Abstract
Aminoacyl–tRNA synthetases (aaRSs) are remarkable enzymes that are in charge of the accurate recognition and ligation of amino acids and tRNA molecules. The greatest difficulty in accurate aminoacylation appears to be in discriminating between highly similar amino acids. To reduce mischarging of tRNAs by non-cognate amino acids, aaRSs have evolved an editing activity in a second active site to cleave the incorrect aminoacyl–tRNAs. Editing occurs after translocation of the aminoacyl–CCA76 end to the editing site, switching between a hairpin and a helical conformation for aminoacylation and editing. Here, we studied the consequence of nucleotide changes in the CCA76 accepting end of tRNALeu during the aminoacylation and editing reactions. The analysis showed that the terminal A76 is essential for both reactions, suggesting that critical interactions occur in the two catalytic sites. Substitutions of C74 and C75 selectively decreased aminoacylation keeping nearly unaffected editing. These mutations might favor the regular helical conformation required to reach the editing site. Mutating the editing domain residues that contribute to CCA76 binding reduced the aminoacylation fidelity leading to cell-toxicity in the presence of non-cognate amino acids. Collectively, the data show how protein synthesis quality is controlled by the CCA76 homogeneity of tRNAs.
INTRODUCTION
Protein synthesis is a central process in organisms from all three domains of life, providing the link between the genetic information encoded in DNA and functional proteins. This process critically relies on the correct formation of aminoacyl–tRNA (aa–tRNA) by aminoacyl–tRNA synthetase (aaRS) to establish the genetic code through rigorous control of the two-step aminoacylation (1,2). The amino acid is first activated with ATP to synthesize the aa-AMP intermediate with the release of pyrophosphate; the amino acid moiety of the intermediate is subsequently transferred to the tRNA bearing the cognate nucleotide triplet (1). Mis-translation arising from disruption in the fidelity of these interactions has profound consequences (3). However, the presence of various types of amino acids and their analogs, and the fact that amino acids only differ in the side chain, greatly challenge the specificity of aaRS. About half of the aaRSs misactivate non-cognate amino acids (4). To solve this problem, the proofreading (editing) mechanism of aaRS has evolved to hydrolyze the mis-products. Editing can occur at the aa-AMP level (pre-transfer editing) and/or mischarged tRNA level (post-transfer editing), depending on the specific aaRS (4).
Leucyl–tRNA synthetase (LeuRS), isoleucyl–tRNA synthetase (IleRS) and valyl–tRNA synthetase (ValRS) belong to the class Ia of aaRSs, characterized by the connective peptide 1 (CP1) and 2 (CP2) in the Rossmann fold nucleotide binding domain where the synthetic active site is located (5). The CP1 domain of LeuRS is located 35 Å away from the Rossmann fold domain and is responsible for post-transfer editing (6,7). Recent studies have revealed that Escherichia coli, Aquifex aeolicus and human cytoplasmic LeuRSs (EcLeuRS, AaLeuRS and hcLeuRS) all employ three different pathways (tRNA-independent, tRNA-dependent pre-transfer editing and post-transfer editing), but in different proportions relative to the total editing activity, to remove non-cognate amino acids (8,9). Similar results were also found with E. coli IleRS and ValRS (10).
X-ray crystal structures of LeuRS and tRNALeu in aminoacylation and post-transfer editing states clearly revealed that the tRNALeu main body conformation is indistinguishable between the two states (6,11). However, the CCA76-end shifts to the CP1 domain from the aminoacylation active site and is specifically recognized by several conserved residues, including Lys300, Tyr330, Arg344 and Leu327 (6,11). Therefore, these residues in CP1 collectively constitute the tRNA entrance pathway in post-transfer editing (6). In vitro studies have shown that the interaction between the Tyr330 and tRNALeu CCA76-end is critical for both tRNA-dependent pre-transfer editing and post-transfer editing (8). However, the role of other crucial resides in editing and their significance in vivo are unclear. In addition, the contribution of the CCA76-end to the aminoacylation and editing still remains elusive.
The CCA76 sequence is conserved at the 3′-end of all mature tRNA molecules to function as the site of amino acid attachment (12). This sequence is gene encoded in E. coli and related bacteria or acquired in eukaryotes and maintained by stepwise nucleotide addition by the ubiquitous CCA-adding enzyme (tRNA nucleotidyltransferase, CCase), which is an unusual RNA polymerase that does not use a nucleic acid template for nucleotide addition (13,14).
The CCA76 sequence is a universal ligand during several critical steps of protein biosynthesis. It is successively recognized by aaRSs (1,12), elongation factor (EF-Tu) (15) and rRNA (16,17). In F. Crick's adaptor hypothesis, the CCA76 sequence is the ultimate adaptor group that carries the amino acid to the decoding center of the ribosome, whereas at the other end of the molecule, the anticodon triplet is needed to fit with the codon triplet of the mRNA. Because all the tRNAs have to fit to the unique ribosome decoding center, evolution has selected a unique CCA76 sequence shared by all tRNAs. Additional constraints for CCA76 conservation result from the interactions with the EF-Tu and the 20 aaRSs. With the later enzymes, the CCA76 acceptor end is the substrate of two successive reactions starting with the aminoacylation reaction then followed by a proofreading reaction catalyzed in a second catalytic site (6,11). A critical conformational change, resulting from the flexibility of the single-stranded CCA76 sequence, is required to allow transition from the synthetic to the editing site (6,11).
The starting point of the present study was to clarify the role of the ubiquitous CCA76-end in the leucine aminoacylation system. Therefore, we generated a series of CCA76-end mutants and tested their impact on the aminoacylation and editing reactions catalyzed by E. coli LeuRS (EcLeuRS). Furthermore, mutants were generated in the CP1 editing domain of LeuRS in order to disrupt the interaction with the CCA76-end during the editing process. The in vivo effect of the mutations was then analyzed in a LeuRS-deficient strain in stress conditions under the pressure of elevated concentrations of non-cognate amino acid. Combining in vitro and in vivo results of protein and tRNA mutagenesis showed how life can regulate protein synthesis and control the aminoacylation quality by using the universally conserved sequence.
MATERIALS AND METHODS
Materials
l-leucine, dithiothreitol, NTP, 5′-GMP, tetrasodium pyrophosphate, inorganic pyrophosphate, ATP, Tris–HCl, MgCl2, NaCl, mouse anti-His6 antibody and activated charcoal were purchased from Sigma (USA). [3H]leucine, [3H]isoleucine and [α-32P]ATP were obtained from Amersham Biosciences (England). Pfu DNA polymerase, the DNA fragment rapid purification kit and a plasmid extraction kit were purchased from Biotech Company (China). KOD-plus mutagenesis kit was obtained from TOYOBO (Japan). T4 ligase and restriction endonucleases were obtained from MBI Fermentas. Phusion high-fidelity DNA polymerase was purchased from New England Biolabs (USA). Ni2+–NTA Superflow was purchased from Qiagen, Inc. (Germany). Polyethyleneimine cellulose plates were purchased from Merck (Germany). Pyrophosphatase was obtained from Roche Applied Science (China). Pyrobest DNA polymerase and the dNTP mixture were obtained from Takara (Japan). Oligonucleotide primers were synthesized by Invitrogen (China). Escherichia coli BL21 (DE3) cells were purchased from Stratagene (USA). The E. coli KL231 strain [F−, leuS31(ts), thyA6, rpsL120(strR), deoC1] was purchased from the E. coli genetic stock center (CGSC, Yale University, USA) (18). T7 RNA polymerase was purified from an overproduction strain in our laboratory (19).
Gene cloning and mutagenesis
The plasmid pET30a-EcleuS encoding EcLeuRS with N-terminal His6-tag previously constructed by our lab (8) was used as the template to construct genes encoding various EcLeuRS mutants using KOD-plus mutagenesis kit for the in vitro activity measurements. EcleuS was amplified, cleaved by NcoI and HindIII and cloned into pTrc99a (pre-cleaved by NcoI and HindIII) to produce pTrc99a-EcleuS. The 18 nucleotides encoding His6-tag at the N-terminus were incorporated into the forward primer during the construction of pTrc99a-EcleuS. The pTrc99a-EcleuS construct was used as the template to construct genes encoding various EcLeuRS mutants using the KOD-plus mutagenesis kit for the in vivo complementation assays in the KL231 strain.
