on the scale bar show that the 5S rRNA gene in Candidatus Pelagibacter sp. HTCC7211 is found in another part of the genome from the adjacent 16S and 23S rRNA genes.
Visualization of historical rRNA annotations
We used the Web Site for the GOLD Database, http://www.genomesonline.org/cgi-bin/GOLD/bin/gold.cgi, to obtain a list of complete genomes sorted in sequential order of creation. Starting with GOLD identifier ‘Gc00001’, we navigated to the summary page for the genome in the Integrated Microbial Genomes (IMG) database (27). We added all rRNA genes to the Gene Cart and used the ‘Show Neighborhood’ feature to visualize the gene contexts of all rRNA genes in the genome.
Determination of original misannotation of ‘cell wall hydrolase’ in PF10695
We searched for PF10695 on the UniProtKB/Swiss-Prot web site (http://www.uniprot.org/), and sorted all accessions for PF10695 by ‘Date of creation’. The first record displayed (Q8CME1, created 2003-03-01) had a protein name of ‘Cell wall-associated hydrolase’. The ‘gene names’ column for this accession, listed nine gene loci (VV1_0473, VV1_0917, VV1_0925, VV1_0970, VV1_1072, VV1_1190, VV1_1418, VV1_1502, VV2_1450), all for the organism Vibrio vulnificus. Using the link for accession Q8CME1, we navigated to each GenBank protein accession for these loci (AAO08321.1, AAO08995.1, AAO09418.1, AAO09424.1, AAO09463.1, AAO09551.1, AAO09653.1, AAO09859.1, AAO09933.1). GenBank reported that all records had been removed, but the obsolete versions were accessible. Each obsolete accession had a note saying ‘similar to invasion-associated proteins; COG0791’. In order to determine when the records were deleted, we clicked on the ‘Revision History’ radio button under ‘Display Settings’ for the protein record display. It showed that the records were removed on 4 January 2006, having been first seen on 22 December 2002.
We determined that the similarity to COG0791 reported in the obsolete protein records was an error. To do this, we searched the Clusters of Orthologous Groups (COG) database for the amino acid sequences using NCBI's Conserved Domain web site (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and found no match to any COG using the default cut-off of E = 0.01. Loosening the cut-off to E = 100, we found poor matches (E > 0.84) to four COGs, none of which were COG0791.
To confirm that the genome context for all the obsolete proteins were within the rRNA operons of V. vulnificus, we obtained their nucleotide accessions and coordinates from the ‘coded_by’ feature of their CDS. We downloaded the FASTA nucleotide records and verified that the nucleotide sequences for all loci were identical. We then performed a blastn search of each nucleotide sequence against the V. vulnificus CMCP6 genome using the NCBI Web site, and they all returned one match to rRNA-23S ribosomal RNA and no other genome feature. This confirmed that all nine obsolete loci were annotated as embedded, overlapping Open Reading Frames (ORFs) with an rRNA 23S sequence before they were deleted from GenBank.
Analysis of spurious ORFs in E. coli rRNA sequences
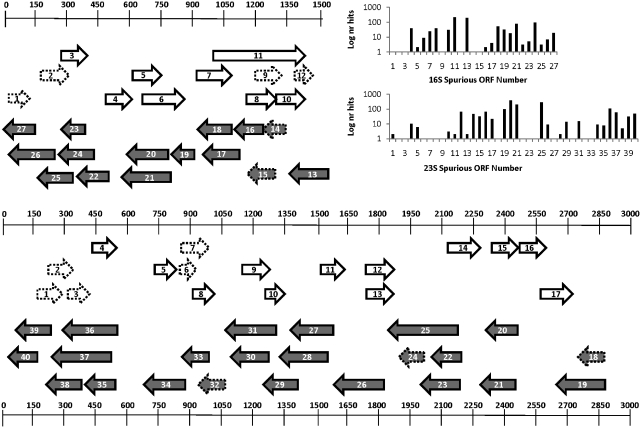
The EMBOSS getorf utility was used to generate spurious ORFs in the rrsH and rrlH genes of E. coli, using a permissive parameter of 100 nt from any methionine codon to any stop codon. The translated protein sequences from the spurious ORFs were compared to a copy of the NCBI non-redundant (nr) protein database (January 2011) with a cut-off of E = 0.001. Custom Perl scripts for parsing the blast output and for retrieving GenBank data were used to identify and fetch the nucleotide sequences for the protein matches to the spurious ORFs. These nucleotide sequences were compared to a copy of the SILVA (28) rRNA database (June 2010) using blastn. The blastn output was parsed with a custom Perl script. If the nucleotide sequence for a protein matched a SILVA rRNA sequence at 90% nucleotide identity over 90% of its length, it was considered a misannotated protein. The misannotated proteins were mapped back to their corresponding spurious ORF from E. coli rRNA in an Excel spreadsheet, which is included in Supplementary Data. The results were visualized as shown in Figure 4.
