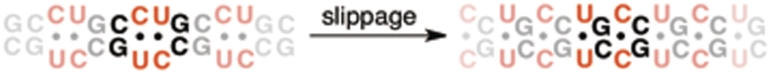Abstract
Myotonic dystrophy type 2 (DM2) is an incurable neuromuscular disease caused by expanded CCUG repeats that may exhibit toxicity by sequestering the splicing regulator MBNL1. A series of triaminotriazine- and triaminopyrimidine-based small molecules (ligands 1–3) were designed, synthesized and tested as inhibitors of the MBNL1–CCUG interaction. Despite the structural similarities of the triaminotriazine and triaminopyrimidine units, the triaminopyrimidine-based ligands bind with low micromolar affinity to CCUG repeats (Kd ∼ 0.1–3.6 µM) whereas the triaminotriazine ligands do not bind CCUG repeats. Importantly, these simple and small triaminopyrimidine ligands exhibit both strong inhibition (Ki ∼ 2 µM) of the MBNL1–CCUG interaction and high selectivity for CCUG repeats over other RNA targets. These experiments suggest these compounds are potential lead agents for the treatment of DM2.
INTRODUCTION
Myotonic dystrophy (DM) is an RNA-mediated disorder that affects both skeletal and smooth muscles as well as internal organs such as the heart, lung and eye lens (1). Two major types of DM have been identified: types 1 (DM1) and 2 (DM2). It was discovered that both DM1 and DM2 are caused by unstable nucleotide repeat sequences in DNA. DM1 is caused by expansion of the CTG repeat (from 80 to >2000 repeats) in the 3′-untranslated region (3′-UTR) of the dystrophia myotonin protein kinase (DMPK) gene on chromosome 19 (2), whereas DM2 is caused by expansion of the CCTG (from 75 to 11 000 repeats) in intron 1 of the ZNF9 (zinc-finger protein 9) gene on chromosome 3 (3). The corresponding expanded transcripts, r(CUG)n and r(CCUG)n for DM1 and DM2, respectively, form inclusion complexes that sequester muscleblind-like (MBNL) proteins in the nucleus. A member of a family of structurally and functionally similar proteins, MBNL is expressed at high level in skeletal muscles and is known to be directly involved in causing DM. MBNL1 is involved in the regulation of RNA splicing, editing and translation (4). The sequestration of MBNL1 proteins results in misregulated, alternative splicing of several pre-mRNAs, including the cardiac troponin T (cTNT), fast skeletal troponin T (Tnnt3), insulin receptor (IR) and chloride channel (ClC-1) pre-mRNAs. It is proposed that these errors in alternative splicing, in turn, cause the multisystemic clinical features of DM (5). It has been suggested that agents that bind selectively to the pathogenic RNAs might reverse the phenotype by inhibiting MBNL1 protein from binding, thereby allowing the protein and mRNAs to function normally (6,7). Indeed, antisense oligonucleotides that target the CUG repeat were shown to reverse the phenotype of DM1 in a mouse model (8,9).
With the knowledge that disrupting the MBNL1–CUG repeat complex might provide a therapeutic strategy to treat DM1, recent efforts have focused on small molecules that target CUG repeats (10–13). An ideal candidate should also reverse the splicing defects observed in DM. For example, pentamidine is capable of rescuing four different pre-mRNAs affected by CUG repeats as well as displacing MBNL1 from CUG ribonuclear foci (10). In this regard, we have developed a simple, low molecular weight ligand (ligand 1) that exhibits high selectivity in binding the CUG base triplet and, further, inhibits the MBNL1–CUG interaction with micromolar IC50 values (Figures 1 and 2) (12). As an alternative-splicing regulator, MBNL1 binds to other stem–loop structures containing pyrimidine–pyrimidine mismatches, including the pre-mRNAs discussed above. Therefore, it is essential to develop ligands that bind selectively to the pathogenic RNAs in preference to the natural targets of the MBNL1 and other nucleic acids. Indeed, achieving RNA and sequence selectivity is particularly difficult given the presence of a large amount of genomic DNA in the nucleus. The majority of work to date has focused on DM1, however recently, Disney and coworkers (14) reported oligomeric ligands that target expanded CCUG repeats in DM2. Specifically, a trimeric ligand with three kanamycin A units was shown to bind strongly to CCUG RNA and inhibit the MBNL1–CCUG interaction. A subsequent report showed that the flexibility of the ligand backbone plays a critical role in determining affinity and selectivity (15). Despite these important advances, alternative approaches are likely needed given the challenges in developing clinically useful therapeutic agents.