In vitro transcription of tRNAs
Six DNA fragments covering the T7 promoter and tRNA gene double strands were synthesized by Invitrogen, phosphorylated and ligated into pUC19 (pre-cleaved by EcoRI and BamHI) to construct the plasmid pUC19-tRNA. To prepare the wild-type (WT) 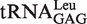 transcript, the forward primer (5′-ctgcagtaatacgactcactatagccgaggtggtgg-3′, with T7 promoter sequence in italics) and the reverse primer (5′-tggtaccgaggacgggacttgaacccgtaagccctattg-3′) were synthesized to amplify the T7 promoter and the gene encoding
transcript, the forward primer (5′-ctgcagtaatacgactcactatagccgaggtggtgg-3′, with T7 promoter sequence in italics) and the reverse primer (5′-tggtaccgaggacgggacttgaacccgtaagccctattg-3′) were synthesized to amplify the T7 promoter and the gene encoding 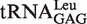 using the Phusion high-fidelity DNA polymerase, which does not add an additional adenosine to the PCR product. The PCR product was separated on a 2% agarose gel, extracted by phenol/chloroform and precipitated in the presence of cold ethanol and 0.3 M NaAc (pH 5.2). The T7 in vitro transcription was carried out at 37°C in a 100 µl reaction mixture containing 40 mM Tris–HCl (pH 8.0), 22 mM MgCl2, 1 mM spermidine, 5 mM DTT, 0.5% Triton X-100, 60 ng μl−1 tDNA template, 5 mM ATP, 5 mM CTP, 5 mM GTP, 5 mM UTP, 0.8 U μl−1 ribonuclease inhibitor, 20 mM GMP and 500 U μl−1 T7 RNA polymerase. One hour later, 2 U of pyrophosphatase was added to remove the pyrophosphate for 30 min, and then 5 U of DNase I (RNase I free) was added and incubated for 1 h to digest the transcription template. The transcript was then loaded into a 15% PAGE–8M Urea gel of 1-mm thickness and 40 cm length, and the gel was run at constant 25 W for 10 h to carefully remove any non-specific bands. The tRNA was cut from the gel and eluted with 0.5 M NaAc (pH 5.2) at room temperature, ethanol precipitated at −20°C after phenol/chloroform extraction two times and dissolved in 5 mM MgCl2. The tRNA was denatured at 80°C for 5 min and slowly cooled down to 30°C. The tRNALeu mutants were prepared by the same procedure, except that the different reverse primer was synthesized and used in preparing the DNA template.
using the Phusion high-fidelity DNA polymerase, which does not add an additional adenosine to the PCR product. The PCR product was separated on a 2% agarose gel, extracted by phenol/chloroform and precipitated in the presence of cold ethanol and 0.3 M NaAc (pH 5.2). The T7 in vitro transcription was carried out at 37°C in a 100 µl reaction mixture containing 40 mM Tris–HCl (pH 8.0), 22 mM MgCl2, 1 mM spermidine, 5 mM DTT, 0.5% Triton X-100, 60 ng μl−1 tDNA template, 5 mM ATP, 5 mM CTP, 5 mM GTP, 5 mM UTP, 0.8 U μl−1 ribonuclease inhibitor, 20 mM GMP and 500 U μl−1 T7 RNA polymerase. One hour later, 2 U of pyrophosphatase was added to remove the pyrophosphate for 30 min, and then 5 U of DNase I (RNase I free) was added and incubated for 1 h to digest the transcription template. The transcript was then loaded into a 15% PAGE–8M Urea gel of 1-mm thickness and 40 cm length, and the gel was run at constant 25 W for 10 h to carefully remove any non-specific bands. The tRNA was cut from the gel and eluted with 0.5 M NaAc (pH 5.2) at room temperature, ethanol precipitated at −20°C after phenol/chloroform extraction two times and dissolved in 5 mM MgCl2. The tRNA was denatured at 80°C for 5 min and slowly cooled down to 30°C. The tRNALeu mutants were prepared by the same procedure, except that the different reverse primer was synthesized and used in preparing the DNA template.
Protein expression and purification
The WT LeuRS (EcLeuRS) and its mutants were produced by transformation of E. coli BL21 (DE3) cells with the corresponding plasmids. A single colony from each of the transformants was chosen and cultured in 500 ml of 2× YT medium at 37°C. When the cells were grown to mid-log phase (A600 = 0.6), IPTG (isopropyl-1-thio-β-d- galactopyranoside) was added to a final concentration of 0.2 mM, and cultivation continued for 6 h at 22°C. The proteins were purified by Ni2+–NTA chromatography, as described previously (8).
Aminoacylation, mis-aminoacylation and deacylation
The kinetic constants of EcLeuRS for WT tRNA and its variants were tested in a reaction mixture containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 0.5 mM dithiothreitol, 4 mM ATP, 40 μM [3H]leucine, 0.25–12 μM EctRNALeu and 5 nM EcLeuRS at 37°C. Mis-aminoacylation assays were carried out at 37°C in a reaction mixture containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 0.5 mM dithiothreitol, 4 mM ATP, 5 µM EctRNALeu, 40 µM [3H]isoleucine and 1 µM EcLeuRS. Preparation of mischarged Ile-tRNALeu was carried out in a similar system with mis-aminoacylation, except that 20 µM EctRNALeu and 1 µM EcLeuRS-T252E were used (20). Hydrolytic editing assays of EcLeuRS or its mutants were performed at 37°C in 100 mM Tris–HCl (pH 7.5), 30 mM KCl, 12 mM MgCl2, 0.5 mM dithiothreitol and 1 µM [3H]Ile-tRNALeu, and the reactions were initiated with 5 nM enzyme. Nine microliter aliquots of the reaction solution were added to Whatman filter pads and quenched with cold 5% trichloroacetic acid (TCA) at various time intervals. The pads were washed three times for 15 min each with cold 5% TCA and then three times for 10 min each with 100% ethanol. The pads were dried under a heat lamp. The radioactivities of the precipitates were quantified with a scintillation counter (Beckman Coulter).
Charging plateau measurements
Charging plateau measurements of transcripts were performed in 40 µl reaction mixtures each containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 0.5 mM dithiothreitol, 4 mM ATP, 20 μM [3H]leucine (15 Ci/mmol), 1 µM tRNA (ΔA76, ΔC75A76, ΔC74C75A76, ΔA76/C75A, ΔA76/C75U and ΔA76/C75G) and 100 nM EcLeuRS at 37°C. Aliquots of 9 µl of the reaction solution were removed at various time intervals, quenched on Whatman filter pads and processed as mentioned in the procedures above. The blank experiment without tRNALeu was performed under the same conditions. In another control experiment, RNase I (final 0.25 U/µl) was added to the aliquot at various time intervals to digest the aminoacylation product and incubated at 37°C for another 5 min. The plateau value of WT tRNALeu was >1400 pmol/A260.
AMP formation
In the editing Nva reaction of EcLeuRS, AMP formation was measured in a reaction mixture containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 5 mM dithiothreitol, 5 U ml−1 pyrophosphatase, 3 mM ATP, 20 nM [α-32P]ATP and 15 mM Norvaline (Nva) in the presence or absence of 5 µM tRNALeu or its variants. The reaction was initiated by the addition of 0.2 µM EcLeuRS or its mutants and incubated at 37°C. Aliquots (1.5 µl) were quenched in 6 µl of 200 mM NaAc (pH 5.0). The quenched aliquots (1.5 µl each) were spotted in duplicate on polyethyleneimine cellulose plates pre-washed with water. Separation of Nva-[32P]AMP, [32P]AMP and [32P]ATP was performed by developing thin layer chromatography (TLC) on polyethyleneimine cellulose plates in 0.1 M NH4Ac and 5% acetic acid. The plates were visualized by phosphorimaging, and the data were analyzed using Multi Gauge Version 3.0 software (FUJIFILM). The gray densities of [32P]AMP spots were compared with the gray density of known [32P]ATP concentrations. Rate constants were obtained from graphs of [32P]AMP formation plotted against time (8).