Figure 4.
NCBI nr Hits to Spurious ORFs in E. coli rRNA. Top left, spurious ORFs in E. coli 16S rRNA. Bottom, spurious ORFs in E. coli 23S rRNA. Scales are in base pairs. White arrows, three reading frames on positive strand, gray arrows, three reading frames on negative strand. Inset at upper right shows the log10 of the number of NCBI nr protein hits to the translated amino acids for each spurious ORF in both of the E. coli rRNA sequences. The NCBI nr protein hit was only counted if its nucleotide sequence matched a known rRNA sequence in the SILVA database. The detail for each hit is provided in Supplementary Data.
Generation and analysis of pseudoreads
The nucleotide sequences of the rRNA subunits from E. coli str. K-12, substr. MG1655, Sulfolobus acidocaldarius DNS 639, and S. cerevisiae S288c were retrieved from GenBank in fasta format. A custom Perl script then removed the fasta headers from this file and concatenated all of the sequence data for all of the rRNAs into one long string. A second Perl script generated 10 000 pseudoreads from this long string by choosing a starting point at random, then pulling a randomly-chosen number of base pairs from a file containing the read lengths of an actual pyrosequencing run. The pseudoreads thus generated, were written to a fasta file that was queried against the 28 April 2010 copy of NCBI's nr database, using blastx (version 2.2.21) and a cutoff of E < 0.001, with 25 summaries and alignments retained. The blastx text output was read into MEGAN (29) version 3.9. The phylogeny and function of the proteins matching the pseudoreads were visualized in MEGAN using default parameters.
RESULTS AND DISCUSSION
Search for eIGRs
As described in the ‘Materials and Methods’ section, we searched for known eIGRs from Candidatus Pelagibacter sp. HTCC7211 in our metatranscriptome, in order to compare our results with the study that discovered eIGRs (24). The most commonly occurring Candidatus Pelagibacter sp. HTCC7211 eIGR in our dataset was eIGR1 (Table 1). A blastn comparison of the genome nucleotide coordinates for eIGR1 (GI 254455249:44547; 44776, 230 bp long) to the GenBank nucleotide database revealed that Positions 77–230 of eIGR1 matched with 99% identity to the single 23S rRNA gene of Candidatus Pelagibacter ubique HTCC1062. Positions 118–187 of eIGR1 matched with 83% identity to all seven 23S rRNA genes of E. coli K-12, confirming that the Candidatus Pelagibacter ubique HTCC1062 23S annotation was reasonably accurate and that at least half of eIGR1 contained unannotated 23S rRNA sequence. When we examined the larger context of the eIGR1 region of the Candidatus Pelagibacter sp. HTCC7211 in the NCBI genome browser, we saw that it was flanked by a ‘cell wall hydrolase’ (Pfam PF10695) on one side. The ‘cell wall hydrolase’ was in turn flanked by a long stretch of unannotated sequence. We extracted the nucleotides of the entire unannotated region of Candidatus Pelagibacter sp. HTCC7211 near eIGR1 (NZ_DS995298.1:43539-50898, 7359 bp) and submitted the region to the RNAmmer 1.2 WebServer (30). The predicted 5′-end of the 23S extended well into the eIGR1 region (Figure 1). This meant that gene locus PB7211_763 (‘cell wall hydrolase’) of Candidatus Pelagibacter sp. HTCC7211 overlapped a 23S sequence on the antisense strand, something that has always been considered an annotation error in Bacteria and Archaea. However, the protein sequence of PB7211_763 returned a strong (E = 1.24e-44) match to pfam10695, Cw-hydrolase, using the search function of the NCBI Conserved Domain web site. We found this result surprising and worth investigating further.
Table 1.
Rank order listing of Candidatus Pelagibacter sp. HTCC7211 eIGRs found in marine metatranscriptomes
| eIGR | Count | Comment |
|---|---|---|
| 1 | 22 103 | Unannotated 23S rRNA (this study) |
| 6 | 3257 | Unannotated RNase P [Shi et al. (24)] |
| 9 | 1959 | Unannotated putative tmRNA [Shi et al. (24)] |
| 11 | 243 | Unannotated 5S rRNA (this study) |
| 2 | 73 | Glycine-activated riboswitch |
| 7 | 61 | Glycine-activated riboswitch |
| 10 | 36 | Unknown |
| 5 | 10 | Unknown |
| 8 | 10 | Unknown |
| 3 | 4 | Unknown |
| 4 | 1 | Unknown |
The numbering of the eIGRs is taken from Shi et al. (24). The comment column describes the content of the intergenic region, if known.