Figure 1.
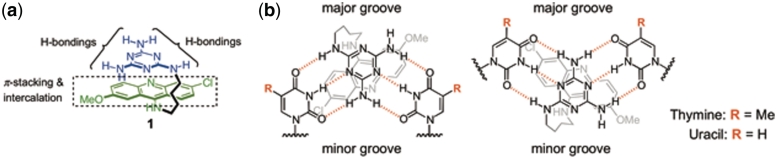
(a) Design principle of ligand 1 and (b) two possible binding modes with different groove preference of ligand 1 binding to T–T/U–U mismatch.
Figure 2.
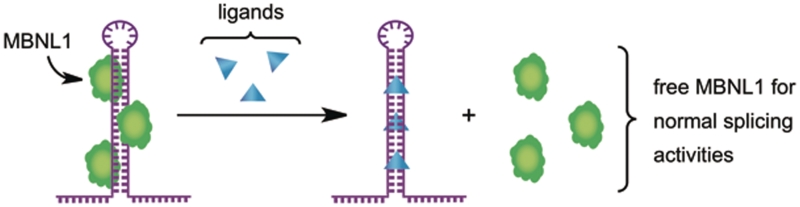
Ligand 1 inhibits the MBNL1–CUG interaction by binding to the U–U mismatches of the pathogenic RNA. MBNL1 is released and resumes its alternative splicing functions for more than 12 known pre-mRNAs.
In examining the interaction of CUG repeats with ligand 1, the triaminotriazine moiety was found to be important for the selective binding of U–U mismatches, whereas the acridine unit provided binding affinity for the CUG RNA (12). Stacking of the two heterocyclic units, which was observed by 1H NMR, is likely to increase the mismatch selectivity by significantly reducing the ability of the acridine to function as an unselective RNA intercalator. Although the binding mode is not known, ligand 1 was designed to intercalate between the U–U and G–C base pairs and simultaneously form a base triplet with the triaminotriazine unit inserted between the U–U mismatch (Figure 1). Therefore, small molecule ligands structurally similar to ligand 1 (IC50 = 43 µM for MBNL1–CUG) were explored. Herein, we report on efforts toward a similar approach, to develop π-stacked intercalators that specifically recognize CCUG repeats and inhibit the MBNL1–CCUG interaction.
MATERIALS AND METHODS
Materials
RNA oligonucleotides were purchased from Dharmacon RNAi Technologies, Inc. with 2′-deprotection, desalting and HPLC purification. DNA sequences were purchased from Integrated DNA Technologies, Inc. with standard desalting. The RNA oligonucleotides were dissolved in Ambion RNA Storage Solution (pH 6.4) to give 1–2 mM stock solutions. DNA sequences were dissolved in TE buffer (pH 7.6) to give 1–2 mM stock solutions. The oligonucleotide concentration was determined by its absorbance at λmax = 260 nm at 25°C on a Shimadzu UV-2501PC spectrophotometer. The concentration of each single-stranded sequence was calculated using Beer's law with extinction coefficient (ε260) provided by the supplier. RNA and DNA duplexes were freshly prepared by mixing the required volumes of the corresponding ssRNA and ssDNA, respectively. The solution was annealed by heating in a water bath at 95°C for 2 min and slow cooling to 25°C. Final duplex solutions were prepared by adding MOPS buffer (100 mM; pH 7.0), NaCl solution (5.0 M) and diluting to the required concentrations by adding AccuGENE Molecular Biology Grade Water and 1 or 10% (v/v) DMSO to give 20 mM MOPS and 300 mM NaCl solutions.
Protein expression and purification
An expression vector for a truncated MBNL1 comprised of amino acids 1–272 was obtained from Maurice S. Swanson (University of Florida, College of Medicine, Gainesville, FL, USA). This MBNL1 construct is comprised of the four zinc-finger motifs of MBNL1 and a hexahistidine tag (C-terminus) and binds RNA with similar affinity as the full-length MBNL1. The protein was expressed and purified as described previously (16). The molecular weight was confirmed by MALDI-TOF mass spectrometry, the concentration was determined by Bradford protein assay (Bio-Rad), and the purity determined by silver-stained SDS–PAGE.