KL231 complementation assay
Competent KL231 cells were electro-transformed with various plasmids, including pTrc99a (negative control), pTrc99a-EcleuS, pTrc99a-EcleuS-L327G, -L327R, -K300E, -Y330D, -R344D, -K300E/Y330D, -K300E/R344D, -Y330D/R344D and -K300E/Y330D/R344D. Transformants were grown on solid LB plates supplemented with 100 μg ml−1 ampicillin and 200 μg ml−1 thymine at 30°C. A single colony was selected and grown in liquid LB medium supplemented with 100 μg ml−1 ampicillin, 200 μg ml−1 thymine and 0.1 mM IPTG at 30°C. The cells harboring these plasmids were diluted and adjusted to the same A600 value of 0.2. To test the effect of non-cognate Nva on cell growth, 5 μl of the diluted mixture were dropped on a solid LB plate (supplemented with 100 μg ml−1 ampicillin, 200 μg ml−1 thymine and 0.1 mM IPTG) containing 0, 5, 10, 20, 50, 100 or 200 mM Nva, and the plates were incubated at 42°C to observe the growth of cells.
RESULTS
Mutation of the CCA76-end impacts the tRNA aminoacylation reaction
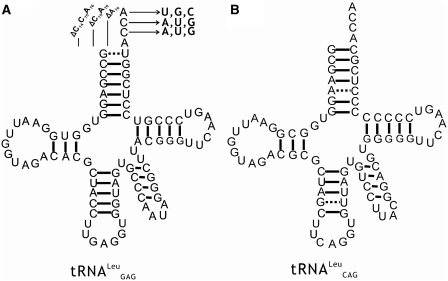
All tRNALeu isoacceptors from various species contain the absolutely conserved A73 (12), which functions as the discriminator during recognition by LeuRSs in different species (12,21). However, the effect of the CCA76-end on tRNA aminoacylation has not been reported. Here, each nucleotide of the CCA76-end of 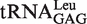 was individually mutated to one of the three other nucleotides (Figure 1A), and the aminoacylation kinetic constants of EcLeuRS for these tRNALeus were assayed. In aminoacylation of the tRNALeu A76U and A76C mutants, the catalytic efficiency of EcLeuRS was obviously decreased (155.8 and 87.2 s−1 mM−1, respectively) as compared with that of the WT tRNA transcript (923.1 s−1 mM−1). The amino acid-accepting activity of the A76G mutant was too low to accurately measure the kinetics. Similarly, the three C75 mutants reduced the amino acid-accepting activity to 2–46% of the native tRNALeu, with the C75G mutant showing the greatest decrease (14.6 s−1 mM−1). Additionally, the three tRNA variants from C74 showed obvious decreases in amino acid-accepting activity; their catalytic constants for EcLeuRS ranged from 8% (C74G, 69.4 s−1 mM−1), 41% (C74U, 381.8 s−1 mM−1) to 50% (C74A, 460.3 s−1 mM−1) of that with the WT tRNALeu (Table 1). All the decreases were mainly due to a reduced kcat value of EcLeuRS and/or changed Km for these tRNALeu mutants. These results clearly showed that the CCA76-end mutations negatively impacted the aminoacylation of these mutants to various levels, and the presence of guanosine at each of the 3-nt positions resulted in the lowest level of tRNA aminoacylation.
was individually mutated to one of the three other nucleotides (Figure 1A), and the aminoacylation kinetic constants of EcLeuRS for these tRNALeus were assayed. In aminoacylation of the tRNALeu A76U and A76C mutants, the catalytic efficiency of EcLeuRS was obviously decreased (155.8 and 87.2 s−1 mM−1, respectively) as compared with that of the WT tRNA transcript (923.1 s−1 mM−1). The amino acid-accepting activity of the A76G mutant was too low to accurately measure the kinetics. Similarly, the three C75 mutants reduced the amino acid-accepting activity to 2–46% of the native tRNALeu, with the C75G mutant showing the greatest decrease (14.6 s−1 mM−1). Additionally, the three tRNA variants from C74 showed obvious decreases in amino acid-accepting activity; their catalytic constants for EcLeuRS ranged from 8% (C74G, 69.4 s−1 mM−1), 41% (C74U, 381.8 s−1 mM−1) to 50% (C74A, 460.3 s−1 mM−1) of that with the WT tRNALeu (Table 1). All the decreases were mainly due to a reduced kcat value of EcLeuRS and/or changed Km for these tRNALeu mutants. These results clearly showed that the CCA76-end mutations negatively impacted the aminoacylation of these mutants to various levels, and the presence of guanosine at each of the 3-nt positions resulted in the lowest level of tRNA aminoacylation.
Figure 1.
Secondary structures of tRNALeus and location of the mutations. Cloverleaf structures of 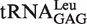 (A) and
(A) and 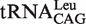 (B) used in the present work. The point and deletion mutations on the
(B) used in the present work. The point and deletion mutations on the 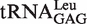 are shown.
are shown.
Table 1.
Aminoacylation kinetics of transcripts of 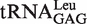 by EcLeuRS
by EcLeuRS
| tRNA | Km (µM) | kcat (s−1) | kcat/Km (s−1 mM−1) | kcat/Km (relative) |
|---|---|---|---|---|
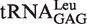 |
1.95 ± 0.23 | 1.80 ± 0.27 | 923.1 | 1 |
| A76U | 1.99 ± 0.30 | 0.31 ± 0.04 | 155.8 | 0.17 |
| A76G | – | <<0.002 | – | – |
| A76C | 3.90 ± 0.48 | 0.34 ± 0.03 | 87.2 | 0.09 |
| C75A | 0.38 ± 0.05 | 0.16 ± 0.03 | 421.1 | 0.46 |
| C75U | 1.18 ± 0.17 | 0.10 ± 0.02 | 84.7 | 0.09 |
| C75G | 1.78 ± 0.22 | 0.026 ± 0.004 | 14.6 | 0.02 |
| C74A | 0.63 ± 0.05 | 0.29 ± 0.04 | 460.3 | 0.50 |
| C74U | 0.55 ± 0.07 | 0.21 ± 0.03 | 381.8 | 0.41 |
| C74G | 0.36 ± 0.05 | 0.025 ± 0.002 | 69.4 | 0.08 |
LeuRS can aminoacylate tRNALeu lacking the terminal adenosine
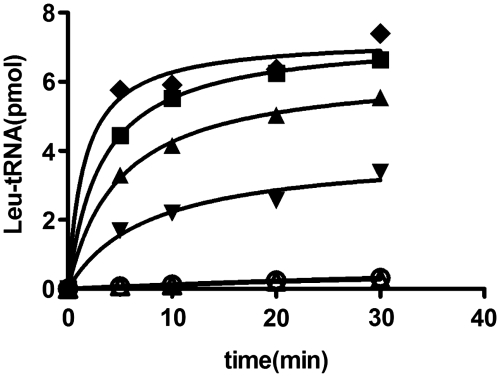
Besides point mutations, we also performed progressive deletions from the CCA76-end of tRNA in order to obtain the following three mutants: ΔA76 (ending with C75), ΔC75A76 (ending with C74) and ΔC74C75A76 (ending with A73) (Figure 1A). In initial charging plateau measurements for accepting activity with a large excess of 1 µM enzyme, it was surprising to observe that LeuRS could charge ΔA76 mutant obviously. As it has been reported that some aaRSs can ligate their cognate amino acids to themselves without tRNA (by self-aminoacylation occurring at high enzyme concentration and excess of adenylate formation) (22–24), we tested the self-aminoacylation of EcLeuRS at different concentrations without tRNA. We found that self-aminoacylation of EcLeuRS was negligible at low concentrations (<300 nM). However, EcLeuRS at higher concentrations >500 nM could be self-aminoacylated and was resistant to RNase I (Supplementary Figure S1). Therefore, we tested the ΔA76 charging plateau value by using 100 nM EcLeuRS and found that 70% of ΔA76 could be charged with 3H-Leu. In the same conditions, no detectable labeled product could be measured at the absence of tRNA (Figure 2). To further confirm that, the tRNA but not the enzyme itself was ligated with 3H-Leu, we treated the aminoacylation product in the presence of RNase I, and the values dropped down to the same level with that in the absence of tRNA (Figure 2). These results confirmed that LeuRS could aminoacylate tRNALeu with the terminal A76 deleted. However, reliable aminoacylation kinetics could not be measured because of the very low kcat. However, the other two tRNA mutants, ΔC75A76 and ΔC74C75A76, could not be aminoacylated at 100 nM or higher concentrations of EcLeuRS (data not shown).