Analysis of seed sequences for PF10695
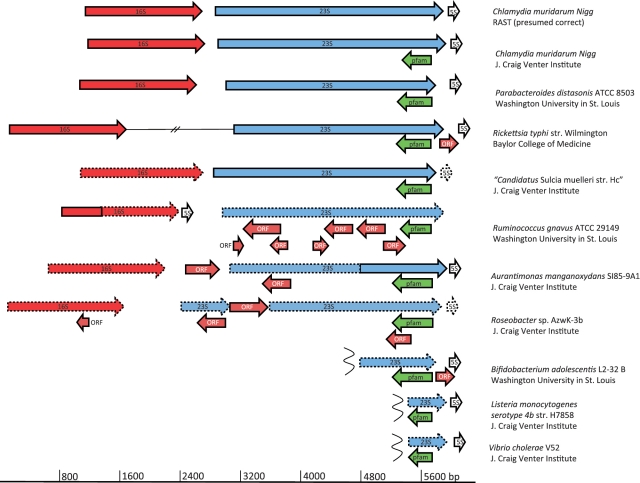
In order to determine how well PF10695 was characterized, we obtained all 10 of its seed sequences from the NCBI Conserved Domain web site. We then examined the gene contexts for each seed sequence as described in the ‘Materials and Methods’ section and found that all 10 seed sequences were in fact embedded and overlapping ORFs within rRNA operons (Figure 2), just as PB7211_763 of Candidatus Pelagibacter sp. HTCC7211 was. Clearly, PF10695 had been created in error from misannotations. We asked how this had come about.
Figure 2.
Gene context for seed sequences of Pfam 10695. The seed sequences are shown in green. The 16S, 23S and 5S sequences are shown in red, blue and white with dotted outlines for those sequences that were not annotated by the sequencing center shown, but were found in this study using the RNAmmer 1.2 Web server. Other embedded ORFs within the rRNA operon are shown in brown.
The ‘Materials and Methods’ section describes how we determined the first misannotation of an embedded and overlapping ORF annotated ‘cell wall hydrolase’ within an rRNA operon. The first misannotation was made in nine copies of rRNA operons in V. vulnificus CMCP6. The misannotation was eventually corrected by deleting all nine protein sequences from GenBank; however they were active for ∼3 years in GenBank before being deleted and are still active in SwissProt. The ‘Materials and Methods’ section describes exactly how we found the original misannotation of ‘cell wall hydrolase’ using the SwissProt (UniProtKB) database. During the course of that investigation, we noted that the only ‘reviewed’ record for PF10695 was locus TC_0114 of Chlamydia muridarum, one of the seed sequences for PF10695. Since SwissProt curators had reviewed this locus, we examined Revision Histories in GenBank and SwissProt (UniProtKB) in detail to see what basis they might have found for this being a valid protein.
It appears that at some point, a SwissProt curator might have thought that there was some experimental evidence for TC_0114, even though none of the 41 versions of it in SwissProt (accession Q9PLI5) contain such a notation. The indication of experimental evidence comes from GenBank's record of the Swiss-Prot protein sequence for TC_0114 (protein accession Q9PLI5, GI 30316295). It currently shows a feature of ‘/experiment = “experimental evidence, no additional details recorded”’ added 13 April 2006. However, GenBank's protein accession for TC_0114 (protein accession AAF38993, GI 29251569) has a note saying ‘identified by Glimmer2; putative’ and makes no mention of experimental evidence. We could find no literature supporting experimental evidence for TC_0114 and conclude that it in fact was a spurious prediction of Glimmer2 and was incorrectly reviewed by SwissProt.
A likely origin of protein family, PF10695 was now discernable. From late 2002 to early 2006, the spurious ORFs in the unannotated 23S rRNA operon of V. vulnificus were stored in GenBank with a product of ‘cell wall hydrolase’. They were very similar (74% amino acid identity) to the incorrectly reviewed SwissProt entry for TC_0114 of C. muridarum. Annotators or pipelines using Glimmer2 for gene finding would have found a SwissProt ‘reviewed’ protein and a protein annotated ‘cell wall hydrolase’ embedded and overlapping 23S rRNA sequences. On the basis of this evidence, some annotators or pipelines evidently called their spurious ORF ‘cell wall hydrolase’, while others called them ‘conserved hypothetical’. Others saw weak or erroneous matches to other proteins and annotated them from those matches. Thus, a variety of annotations arose for spurious embedded, overlapping proteins within rRNA operons, the most common of which was ‘cell wall hydrolase’. When these annotations accumulated to sufficient levels, it apparently prompted Pfam to create the protein family PF10695, ‘Cw-hydrolase’. As of this study, PF10695 contains 1780 NCBI proteins and 1653 metagenomic fragments.