Electrophoretic mobility shift assay
RNAs were labeled with [γ-32P]-ATP using T4 polynucleotide kinase (New England Biolabs) and were purified by phenol extraction and precipitation from ethanol. Labeled RNA (5 µl, 5 nM) was heated at 95°C for 2 min, placed on ice for 10 min and diluted to 125 µl in RNA storage buffer [66 mM NaCl, 6.7 mM MgCl2, 27 mM Tris–HCl (pH 7.5)]. For assays with competitor tRNA, bulk yeast tRNA was added to the RNA solution to give a final concentration of ∼100 nM. MBNL1 was serially diluted in binding buffer [175 mM NaCl, 5 mM MgCl2, 20 mM Tris–HCl (pH 7.5), 1.25 mM 2-mercaptoethanol (BME), 12.5% glycerol, 2 mg/ml bovine serum albumin (BSA), 0.1 mg/ml heparin]. Protein (5 µl) and RNA (5 µl) solutions were combined. The reaction mixture was incubated at room temperature for 25 min and loaded onto a 6% polyacrylamide gel (80 : 1) at 4°C. The gels were run for 1 h at 360 V in 22.5 mM Tris–borate buffer (pH 8). Gels were visualized on a Molecular Dynamics Storm PhosphorImager. The apparent Kd values (assuming a 1:1 stoichiometry) were obtained by fitting fraction RNA bound versus protein concentration using the following equation: fraction RNA bound = 1/[1 + (Kd/[protein]total)]. All measurements were performed with >10-fold excess of protein over RNA so that [protein] would be approximately equal to [protein]total.
Inhibition assays using gel electrophoretic mobility shift assay
The inhibition of the MBNL1–RNA complex was investigated using the above procedure, but with addition of the small molecule to the RNA–protein complex after 25 min of incubation. The reaction mixture was incubated for an additional 10–15 min at room temperature. The inhibition assays were performed in the presence of 1 or 10% (v/v) DMSO (depending on the solubility of ligand). The IC50 values were determined by fitting to the equation: B = ΔB exp(−0.69[ligand]/IC50) + Binitial, where B is the radioactivity (cpm) of the RNA, ΔB = Bfinal − Binitial, [ligand] is the ligand concentration, IC50 is the concentration of ligand, at which (F = 0.5ΔB + Binitial). The apparent inhibition constant (Ki) was determined using the equation: Ki = IC50 × (Kd/[protein]total), where Kd is the dissociation constant of the MBNL1–RNA complex and [protein] is at least 7-fold greater than the Kd.
Thermal denaturation studies
The melting temperatures of the duplexes were measured on a Shimadzu UV-2501PC spectrophotometer equipped with a temperature controller. The absorbance of each RNA or DNA solution (10 µM duplex) with 20 mM MOPS (pH 7.0), 300 mM NaCl and 10 mM EDTA was annealed and cooled to 0°C. The absorbance of each sample was monitored at 260 nm from 0°C to 90°C at a ramp rate of 1°C/min. The melting temperature (Tm) of each sample was determined from the maximum point of the first derivative of the melting curve with Origin 7.0.
Isothermal titration calorimetry studies
Isothermal titration calorimetry (ITC) measurements were performed at 25°C on a MicroCal VP-ITC calorimeter. A typical experiment consisted of titrating 10 µl of a ligand solution (500 µM) from a 250 µl syringe (stirred at 300 rpm) into a sample cell containing 1.42 ml of a RNA or DNA solution (10–20 µM) with a total of 30 injections (1 µl for the first injection and 10 µl for the remaining injections). The initial delay prior to the first injection was 60 s. The duration of each injection was 24 s and the delay between injections was 400 s. All experiments were performed in triplicate. Data analysis was carried out with Origin 5.0 software (MicroCal). Binding parameters, such as the dissociation constant (Kd), enthalpy change (ΔH) and entropy change (ΔS), were determined by fitting the experimental binding isotherms with appropriate models. The ligand stock solution was 50 mM in DMSO. The buffer solution for ITC experiments was MOPS (20 mM; pH 7.0), NaCl (300 mM) and 1% (v/v) DMSO to balance the DMSO in the ligand solution.