Figure 2.
Aminoacylation of tRNALeu-ΔA76, -ΔA76/C75A, -ΔA76/C75U or -ΔA76/C75G by EcLeuRS.Charging plateaus of EctRNALeu-ΔA76 (filled diamond), -ΔA76/C75A (filled square), -ΔA76/C75U (filled triangle) or -ΔA76/C75G (filled inverted triangle) were measured as described in the Materials and methods section. The control (unfilled circle) was performed without tRNA. The charging plateaus were determined using GraphPad Prism software. According to the plateau values (to calculate the amount of tRNALeu charged leucine) and total tRNALeu used in the reaction, EctRNALeu-ΔA76, -ΔA76/C75A, -ΔA76/C75U and -ΔA76/C75G are aminoacylated to 70, 70, 61 and 35%, respectively. Digestion of aminoacyl–EctRNALeu-ΔA76 by RNase I digestion (unfilled triangle) and no product accumulation in control excluded the self-aminoacylation of the enzyme.
To further analyze the ΔA76 mutant, we mutated the terminal C75 to the three other nucleotides in order to identify potential interactions that could be more productive in aminoacylation. The resulting mutants, ΔA76/C75A, ΔA76/C75U and ΔA76/C75G, could be aminoacylated by EcLeuRS to plateau levels of 70, 61 and 35%, respectively (Figure 2). Therefore, more productive interactions were not induced by these nucleotides, and here again, the mutant harboring a guanosine mutation was the least active compared to those with the other nucleotides.
Role of CCA76-end in total editing
Usually, more than one ATP molecule is consumed by an aaRS in the presence of a non-cognate amino acid due to repetitive cycles of synthesis–hydrolysis of the non-cognate products. The excess of ATP consumption can be determined by measuring the release of AMP in the TLC assay (25). In the presence of tRNA, the TLC assay measures the global editing activity, including the tRNA-independent and tRNA-dependent pre-transfer editing in addition to the post-transfer editing (8,26). In the absence of tRNA, the assay measures the AMP produced from the sole tRNA-independent pre-transfer editing activity (26).
In the presence of non-cognate Nva, the observed rate constant (kobs) of WT LeuRS for AMP formation without tRNA in the editing reaction was 0.56 ± 0.07 s−1, accounting for 13% of that in the presence of tRNA transcript (4.42 ± 0.64 s−1) (Table 2). The tRNA transcript without the modified bases was tested to determine if it was as efficient as the native tRNA in stimulating the editing activity. The kobs for AMP formation in the presence of 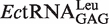 purified from the overproducing E. coli strain (27) was 5.59 ± 0.76 s−1. This value was only slightly larger than that in the presence of transcript (4.42 ± 0.64 s−1), indicating that the modified bases in EctRNALeu did not play a critical role in stimulating editing activity (Table 2).
purified from the overproducing E. coli strain (27) was 5.59 ± 0.76 s−1. This value was only slightly larger than that in the presence of transcript (4.42 ± 0.64 s−1), indicating that the modified bases in EctRNALeu did not play a critical role in stimulating editing activity (Table 2).
Table 2.
kobs of AMP formation by EcLeuRS in the presence of Nva and 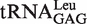 or its variants
or its variants
| tRNA | kobs (s−1) | Relative kobs |
|---|---|---|
No 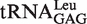
|
0.56 ± 0.07 | 0.13 |
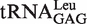 (in vivo)a (in vivo)a
|
5.59 ± 0.76 | 1.27 |
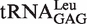 (transcript)b (transcript)b
|
4.42 ± 0.64 | 1 |
| A76U | 0.24 ± 0.04 | 0.05 |
| A76G | 0.20 ± 0.04 | 0.05 |
| A76C | 0.48 ± 0.06 | 0.11 |
| ΔA76 | 0.71 ± 0.08 | 0.16 |
| C75A | 8.34 ± 1.03 | 1.89 |
| C75U | 6.26 ± 0.81 | 1.42 |
| C75G | 0.64 ± 0.07 | 0.15 |
| C74A | 7.48 ± 0.98 | 1.69 |
| C74U | 8.87 ± 1.34 | 2.01 |
| C74G | 2.65 ± 0.32 | 0.60 |
a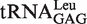 obtained from the overproducing E. coli strain (27).
obtained from the overproducing E. coli strain (27).
b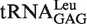 obtained by T7 in vitro transcription.
obtained by T7 in vitro transcription.
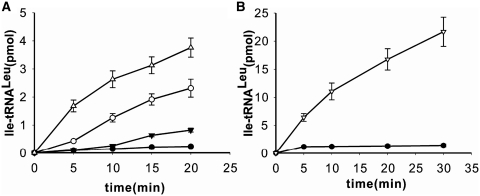
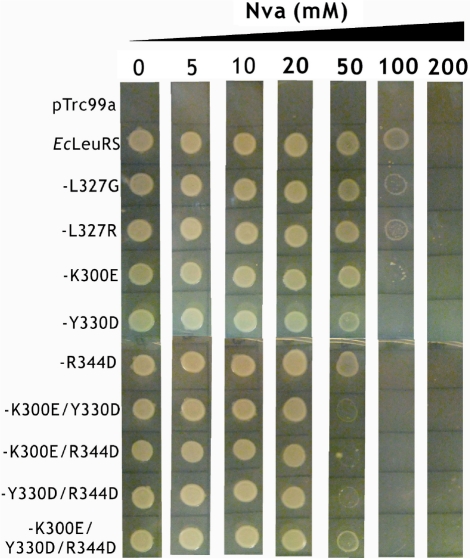
Different variants of the CCA76-end were also tested for their abilities to stimulate AMP formation in editing reactions with EcLeuRS. The AMP formation rates for A76U, A76G and A76C mutants dropped down to a level similar to that without tRNA, indicating only tRNA-independent pre-transfer editing remained and the tRNA-dependent editing was abolished (Table 2). As a consequence of this loss of editing ability, the two A76-mutants of tRNALeu (A76U and A76C) were obviously mischarged with Ile (Figure 3A). The decreased formation of Ile-tRNALeu of A76G may be due to its deficiency in accepting the amino acid. Among the other six variants of tRNALeu, only C75G with a kobs value of 0.64 ± 0.07 s−1 had tRNA-independent pre-transfer editing and lost the tRNA-dependent editing, indicating that C75G could not play a role in the tRNA-dependent editing; C74G with a kobs value of 2.65 ± 0.32 s−1 decreased the tRNA-dependent editing to nearly half of that of the WT tRNALeu; the four mutants of tRNALeu, C75A, C75U, C74A and C74U stimulated more AMP formation than WT tRNALeu, with the kobs values of 8.34 ± 1.03, 6.26 ± 0.81, 7.48 ± 0.98 and 8.87 ± 1.34 s−1, respectively (Table 2), indicating that these mutations did not affect the tRNA-dependent editing but rather stimulated it. The six variants of nucleotides 74 and 75 did not form any detectable Ile-tRNALeu in the misacylation assay (data not shown). Additionally, in the presence of the ΔA76 mutant, EcLeuRS had comparable editing activity with that in the absence of tRNA (Table 2). Correspondingly, EcLeuRS catalyzed the synthesis of an obvious amount of mischarged ΔA76 mutant (Ile-tRNALeu-ΔA76) (Figure 3B). These results clearly showed that the terminal A76 is a critical element in the tRNA-dependent editing activity of EcLeuRS, and the nucleotides C74 and C75 crucial for aminoacylation do not play an indispensible role to the tRNA-dependent editing of EcLeuRS.