The staff at Pfam reviewed this article, concurs that PF10695 is spurious and has marked it for deletion in release 26.0 (A. Bateman, personal communication). They informed us that four other families were deleted in the past for the same reason (PF07612, PF07616, PF07630 and PF07633) and another (PF05330) was deleted because it contained spurious human genes based on a repeat.
Additional misannotations of rRNA in GenBank
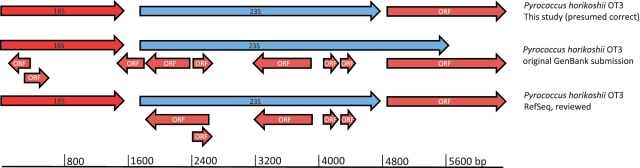
The additional misannotations of embedded, overlapping proteins within rRNA operons shown in brown in Figure 2 indicated that the misannotation of rRNA operons was not confined to pfam10695. Therefore, we inquired into their origin as well. To do this, we obtained a date-sorted list of all microbial genomes in the Genomes OnLine Database (GOLD) (31), and visualized the gene contexts of their rRNA operons, starting with Haemophilus influenza, as described in the ‘Materials and Methods’ section. The first annotated rRNA operon with protein sequences overlapping with and embedded in an rRNA operon appeared in the 1998 genome annotation for Pyrococcus horikoshii OT3 (32), the 14th complete genome sequenced. The authors commented in their original submission to GenBank (BA000001.2) that, ‘All the sequence with length 100 codons or more between ATG and GTG and stop codon are defined as CDS’. They also said that ORFs as small as 50–99 codons long were also considered probable protein-coding regions if they showed some similarity to proteins in public databases. Summarizing their approach, the authors explained, ‘It should be noted that the ORFs mentioned above merely represent the protein-coding potentiality under the defined assumptions’. Nothing was said about eliminating overlapping ORFs; apparently these were retained either deliberately or inadvertently.
There are two potential reasons why the authors might have taken a CDS-finding approach so prone to false positives. First, their study organism was an Archaeon, the least studied domain of life, and they might have preferred to call false positives rather than to miss a novel Archaeon protein. Second, they may have noted reports (33–35) that genes arising from horizontal gene transfer are sometimes missed by CDS-finding algorithms that rely on codon frequencies (36) or Markov chain models (37) of ‘typical’ genes in the genome. Whatever their reasoning, these authors provided a genome with multiple protein coding domains overlapping rRNA genes in rRNA operons (Figure 3). At the same time, these authors erroneously called the end of the 23S rRNA subunit, an error that was discovered and corrected by RefSeq curators. Although they corrected the length of the 23S rRNA subunit, the RefSeq curation staff did not remove all of the overlapping ORFs inside the rRNA operon. Interestingly, they lengthened one of the overlapping ORFs so that it overlapped with another one, creating a triple overlap (Figure 3). As a result, neither the nr database nor the RefSeq database at NCBI contain what we presume to be the correct annotation (Figure 3).
Figure 3.
Comparison of annotations of P. horikoshii OT3. This figure compares the annotations of the 16S–23S rRNA operon of P. horikoshii OT3, the 14th genome annotated. The 5S rRNA sequence is located elsewhere in the genome. Coloring and abbreviations are the same as Figure 1.
We were able to demonstrate that, at least 367 additional genomes in NCBI's nr database have misannotated proteins (Figure 4 and Supplementary Data). To demonstrate this, we intentionally generated a large number of spurious ORFs in E. coli 16S and 23S rRNA sequences (Figure 4, top left and bottom) and counted the close matching nr proteins whose nucleotide sequences also had a very strong match to rRNA sequences in the SILVA database (Figure 4, upper right). The majority of the spurious ORFs had at least one nr protein hit whose nucleotide sequence also matched a SILVA rRNA sequence at >90% nucleotide identity over 90% of its length. Some spurious ORFs had well over 100 such matches to misannotated proteins. When the accessions and associated organism names for all misannotated proteins were combined, it emerged that genome sequences for 367 organisms in nr contained misannotated proteins (Supplementary Data).
Misannotations of rRNA in the SEED
Having found this instance of overlapping ORFs embedded within rRNA operons in only the 14th genome sequenced, we continued looking for them in subsequent genomes. The 27th genome sequenced, Chlamydophila pneumonia AR39 (GOLD identifier Gc00027, completed 15 March 2000), contained an annotation for a putative hypothetical protein at gene locus CP0987, overlapping a 23S rRNA sequence, thus indicating that it coded for protein and rRNA at the same time, exactly as was the case for PF10695 seed sequences. The original GenPept accession for CP0987 (AAF38766.1) has been removed as obsolete but the accession created for it in RefSeq (NP_445524.1) is still active. More significantly, the Gene Detail for CP0987 in IMG (Object 637042263) showed this SEED (38) identifier for CP0987: ‘Retron-type reverse transcriptase, fig|115711.7.peg.950’. As we said in the ‘Introduction’ section, while inserts of protein sequences are known in Bacteria and Archaea, dual coding of protein and rRNA at the same time is not known in Bacteria and Archaea. This indicated that the SEED database also had errors in it.