Synthetic procedures
1H and 13C NMR spectra for structural characterization were recorded on a Varian Unity Inova 500 spectrometer (1H: 500 MHz; 13C: 125 MHz). Unless otherwise stated, all NMR measurements were carried out in CDCl3 at room temperature. Chemical shifts were reported as parts per million (ppm) in scale using residual peaks of DMSO-d6 (1H: 2.50; 13C: 39.52) as internal standard. Coupling constants (J) were reported in Hertz. Mass spectra were obtained by the Mass Spectrometry Laboratory, School of Chemical Sciences, University of Illinois.
All non-aqueous reactions were carried out under dry N2 atmosphere with oven-dried (115°C) glassware. Unless otherwise stated, all solvents and reagents were of reagent quality, purchased commercially and used without further purification. The progress of reaction was monitored by thin layer chromatography (TLC) performed on Merck pre-coated silica gel 60F254 plates, and compounds were visualized by UV and subsequently with a spray of 5% (w/v) phosphomolybdic acid hydrate in ethanol with subsequent heating. Flash chromatography was carried out on columns of Macherey-Nagel MN Kieselgel 60 M (230–400 mesh) silica gel or Sigma-Aldrich Brockmann I, standard grade (∼150 mesh) alumina.
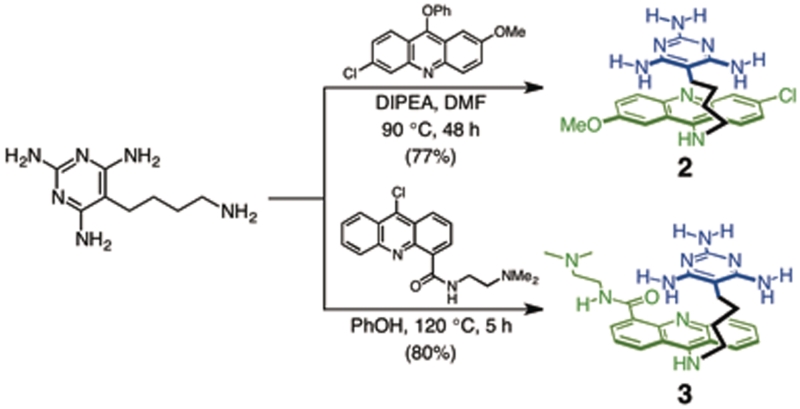
Ligand 2
A mixture of 163 mg (0.830 mmol) of 5-(4-aminobutyl)-2,4,6-triaminopyrimidine (17), 334 mg (0.996 mmol) of 6-chloro-2-methoxy-9-phenoxyacridine and 0.17 ml (0.98 mmol) of DIPEA in 6 ml of DMF was heated to 90°C in an oil bath for 48 h. The reaction mixture was cooled to room temperature and concentrated in vacuo. The residue was purified by flash chromatography on alumina using CH2Cl2 : MeOH mixture (9 : 1 gradient to 4 : 1) to afford 279 mg (0.638 mmol, 77%) of ligand 2 as a yellow solid. 1H NMR (DMSO-d6): δ 8.36 (d, J = 9.5 Hz, ArH, 1H), 7.87 (d, J = 2 Hz, ArH, 1H), 7.83 (d, J = 9.5 Hz, ArH, 1H), 7.60 (d, J = 2 Hz, ArH, 1H), 7.40 (dd, 2.5 and 9.5 Hz, ArH, 1H), 7.32 (dd, 2 and 9 Hz, ArH, 1H), 6.91 (t, J = 5.5 Hz, AcrNH, 1H), 5.40 (s, NH2, 4H), 5.11 (s, NH2, 2H), 3.92 (s, OCH3, 3H), 3.73 (q, J = 7 Hz, NHCH2, 2H), 2.21 (t, J = 7.5 Hz, PymCH2, 2H), 2.78 (quin, J = 7.5 Hz, CH2, 2H) and 1.37 (quin, J = 8 Hz, CH2, 2H). 13C NMR (DMSO-d6): δ 162.0, 160.7, 155.0, 150.4, 148.2, 146.2, 133.4, 130.8, 127.2, 126.5, 124.2, 122.6, 116.9, 114.5, 100.6, 85.6, 55.7, 49.6, 30.4, 25.4 and 22.8. ESI-MS (m/z): 438.2 ([M + H]+, 100%). ESI-HRMS (m/z): [M + H]+ calcd for C22H25N7OCl, 438.1809, found, 438.1814.