Figure 3.
Misacylation of tRNA-A76 mutants with Ile. (A and B) WT tRNALeu (filled circle) and mutated derivatives A76U (unfilled circle), A76G (filled inverted triangle) or A76C (unfilled triangle) and ΔA76 (unfilled inverted triangle) were mischarged with non-cognate Ile.
Role of CCA76-end in post-transfer editing
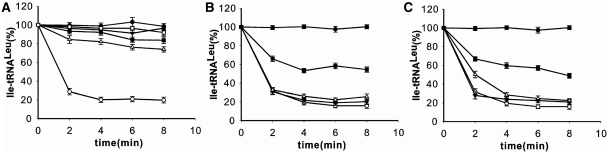
It is now universally accepted that after aminoacylation, the CCA76-end of the tRNA is translocated ∼35 Å from the aminoacylation active site to the editing active site embedded in the CP1 domain (6). Both tRNA CCA76-end and mischarged amino acid moieties are recognized by the active site of post-transfer editing within the CP1 domain of LeuRS. To test the contribution of the CCA76-end to the post-transfer editing, we first mischarged 10 variants of the tRNA with non-cognate 3H-Ile using an editing-deficient LeuRS-T252E mutant (20). The deacylation of the mischarged Ile-tRNALeu by WT LeuRS was then measured. The mutants of tRNALeu could be mischarged with Ile by the LeuRS-T252E mutant. However the three mis-acylated mutants of A76 (A76C, A76G and A76U) were resistant to deacylation catalyzed by LeuRS (Figure 4A), indicating the crucial role of this nucleotide in post-transfer editing. This result was also consistent with their inability to stimulate total editing in the TLC assay (Table 2). In addition, Ile-tRNALeu-ΔA76 could not be hydrolyzed by the post-transfer editing (Figure 4A). This is consistent with the fact that the A76 base is specifically recognized by the main chain carbonyl- and amino-groups of EcLeuRS Leu327 in the Nva2AA-containing structure (analog of the post-transfer editing substrate) (28) and Thermus thermophilus LeuRS-tRNALeu structure (in post-transfer editing conformation) (6). Obviously, changing the terminal base affects the proper positioning of the CCA76-end into the CP1 domain and thus blocks the hydrolytic editing reaction.
Figure 4.
Effect of the CCA76 mutations on deacylation of mischarged tRNALeu. Mischarged tRNA mutants from A76, C75 or C74 were prepared by editing-deficient EcLeuRS-T252E (20). Mischarged tRNA-ΔA76 mutant was obtained with WT EcLeuRS. Deacylation of mischarged WT tRNALeu was indicated (unfilled circle). The deacylation assay of mischarged ΔA76 (unfilled square), A76U (filled inverted triangle), A76G (unfilled triangle), A76C (filled square) in (A), C75A (filled inverted triangle), C75U (unfilled triangle), C75G (filled square) in (B) or C74A (filled inverted triangle), C74U (unfilled triangle), C74G (filled square) in (C) was performed at 37°C with 1 µM [3H]Ile-tRNALeu and 5 nM EcLeuRS. The spontaneous hydrolysis of each mischarged tRNA (control) was carried out without enzyme. For clarity, a single representative control is shown (filled circle).
In general, mutations of nucleotides 75 and 74 did not influence the tRNA deacylation properties, except when the guanosine mutations C75G and C74G were introduced (Figure 4B and C). The result suggested that guanine residues at either position of CCA76-end may prevent proper interaction with the enzyme and/or tRNA end translocation.
The tRNA entrance pathway critically contributes to quality control
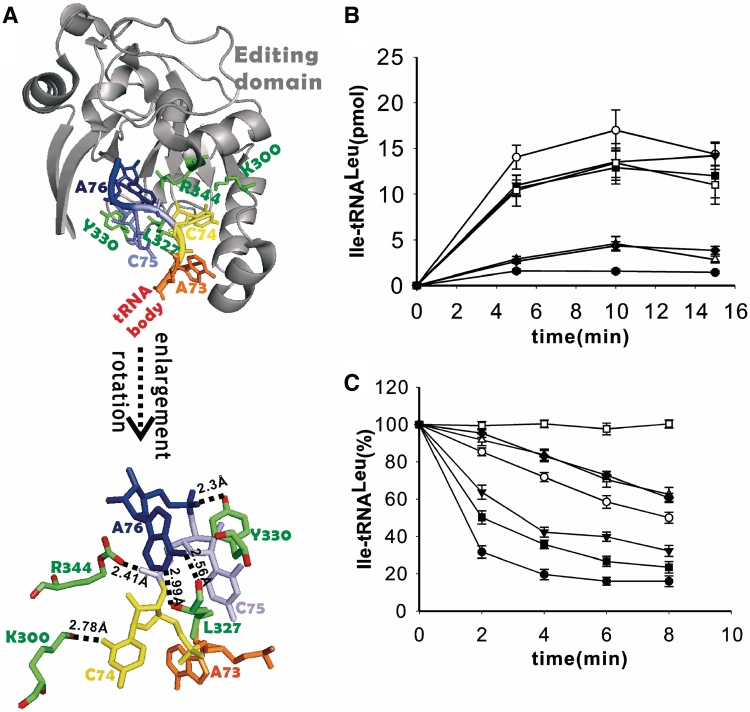
According to the crystal structure, the CP1 editing domain binds only with CCA76-end residues without any interaction with the main tRNA body. In detail, the equivalent side chains in EcLeuRS interacting specifically with the C74 base and the phosphates of C75 and A76 are Lys300, Arg344 and Tyr330, respectively (6). Additionally, the main chain carbonyl- and amino-group of Leu327 interact with the A76 base (Figure 5A). These amino acid residues are absolutely conserved in prokaryotic LeuRSs and thus constitute an entrance pathway to orient the aminoacylated CCA76-arm into the editing active site. Previous studies have demonstrated the essential role of Tyr330 of EcLeuRS in tRNA-dependent editing (8). However, other sites (Leu327, Lys300 and Arg344) and their functions in vivo have not been determined. We studied here the roles of residues Leu327, Lys300, Tyr330 and Arg344 by mutating them individually or in combination. The in vitro and in vivo assays were then performed in order to analyze the effects of these mutations.
Figure 5.
The tRNA entrance pathway contributes critically to the quality control. (A) View of tRNALeu CCA76-end bound to the CP1 domain (gray) (6), showing the binding pattern between C74 (yellow), C75 (cyan), A76 (blue) and the key amino acids (green) in the CP1 editing domain (upper). The tRNA main body before nucleotide A73 (orange) and LeuRS main body except for the editing domain were omitted for clarity. The lower panel shows the closer protein–CCA76 interaction network. Distances between the amino-acid residues here studied and the nucleotides are shown. (B) Mischarging of 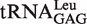 with non-cognate Ile by WT EcLeuRS (filled circle), -Y330D (unfilled circle), -K300E/Y330D (filled inverted triangle), -K300E/R344D (unfilled triangle), -Y330D/R344D (filled square), -K300E/Y330D/R344D (unfilled square) and -R344D (filled diamond) with significant misaminoacylation ability. Mutants that did not accumulate Ile-tRNALeu are not shown. (C) Deacylation curves of Ile-tRNALeu by WT EcLeuRS (filled circle), -L327G (unfilled circle), -L327R (filled inverted triangle), -K300E/R344D (unfilled triangle), -K300E (filled square) and -R344D (filled diamond). Editing-defective mutants (EcLeuRS-Y330D, -K300E/Y330D, -Y330D/R344D and -K300E/Y330D/R344D) are not shown for clarity. Spontaneous hydrolysis without enzyme addition was carried out as a control (unfilled square) as indicated.
with non-cognate Ile by WT EcLeuRS (filled circle), -Y330D (unfilled circle), -K300E/Y330D (filled inverted triangle), -K300E/R344D (unfilled triangle), -Y330D/R344D (filled square), -K300E/Y330D/R344D (unfilled square) and -R344D (filled diamond) with significant misaminoacylation ability. Mutants that did not accumulate Ile-tRNALeu are not shown. (C) Deacylation curves of Ile-tRNALeu by WT EcLeuRS (filled circle), -L327G (unfilled circle), -L327R (filled inverted triangle), -K300E/R344D (unfilled triangle), -K300E (filled square) and -R344D (filled diamond). Editing-defective mutants (EcLeuRS-Y330D, -K300E/Y330D, -Y330D/R344D and -K300E/Y330D/R344D) are not shown for clarity. Spontaneous hydrolysis without enzyme addition was carried out as a control (unfilled square) as indicated.