In order to find misannotations in The SEED Viewer, we did an identifier search for fig|115711.7.peg.950 and asked for 64 comparable regions on the Annotation Overview page. The system returned 30 gene contexts, all showing a gene of similar length to CP0987, all completely overlapping a 23S rRNA gene in the exact same manner as the seed sequences for PF10695. All 30 genes were annotated ‘Retron-type reverse transcriptase’. The Web page said that these features were part of a subsystem called ‘Group II intron-associated genes’, however the subsystem had not been classified for the organism. We clicked on the link to ‘Group II intron-associated genes’, but there was no diagram of a subsystem and no literature listed in the ‘Functional Roles’ tab. We visually inspected the 30 gene contexts and found additional embedded and overlapping ORFs in the 23S rRNA sequences displayed. Of the 30 contexts, 12 also showed a conserved hypothetical protein that also completely overlapped the 23S rRNA, just down-stream of the misannotated ‘Retron-type reverse transcriptase’. Three of the 30 contexts also showed a different conserved hypothetical protein that also completely overlapped the 23S rRNA sequence, and three more contexts showed from one to three very small (<53 amino acids) conserved hypothetical proteins completely overlapping a 23S rRNA sequence. By ‘completely overlapping’ we mean ‘coding for rRNA and protein at the same time’.
This confirmed that there are errors in the SEED, despite the care taken by the current Rapid Annotations using Subsystems Technology (RAST) pipeline not to add any more errors. The RAST pipeline begins by calling tRNA and rRNA genes and ‘the server will not consider retaining any protein-encoding genes that are embedded in rRNAs. These gene calls are almost certainly artefacts of the period in which groups were learning how to develop proper annotations, and RAST attempts to avoid propagating these errors’. Still, we assert that existing misannotations should be found and corrected.
Eukaryotic pseudo rRNA genes and antisense transcripts to rRNA genes
Having fully addressed the overrepresented eIGRs in our dataset, we now looked for overrepresented protein sequences. We found them in some Eukaryotic sequences (Table 2). Again, the reason for the overrepresentation was rooted in sequence similarity between the protein sequences and known rRNA sequences. However, we discovered that homology between ‘senescence associated proteins’ and rRNA sequences have in fact been reported in studies of Eukaryotes (22), along with the other examples of protein–rRNA homology noted in the ‘Introduction’ section (18–21). We could not determine whether the protein in Chlamydomonas with similarity to 18S rRNA was a misannotation or a confirmed protein with homology to 18S rRNA, due to difficulties in annotation of Eukaryotic rRNA sequences have been discussed in the literature (30). However, it was clear that at least some real Eukaryotic proteins with rRNA homology do in fact exist, and therefore have the potential to generate false positives in metatranscriptomic studies.
Table 2.
Amino acid and nucleotide comparison of highly represented putative mRNAs
| Putative mRNAs | Organism | KEGG annotation | Base pairs/ AA | Prog | ID (%) | Len (%) | NCBI nt/nr best specific hit |
|---|---|---|---|---|---|---|---|
| 42 003 | Phaeodactylum | Hypothetical protein (pti:PHATRDRAFT_37403) | 324 | blastn | 99 | 96 | Uncultured organism 28S rRNA |
| 107 | blastp | 90 | 84 | Senescence-associated protein | |||
| 19 678 | Ostreococcus | Predicted protein (olu:OSTLU_9775) | 264 | blastn | 99 | 99 | Uncultured organism 28S rRNA |
| 88 | blastp | 90 | 102 | Senescence-associated protein | |||
| 13 880 | Chlamydomonas | Hypothetical protein (cre:CHLREDRAFT_155068) | 264 | blastn | 100 | 88 | Uncultured organism 28S rRNA |
| 87 | blastp | 26 | 30 | Unknown protein (Glycine max) |
The blastn and blastp matches are shown for the three most abundant putative mRNA transcripts, which accounted together for (75 561/904 042)= 8.36% of total putative mRNA. AA, amino acids; Prog, program; ID, identity of best match; Len, length of alignment to best match.
MEGAN analysis of false positive protein matches to pseudoreads of rRNA
In order to gauge the full scope of potential false positives due to misannotations of rRNA operons in Bacteria and Archaea and to Eukaryotic proteins with rRNA homology, we used MEGAN software to perform phylogenetic and functional analysis of spurious protein matches to ‘pseudoreads’ of rRNA (Figure 5). Nearly 90% of the pseudoreads had hits to which phylogeny could be assigned. Despite the fact that the pseudoreads of rRNA came from only three model organisms (E. coli, S. acidocaldarius and S. cerevisae), the spurious phylogenetic analysis included Bacteroidetes, Firmicutes, Alpha proteobacteria and a striking diversity of Eukaryotic taxa, including pine tree and chicken. Despite the fact that all the pseudoreads were rRNA sequences with no protein function, functional analysis in MEGAN was possible on (1807/10 000) = 18% of the reads (Table 3). Not surprisingly, the most frequent functional category was ‘cell wall hydrolase’.