The following single, double and triple mutants were constructed in EcLeuRS to form EcLeuRS-L327G, -L327R, -K300E, -Y330D, -R344D, -K300E/Y330D, -K300E/R344D, -Y330D/R344D and -K300E/Y330D/R344D. All these mutants displayed intact synthetic activities (Leu activation and aminoacylation) as expected from their location in the discrete CP1 editing domain (data not shown). However, EcLeuRS-Y330D, -K300E/Y330D, -Y330D/R344D and the triple mutant were completely defective in post-transfer editing (data not shown), and they produced significant amounts of Ile-tRNALeu (Figure 5B). The other mutants were impaired in post-transfer editing to different extents (Figure 5C), and only EcLeuRS-R344D and EcLeuRS-K300E/R344D synthesized mischarged tRNALeu, but obviously at lower levels than that with EcLeuRS-Y330D and the derived double and triple mutants (Figure 5B). In the TLC assay, EcLeuRS-Y330D, its derived double and triple mutants and EcLeuRS-K300E/R344D only retained the tRNA-independent pre-transfer editing activity (Table 3). Altogether, the data showed that the Tyr330 residue plays a major role in tRNA-dependent editing as previously demonstrated (8). The negative effect of Y330D on editing was dominant, and the derived multiple mutants exhibited similar defects. Only the combination mutant EcLeuRS-K300E/R344D could reach the same level of tRNA-independent editing of the single EcLeuRS-Y330D mutation. Other individual mutants showed more limited decreases in post-transfer editing activity, inducing no measurable tRNA mischarging (data not shown). These in vitro data confirmed that mutating the CCA76 entrance pathway of the CP1 domain has a negative effect on the accuracy of tRNALeu aminoacylation.
Table 3.
kobs of AMP formation by EcLeuRS or mutated derivatives in the presence of Nva
| EcLeuRS |
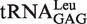 (transcript) (transcript) |
kobs (s−1) | Relative kobs |
|---|---|---|---|
| WT | + | 4.42 ± 0.64 | 1 |
| WT | – | 0.56 ± 0.07 | 0.13 |
| -L327G | + | 1.86 ± 0.26 | 0.42 |
| -L327R | + | 3.60 ± 0.45 | 0.81 |
| -K300E | + | 2.54 ± 0.41 | 0.57 |
| -Y330D | + | 0.59 ± 0.06 | 0.14 |
| -R344D | + | 1.26 ± 0.19 | 0.29 |
| -K300E/Y330D | + | 0.46 ± 0.03 | 0.10 |
| -K300E/R344D | + | 0.82 ± 0.07 | 0.19 |
| -Y330D/R344D | + | 0.49 ± 0.06 | 0.11 |
| -K300E/Y330D/R344D | + | 0.59 ± 0.06 | 0.14 |
To evaluate if these losses of accuracy could affect the fidelity of protein synthesis in vivo, complementation assays using the leuS temperature-sensitive strain KL231 were performed (18). The strain KL231 was transformed with various plasmids encoding EcLeuRS and different mutants. At 42°C, all the transformants grew on medium supplemented with 100 μg ml−1 ampicillin and 200 μg ml−1 thymine (Figure 6), and the mutants were well expressed at a level similar to that of WT EcLeuRS (Supplementary Figure S2). The growth was then observed at 42°C in the presence of increasing concentrations of Nva in the growth medium. No growth difference could be observed below 20 mM of Nva. However, in the presence of 50 mM Nva, the strains harboring the genes-encoding EcLeuRS-Y330D, -K300E/Y330D, -K300E/R344D, -Y330D/R344D and -K300E/Y330D/R344D grew slowly or were inhibited completely, especially those containing the last double and triple mutants (Figure 6). With a 100 mM of Nva no strains containing mutants were able to grow, except for the strain expressing WT enzyme which only grew at a very slow rate. All strains died in the presence of 200 mM Nva.
Figure 6.
In vivo toxicity resulting from mutations in the CP1 tRNA entrance pathway of EcLeuRS. The complementation assay was performed using E. coli thermosensitive strain KL231 at 42°C, on solid LB plates supplemented with 100 μg ml−1 ampicillin, 200 μg ml−1 thymine, 0.1 mM IPTG and increasing concentrations of non-cognate Nva. KL231 was transformed with mutated copies of LeuRS-encoding genes, and growth was compared with the negative control (empty pTrc99a vector) and positive control (WT EcLeuRS). Expression of all LeuRSs proteins was controlled by Western blot (Supplementary Figure S2).
Altogether, the in vivo data are consistent with the in vitro results, demonstrating that perturbing the tRNA CCA76-end entry pathway in the CP1 domain has a significantly negative effect on the fidelity of protein biosynthesis.
Different tRNALeu isoacceptors stimulate LeuRS editing with the same efficiency
In E. coli, there are five different tRNALeu isoacceptors with two types of first base pairs in its accepting stem (Figure 1). 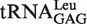 and
and 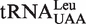 harbor the wobble base pair (G1-U72); while
harbor the wobble base pair (G1-U72); while 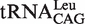 ,
, 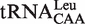 and
and 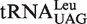 contain the standard Watson–Crick one (G1-C72). It has been shown that EcLeuRS has the same specificity and activity toward different isoacceptors in aminoacylation (29). We transcribed the
contain the standard Watson–Crick one (G1-C72). It has been shown that EcLeuRS has the same specificity and activity toward different isoacceptors in aminoacylation (29). We transcribed the 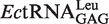 (with G1-U72) and
(with G1-U72) and 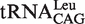 (with G1-C72) to represent two types of tRNALeu and compared their capacity to stimulate editing by EcLeuRS. The data showed that they stimulated EcLeuRS editing activity with nearly identical efficiency (4.42 ± 0.64 and 4.54 ± 0.36 s−1, respectively) (Table 4). To explore the possible role of the first base pair in EcLeuRS editing, we mutated the U72 to C72 of
(with G1-C72) to represent two types of tRNALeu and compared their capacity to stimulate editing by EcLeuRS. The data showed that they stimulated EcLeuRS editing activity with nearly identical efficiency (4.42 ± 0.64 and 4.54 ± 0.36 s−1, respectively) (Table 4). To explore the possible role of the first base pair in EcLeuRS editing, we mutated the U72 to C72 of 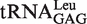 or the C72 to U72 of
or the C72 to U72 of 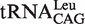 to change the first base pair pattern. Measurements of AMP formation showed that the
to change the first base pair pattern. Measurements of AMP formation showed that the 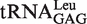 (G1-C72) had the editing kobs of 3.75 ± 0.50 s−1, while the
(G1-C72) had the editing kobs of 3.75 ± 0.50 s−1, while the 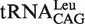 (G1-U72) mutant stimulated the editing with kobs of 6.10 ± 0.81 s−1 (Table 4). The above results suggested that EcLeuRS editing has no preference among WT isoacceptors with different first base-pairs. However, changing the first base-pair obviously led to distinct editing-stimulating capacities among isoacceptors. Therefore, the first base pair pattern may cooperate with other sequence differences between the two tRNALeus to confer the same level of editing-stimulating capacity of the two isoacceptors of WT tRNALeu.
(G1-U72) mutant stimulated the editing with kobs of 6.10 ± 0.81 s−1 (Table 4). The above results suggested that EcLeuRS editing has no preference among WT isoacceptors with different first base-pairs. However, changing the first base-pair obviously led to distinct editing-stimulating capacities among isoacceptors. Therefore, the first base pair pattern may cooperate with other sequence differences between the two tRNALeus to confer the same level of editing-stimulating capacity of the two isoacceptors of WT tRNALeu.
Table 4.
kobs of AMP formation in the presence of different transcripts of tRNALeu isoacceptors or mutants
| tRNA | kobs (s−1) | Relative kobs |
|---|---|---|
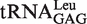 |
4.42 ± 0.64 | 1 |
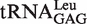 (G1-C72) (G1-C72) |
3.75 ± 0.50 | 0.85 |
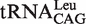 |
4.54 ± 0.36 | 1 |
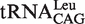 (G1-U72) (G1-U72) |
6.10 ± 0.81 | 1.34 |
DISCUSSION
The CCA76-end sequence is critical for both aminoacylation and editing reactions
The CCA76 sequence is conserved at the 3′-end of all mature tRNA molecules. Although it is an indispensable prerequisite for a functional tRNA, very few organisms like E. coli encode the CCA76 triplet in their tRNA genes. Most of the time, the CCA76 tail is to be added post-transcriptionally in eukaryotes, archaea and many bacteria, where the tRNA genes do not encode the CCA76 terminus (30). This sequence is acquired and maintained by step-wise nucleotide addition by the ubiquitous tRNA nucleotidyltransferase that synthesizes this specific triplet without a nucleic acid template. The CCA sequence plays key roles during several steps of protein biosynthesis. During tRNA charging catalyzed by aaRSs, the 2′ or 3′ hydroxyl of ribose in A76 is the group involved in the esterification reaction with the carboxyl of the amino acid. Although the CCA76-end does not play a critical role in recognition between tRNA and aaRS, it is essential in terms of catalytic efficiency for the aminoacylation reaction (31–33). The universal CCA76 sequence is also involved in EF-Tu binding and during peptide bond formation by interacting with specific nucleotides of the 23S rRNA (17). Little is known about the precise role of the CCA76 nucleotides during the editing reaction catalyzed by aaRSs. It has been shown that only chargeable tRNAVal mutants are able to stimulate the editing reaction of ValRS, suggesting that the enzyme required prior charging of tRNA (31,32,34). Although tRNA is not a strict prerequisite for the editing of both classes I and II aaRSs (26,35–37), its presence strongly stimulates editing (38). Remarkably, both classes of aaRSs use effectively the ability of the CCA76-end of tRNA to switch between a hairpin and a helical conformation during the aminoacylation and editing reactions. Our previous investigations have shown that the interaction between the tRNALeu CCA76-end and the CP1 domain of LeuRS is critical for tRNA-dependent editing (post-transfer and pre-transfer editing) (8). However, the molecular basis of the phenomenon is not yet understood in detail. Here, we extended the investigations to the three CCA76 nucleotides and assayed the effect of mutations on the tRNA-dependent pre-transfer editing and post-transfer editing.
We showed that the universally conserved A76 is essential for tRNALeu to trigger both aminoacylation and editing reaction of LeuRS. The mutants of tRNALeu with any base substituted for A76 showed significant decreases of aminoacylation properties and total loss of the ability to stimulate the editing activity and catalyze deacylation of pre-formed Ile-tRNALeu. As a consequence of this loss of editing activity, two of these mutants (A76U, A76C) were misacylated by Ile. Mutations of the other bases of the CCA76-end had negative effects on the aminoacylation properties, ranging between 2- and 50-fold decreases of the catalytic efficiency of EcLeuRS, with the strongest effect being observed when C75 and C74 were substituted by G75 and G74, respectively. Correspondingly, the mutant A76G was totally inactive, showing that the bulky guanosine substitutions were the least accepted in the synthetic active site. The three guanosine mutants also decreased the deacylation of the mischarged Ile-tRNALeu, but failed in misacylating tRNALeu with Ile. Comparison of the global editing activity of EcLeuRS in the presence of the tRNALeu mutants revealed that four mutations (C75A, C75U, C74A and C74U) led to an increase in the editing up to 2-fold despite the fact that their accepting capacity with Leu were impaired during the aminoacylation reaction. EcLeuRS deacylated these mischarged mutants of tRNALeu with Ile with unchanged ability, suggesting that these mutants of tRNALeu preferred binding at the editing active site rather than at the synthetic site. Previous studies have shown that both classes of aaRSs use effectively the ability of the CCA76-end of tRNA to switch between a hairpin and a helical conformation of aaRSs for aminoacylation and editing. In class I aaRSs, their cognate tRNA CCA76-ends adopt a regular helical conformation into the editing site of aaRSs, and a distorted hairpin conformation into the synthetic site (6). A mirror image is observed in class II aaRSs, as already seen for tRNA binding and amino acid activation. The stimulation of the editing activity of C75A, C75U, C74A and C74U mutants strongly suggests that these mutations may favor the helical conformation, which is more suitable for the editing reaction. Moreover, the fact that these mutated tRNAs are poorly aminoacylated in the synthetic active site may favor stagnation of the non-cognate adenylate into the synthetic active site and its subsequent editing by the tRNA-dependent pre-transfer editing also measured in the global editing assay.
The CCA76-end entrance pathway into editing domain of EcLeuRS critically contributes to protein synthesis fidelity
Despite the importance of the interaction between CCA76-end of tRNA and the CP1 domain of class Ia aaRSs revealed by in vitro methods, the corresponding in vivo evidence is generally lacking. In the present work, we developed an efficient system with the combination of non-cognate Nva and leuS temperature-sensitive E. coli strain to assess the significance of LeuRS tRNA-dependent editing in vivo. Our evaluation of the tRNA CCA76-end entrance pathway using the leuS temperature-sensitive KL231 strain revealed the following aspects. (i) Without or at a low concentration of non-cognate amino acid (e.g. 20 mM Nva here), mutation of pivotal residues controlling protein biosynthesis fidelity had no obvious effect on the bacterial cell viability. The discrimination by EF-Tu and/or the ribosome may have prevented Nva mis-incorporation, or the E. coli cells could endure low level of Nva mis-incorporation at Leu codons; (ii) at extremely high concentrations of non-cognate amino acids, even the WT enzyme had no ability to prevent all the mis-activated amino acids from entering the newly synthesized polypeptide on the ribosome, and the mis-translation led to an obvious inhibitory effect on cell growth; (iii) our in vitro results were consistent with in vivo data, demonstrating the importance of conserved amino acid residues in aaRS and nucleotides in tRNA in protein synthesis quality control, especially under severe environmental stress.
Multiple steps to monitor and control the CCA acceptor sequence
As mentioned above, the trinucleotide CCA76 sequence is present at the 3′ terminus of all mature tRNAs. Despite this high conservation, transcripts of E. coli tRNAVal with altered 3′ termini are readily aminoacylated and can function in polypeptide synthesis (32). Accordingly, the present study performed on LeuRS shows that the aminoacylation reaction admits some sequence flexibility at the CCA76 end. Several CCA-mutated tRNAs can be efficiently aminoacylated, and LeuRS can even aminoacylate a tRNALeu lacking the terminal adenosine, showing a remarkable plasticity of the synthetic site of LeuRS and tRNALeu acceptor end. Several studies suggest that the aminoacylation of similar molecules may occur naturally in vivo. Despite the high fidelity of the CCA-adding enzyme, in some conditions, CCase can add a wrong nucleotide to the 3′-end of the tRNA, leading to CCA-modified tRNAs (39). Moreover, mutations of CCase may exist in vivo with relaxed active sites as those engineered in vitro that lead to nucleotide mis-incorporation at the end of tRNA (40). In some bacteria the CCA-adding activity is naturally split into a CC-adding enzyme and an A-adding enzyme (41). Therefore, a tRNA deprived of the terminal A76 might temporarily exist in vivo and may be the substrate of LeuRS.