Figure 5.
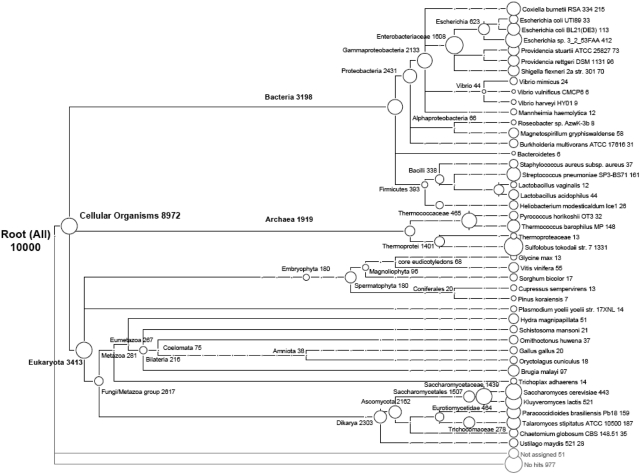
MEGAN phylogenetic analysis of pseudoreads. The phylogeny of proteins in nr matching 10 000 translated pseudoreads of rRNA taken from E. coli, Sulfolobus, and S. cerevisiae is shown, with the number of reads classified to the taxon shown.
Table 3.
MEGAN functional analysis of pseudoreads
| GO term | Number of reads |
|---|---|
| Hydrolase activity | 563 |
| Mitochondrion | 317 |
| Regulation of cellular respiration | 250 |
| Phosphatidylcholine phospholipase C activity | 154 |
| Integral to membrane | 139 |
| Metabolic process | 126 |
| Chloroplast | 117 |
| Catalytic activity | 108 |
| Endonuclease activity | 31 |
| Transferase activity | 2 |
| Total | 1807 |
The functions of proteins in nr matching 10 000 translated pseudoreads of rRNA taken from E. coli, S. acidocaldarius and S. cerevisiae are shown, with the number of reads classified to the Gene Ontology (GO) term shown.
Analysis of standard operating procedures of major sequencing centers
The Standard Operating Procedures (SOPs) of the four major sequencing centers participating in the Human Microbiome Project (available at http://hmpdacc.org/tools_protocols/tools_protocols.php; Supplementary Data) show a complete reliance on Rfam and RNammer to find rRNA genes. None have adapted Niels Larsen's ‘search_for_rnas’ (available from the author) to use blastn searches of known rRNAs. In a recent comparison (39), it was found that two pipelines that have adapated ‘search_for_rnas,’ the Integrated Microbial Genomes Expert Review (IMG_ER) (40) pipeline of the Joint Genome Institute (JGI; Supplementary Data) and the RAST pipeline (17), correctly found all three rRNA subunits of Halorhabdus utahensis. The J Craig Venter Institute (JCVI) pipeline, which relies on RNammer (Supplementary Data), found only one-third of the 16S sequence and no 23S sequence at all. Only the 5S was found correctly by JCVI. Also, RNammer does not even attempt to find Eukaryotic 5.8S and performs poorly on Archaeal 5S and Eukaryotic 18S sequences (30). Additionally, we found in this study that while it works well for complete sequences, it does not work for incomplete sequences, such as are found at the ends of contigs in draft sequences. True, RNammer is rapid and consistent, as the authors claim, but it is not accurate enough in our opinion to be used completely on its own for all draft and completed genome sequences. Therefore, it is our opinion that all of the SOPs of sequencing centers associated with the Human Microbiome Project would be improved by adapting ‘search_for_rnas’, as has been done with the IMG-ER and RAST pipelines, in addition to using RNammer for initial rRNA finding.
An additional improvement to all SOPs associated with the Human Microbiome Project is an explicit manual or automated check to see if in fact any rRNA operons were found at all prior to doing any gene finding and elimination of overlaps. The check should also consider whether the size and organization of the putative rRNA operons depart from the known size and organization for the sequenced organism's domain of life. An automated check could be fashioned by querying the nucleotide sequences of all putative proteins against a known database of rRNA with blastn. A match indicates that the putative protein is likely to be a spurious ORF within an rRNA operon.