However, if under these exceptional circumstances aminoacylation of CCA-mutated tRNAs can occur, these tRNAs would have to face additional discrimination processes based on EF-Tu recognition (42) and interaction with ribosomal RNA during translation (43). Both processes would monitor and exclude these molecules according to different nucleotide recognition specificities (32).
In summary, the study highlighted the significant role of the conserved nucleotides from the CCA76-end and the strong collaboration with amino acid residues of the CP1 editing domain during the aminoacylation and editing catalysis. The CCA76 nucleotides are critical for both aminoacylation and editing; and in combination with the amino acids located in the entrance path of the editing domain, they critically contribute to the fidelity of protein synthesis. It is a remarkable example of a control mechanism that prevent damaged tRNAs from entering the protein synthesis based on the single discrimination of a triplet sequence that is universally found in all tRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China (Nos. 30930022 and 31000355); Committee of Science and Technology in Shanghai (No. 09JC1415900); a Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences (No. 2009S2-19); Programme International de Coopération Scientifique from CNRS (Grant 3606). Funding for open access charge: Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Franck Martin for the help during the experiment.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 3.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 4.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Zhu B, Wang E. The CP2 domain of leucyl-tRNA synthetase is crucial for amino acid activation and post-transfer editing. J. Biol. Chem. 2008;283:36608–36616. doi: 10.1074/jbc.M806745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Guo N, Li T, Wang E, Wang Y. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 8.Tan M, Zhu B, Zhou X, He R, Chen X, Eriani G, Wang E. tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 2010;285:3235–3244. doi: 10.1074/jbc.M109.060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Ma J, Tan M, Yao P, Hu Q, Eriani G, Wang E. Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 2011;39:235–247. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulic M, Cvetesic N, Perona JJ, Gruic-Sovulj I. Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in Class I aminoacyl-tRNA synthetases. J. Biol. Chem. 2010;285:23799–23809. doi: 10.1074/jbc.M110.133553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 12.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan B, Xiong Y, Steitz TA. How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA. Science. 2010;330:937–940. doi: 10.1126/science.1194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou YM. Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA. 2000;6:1031–1043. doi: 10.1017/s1355838200000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 16.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Schimmel P. Trial for peptide bond formation using model molecules based on the interactions between the CCA sequence of tRNA and 23S rRNA. Nucleic Acids Symp. Ser. 2000;44:251–252. doi: 10.1093/nass/44.1.251. [DOI] [PubMed] [Google Scholar]
- 18.Low B, Gates F, Goldstein T, Söll D. Isolation and partial characterization of temperature-sensitive Escherichia coli mutants with altered leucyl- and seryl-transfer ribonucleic acid synthetases. J. Bacteriol. 1971;108:742–750. doi: 10.1128/jb.108.2.742-750.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Wang E, Wang Y. A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif. 1999;16:355–358. doi: 10.1006/prep.1999.1083. [DOI] [PubMed] [Google Scholar]
- 20.Xu M, Li J, Du X, Wang E. Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem. Biophys. Res. Commun. 2004;318:11–16. doi: 10.1016/j.bbrc.2004.03.180. [DOI] [PubMed] [Google Scholar]
- 21.Yao P, Zhu B, Jaeger S, Eriani G, Wang E. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008;36:2728–2738. doi: 10.1093/nar/gkn028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorber B, Kern D, Gieg R, Ebel JP. Covalent attachment of aspartic acid to yeast aspartyl-tRNA synthetase induced by the enzyme. FEBS Lett. 1982;146:59–64. doi: 10.1016/0014-5793(82)80705-9. [DOI] [PubMed] [Google Scholar]
- 23.Rapaport E, Yogeeswaran G, Zamecnik PC, Remy P. Covalent modification of phenylalanyl-tRNA synthetase with phenylalanine during the amino acid activation reaction catalyzed by the enzyme. J. Biol. Chem. 1985;260:9509–9512. [PubMed] [Google Scholar]
- 24.Kovaleva GK, Moroz SG, Favorova OO, Kisselev LL. Tryptophanyl-tRNA synthetase: evidence for an anhydrous bond involved in the tryptophanyl enzyme formation. FEBS Lett. 1978;95:81–84. doi: 10.1016/0014-5793(78)80056-8. [DOI] [PubMed] [Google Scholar]
- 25.Gruic-Sovulj I, Uter N, Bullock T, Perona JJ. tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting Class I aminoacyl-tRNA synthetase. J. Biol. Chem. 2005;280:23978–23986. doi: 10.1074/jbc.M414260200. [DOI] [PubMed] [Google Scholar]
- 26.Zhu B, Yao P, Tan M, Eriani G, Wang E. tRNA-independent pretransfer editing by Class I leucyl-tRNA synthetase. J. Biol. Chem. 2009;284:3418–3424. doi: 10.1074/jbc.M806717200. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Wang E, Wang Y. Overproduction and purification of Escherichia coli tRNALeu. Sci. China, C, Life Sci. 1998;41:225–231. doi: 10.1007/BF02895095. [DOI] [PubMed] [Google Scholar]
- 28.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grøli M, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Li Y, Guo N, Wang E, Wang Y. Discrimination of tRNALeu isoacceptors by the insertion mutant of Escherichia coli leucyl-tRNA synthetase. Biochemistry. 1999;38:9084–9088. doi: 10.1021/bi9901984. [DOI] [PubMed] [Google Scholar]
- 30.Schurer H, Schiffer S, Marchfelder A, Morl M. This is the end: processing, editing and repair at the tRNA 3′-terminus. Biol. Chem. 2001;382:1147–1156. doi: 10.1515/BC.2001.144. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Nameki N, Hasegawa T, Shimizu M, Himeno H. Role of the CCA terminal sequence of tRNA(Val) in aminoacylation with valyl-tRNA synthetase. J. Biol. Chem. 1994;269:22173–22177. [PubMed] [Google Scholar]
- 32.Liu M, Horowitz J. Functional transfer RNAs with modifications in the 3′-CCA end: differential effects on aminoacylation and polypeptide synthesis. Proc. Natl Acad. Sci. USA. 1994;91:10389–10393. doi: 10.1073/pnas.91.22.10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eriani G, Gangloff J. Yeast aspartyl-tRNA synthetase residues interacting with tRNA(Asp) identity bases connectively contribute to tRNA(Asp) binding in the ground and transition-state complex and discriminate against non-cognate tRNAs. J. Mol. Biol. 1999;291:761–773. doi: 10.1006/jmbi.1999.3012. [DOI] [PubMed] [Google Scholar]
- 34.Tardif KD, Horowitz J. Transfer RNA determinants for translational editing by Escherichia coli valyl-tRNA synthetase. Nucleic Acids Res. 2002;30:2538–2545. doi: 10.1093/nar/30.11.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Splan KE, Ignatov ME, Musier-Forsyth K. Transfer RNA modulates the editing mechanism used by class II prolyl-tRNA synthetase. J. Biol. Chem. 2008;283:7128–7134. doi: 10.1074/jbc.M709902200. [DOI] [PubMed] [Google Scholar]
- 36.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of non-cognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 2007;581:5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 37.Minajigi A, Francklyn CS. Aminoacyl transfer rate dictates choice of editing pathway in threonyl-tRNA synthetase. J. Biol. Chem. 2010;285:23810–23817. doi: 10.1074/jbc.M110.105320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dulic M, Cvetesic N, Perona JJ, Gruic-Sovulj I. Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in class I aminoacyl-tRNA synthetases. J. Biol. Chem. 2010;285:23799–23809. doi: 10.1074/jbc.M110.133553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou YM. Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA. 2000;6:1031–1043. doi: 10.1017/s1355838200000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho HD, Verlinde CL, Weiner AM. Reengineering CCA-adding enzymes to function as (U,G)- or dCdCdA-adding enzymes or poly(C,A) and poly(U,G) polymerases. Proc. Natl Acad. Sci. USA. 2007;104:54–59. doi: 10.1073/pnas.0606961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomita K, Weiner AM. Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science. 2001;294:1334–1336. doi: 10.1126/science.1063816. [DOI] [PubMed] [Google Scholar]
- 42.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 43.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.