Gene finding should only proceed after it has been verified that all rRNAs have in fact been found and annotated with accurate starts and ends. If gene finding is done prior to rRNA finding, or after rRNA finding has failed or was done inaccurately, it is almost certain that spurious ORFs will be found with no apparent overlap to any other feature. It does not appear that any of the sequencing centers for the Human Microbiome Project have a quality assurance checkpoint of making sure that rRNA operons have in fact been found and properly annotated prior to proceeding with gene finding. In addition, one center (JCVI) does not mention eliminating protein coding domains overlapping rRNA operons at all in their SOP. All sequencing centers should assure that rRNA operons have been found and properly annotated before finding genes and eliminating overlaps, which is to say putative proteins that also code for rRNA.
Getting GenBank corrections to propagate
The case study of PF10695 demonstrates that errors often propagate, but corrections often do not. The corrections least likely to propagate are deletions of erroneous records. Often, the only record of deletion occurs in text comments; there is no file of deletions for easy programmatic handling. It is cumbersome to write programs that scan the entire history of GenBank, parsing text comments to look for deletions and corresponding replacements. GenBank itself apparently does not write such programs. As we showed above, the original GenPept accession for spurious protein CP0987 (AAF38766.1) has been removed as obsolete, but the accession created for it in RefSeq (NP_445524.1) is still active.
Importance of accurate rRNA operon annotation
Accurate rRNA operon annotation is likely to improve drug discovery and understanding of cellular regulatory processes. There is ample literature describing drug effects on ribosomal metabolism (41–46). Use of this literature certainly requires that all ribosomal subunits be annotated, and effective use requires that the annotations be accurate. In addition, ribosome biosynthesis has long been known to be a major cellular activity, especially in growing cells (47,48). The majority of RNA recovered in metatranscriptomic studies is rRNA, not mRNA. Accurate annotation relating to regulation of ribosome biosynthesis, including promoter locations and binding sites, and further annotation of confirmed antisense proteins such as those found in yeast, is important as well. Future biochemical studies may indeed find that some of the antisense proteins overlapping rRNA sequence in Bacteria and Archaea are in fact expressed and translated, as they are in Eukaryotes. However, this mere potential is no reason to reverse the longstanding, prudent practice of eliminating putative ORFs that overlap Bacterial and Archaeal rRNA sequences and have no confirmed wet or dry lab evidence for their existence, other than their length being over 50–100 codons.
CONCLUSION
Widespread misannotation of spurious ORFs in Bacterial and Archaeal rRNA operons and the existence of Eukaryotic proteins with homology to rRNA combine to create the potential for a false positive rate of 90% in metatranscriptomic studies. Standard Operating Procedures for major sequencing centers should be amended to include a quality assurance checkpoint verifying that rRNA operons of appropriate length have been found before gene finding and elimination of overlapping ORFs proceeds and the JCVI SOP should include elimination of spurious ORFs within Bacterial and Archaeal rRNA operons. Pipelines that do not make use of the ‘search_for_rnas’ program would be improved by adapting it, especially for draft genomes, instead of relying completely on RNammer for all rRNA finding.
The spurious protein family PF10695, whose seed sequences are all misannotations, will be deleted from Pfam in release 26.0. All CDS annotations referring to this protein family (1780 in NCBI alone) need to be deleted from public databases. In addition, all Bacterial and Archaeal proteins whose nucleotide sequences have a significant match to known rRNA sequences need to be deleted. NCBI might consider providing monthly files of deleted proteins to assist in propagating these corrections and indeed all corrections involving deletion of spurious putative protein sequences.
Until the public databases are purged of spurious Bacterial and Archaeal proteins within rRNA operons, metatranscriptomic researchers need to be cognizant of the strong potential for false positives stemming from a failure to completely remove all rRNA from their analysis pipelines prior to translating the putative rRNA and querying a ‘trusted’ protein database. The ‘trusted’ protein database can be queried with pseudoreads of rRNA in order to reveal the thousands of misannotations it will undoubtedly have until rRNA annotation and curation procedures are improved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation (grants EF0424599, OCE0425363); Gordon and Betty Moore Foundation (MEGAMER facility grant to University of California at Santa Cruz, Investigator grant to J.Z.). Funding for open access charge: Gordon and Betty Moore Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This article is dedicated to the memory of Dr. Benjamin R. Munson. We thank Torsten Wendav for programming, and Irina Ilikchyan for useful discussions; Stephan Schuster, Lynn Tomsho and Ji Qi for pyrosequencing.
REFERENCES
- 1.Roberts R, editor. Microsomal Particles and Protein Synthesis. New York: Pergamon Press; 1958. [Google Scholar]
- 2.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn JJ, Studier FW. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc. Natl Acad. Sci. USA. 1973;70:3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsburg D, Steitz JA. The 30S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5S sequences. J. Biol. Chem. 1975;250:5647–5654. [PubMed] [Google Scholar]
- 5.Smitt WW, Vlak JM, Schiphof R, Rozijn TH. Precursors of ribosomal RNA in yeast nucleus. Biosynthesis and relation to cytoplasmic ribosomal RNA. Exp. Cell Res. 1972;71:33–40. doi: 10.1016/0014-4827(72)90259-5. [DOI] [PubMed] [Google Scholar]
- 6.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 7.Brosius J, Dull TJ, Noller HF. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brownlee GG, Sanger F, Barrell BG. Nucleotide sequence of 5S-ribosomal RNA from Escherichia coli. Nature. 1967;215:735–736. doi: 10.1038/215735a0. [DOI] [PubMed] [Google Scholar]
- 10.Carbon P, Ehresmann C, Ehresmann B, Ebel JP. The sequence of Escherichia coli ribosomal 16 S RNA determined by new rapid gel methods. FEBS Lett. 1978;94:152–156. doi: 10.1016/0014-5793(78)80926-0. [DOI] [PubMed] [Google Scholar]
- 11.Georgiev OI, Nikolaev N, Hadjiolov AA, Skryabin KG, Zakharyev VM, Bayev AA. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981;9:6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hindley J, Page SM. Nucleotide sequence of yeast 5S ribosomal RNA. FEBS Lett. 1972;26:157–160. doi: 10.1016/0014-5793(72)80563-5. [DOI] [PubMed] [Google Scholar]
- 13.Rubin GM. The nucleotide sequence of Saccharomyces cerevisae 5.8 S ribosomal ribonucleic acid. J. Biol. Chem. 1973;248:3860–3875. [PubMed] [Google Scholar]
- 14.Rubtsov PM, Musakhanov MM, Zakharyev VM, Krayev AS, Skryabin KG, Bayev AA. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980;8:5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl Acad. Sci. USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl Acad. Sci. USA. 108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coelho PS, Bryan AC, Kumar A, Shadel GS, Snyder M. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauro VP, Edelman GM. rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression. Proc. Natl Acad. Sci. USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chooi WY, Leiby KR. The in vivo expression of pseudo ribosomal RNA genes in Drosophila melanogaster. Mol. Gen. Genet. 1981;182:245–251. doi: 10.1007/BF00269665. [DOI] [PubMed] [Google Scholar]
- 21.Kermekchiev M, Ivanova L. Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation. Mol. Cell Biol. 2001;21:8255–8263. doi: 10.1128/MCB.21.24.8255-8263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharf ME, Wu-Scharf D, Zhou X, Pittendrigh BR, Bennett GW. Gene expression profiles among immature and adult reproductive castes of the termite Reticulitermes flavipes. Insect Mol. Biol. 2005;14:31–44. doi: 10.1111/j.1365-2583.2004.00527.x. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 25.Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2009;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 33.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 34.Medigue C, Moszer I, Viari A, Danchin A. Analysis of a Bacillus subtilis genome fragment using a co-operative computer system prototype. Gene. 1995;165:GC37–GC51. doi: 10.1016/0378-1119(95)00636-k. [DOI] [PubMed] [Google Scholar]
- 35.Medigue C, Rouxel T, Vigier P, Henaut A, Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- 36.Staden R, McLachlan AD. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogh A, Mian IS, Haussler D. A hidden Markov model that finds genes in E. coli DNA. Nucleic Acids Res. 1994;22:4768–4778. doi: 10.1093/nar/22.22.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakke P, Carney N, Deloache W, Gearing M, Ingvorsen K, Lotz M, McNair J, Penumetcha P, Simpson S, Voss L, et al. Evaluation of three automated genome annotations for Halorhabdus utahensis. PLoS ONE. 2009;4:e6291. doi: 10.1371/journal.pone.0006291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 41.Scheunemann AE, Graham WD, Vendeix FA, Agris PF. Binding of aminoglycoside antibiotics to helix 69 of 23S rRNA. Nucleic Acids Res. 38:3094–3105. doi: 10.1093/nar/gkp1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maguire BA. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol. Mol. Biol. Rev. 2009;73:22–35. doi: 10.1128/MMBR.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 44.Mehta R, Champney WS. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob. Agents Chemother. 2002;46: 1546–1549. doi: 10.1128/AAC.46.5.1546-1549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David-Eden H, Mankin AS, Mandel-Gutfreund Y. Structural signatures of antibiotic binding sites on the ribosome. Nucleic Acids Res. 38:5982–5994. doi: 10.1093/nar/gkq411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, Duc AC, Klosi E, Pattabiraman S, Spaller MR, Chow CS. Selection of peptides that target the aminoacyl-tRNA site of bacterial 16S ribosomal RNA. Biochemistry. 2009;48:8299–8311. doi: 10.1021/bi900982t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 48.Kjeldgaard NO, Gausing K. Regulation of biosynthesis of ribosomes. Cold Spring Harb. Monogr. Arch. 1974;4:369–392. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.