Ligand 3
A mixture of 72.3 mg (0.369 mmol) of 5-(4-aminobutyl)-2,4,6-triaminopyrimidine, 145 mg (0.442 mmol) of 9-chloro-N-[2-(dimethylamino)ethyl]-4-acridinecarboxamide (18) and 1.80 g (19.1 mmol) of phenol was heated to 120°C in an oil bath for 5 h. The reaction mixture was cooled to room temperature and diluted with 150 ml of 2 M NaOH solution. The suspension was stirred for 10 min at 0°C. The solid was filtered and purified by flash chromatography on alumina using CH2Cl2 : MeOH mixture (9 : 1 gradient to 4 : 1), to afford 148 mg (0.304 mmol, 80%) of ligand 3 as a yellow solid. 1H NMR (DMSO-d6): δ 12.46 (t, J = 4.5 Hz, CONH, 1H), 8.61 (dd, J = 1 and 7 Hz, ArH, 1H), 8.56 (d, J = 8.5 Hz, ArH, 1H), 8.43 (d, J = 8.5 Hz, ArH, 1H), 7.96 (d, J = 9 Hz, ArH, 1H), 7.76 (t, J = 7.5 Hz, ArH, 1H), 7.52 (s, AcrNH, 1H), 7.45–7.37 (m, ArH, 2H), 5.43 (s, NH2, 4H), 5.13 (s, NH2, 2H), 3.86 (t, J = 6.5 Hz, CONHCH2, 2H), 3.57 (q, J = 5.5 Hz, AcrNHCH2, 2H), 2.55 (t, J = 6 Hz, PymCH2, 2H), 2.31 [s, N(CH3)2, 6H], 2.21 (quin, J = 7.5 Hz, 2H), 1.82 (quin, J = 7 Hz, CH2, 2H), 1.37 (quin, J = 8 Hz, CH2, 2H). 13C NMR (DMSO-d6): δ 165.2, 162.0, 160.7, 153.2, 147.7, 146.7, 133.8, 130.9, 128.5, 128.2, 127.3, 124.2, 122.5, 120.7, 115.6, 114.7, 85.5, 58.0, 49.8, 45.2, 37.2, 30.1, 25.3 and 22.8. ESI-MS (m/z): 488.3 ([M + H]+, 100%). ESI-HRMS (m/z): [M + H]+ calcd for C26H34N9O, 488.2886, found, 488.2888. Anal. calcd for C26H37N9O3·2H2O: C, 59.64, H, 7.12, N, 24.07. Found: C, 60.87, H, 6.85, N, 23.97.
RESULTS
Binding of ligand 1 to CCTG and CCUG sequences
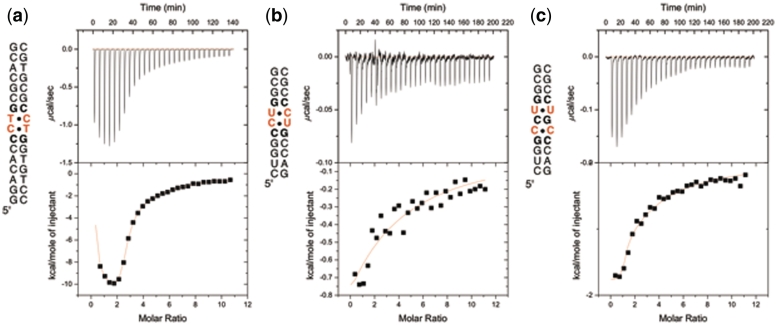
Binding studies of ligand 1 were initially performed with a DNA duplex containing a single (CCTG)2 step (Figure 3a). Using ITC, it was found that ligand 1 binds strongly to the CCTG duplex with a 3 : 1 stoichiometry and Kd(1) = 14 µM, Kd(2) = 0.11 µM and Kd(3) = 37 µM. Job analysis and circular dichroism titration studies support the 3 : 1 stoichiometry (see Supplementary Data). The binding to RNA was examined using the 11-mer shown in Figure 3b. In contrast to the results with the DNA oligonucleotide, ligand 1 binds CCUG comparatively weakly; a single Kd = 79 µM was obtained. The CCUG repeat is capable of forming a slipped structure containing U–U and C–C mismatches separated by C–G base pairs as shown in Figure 4 (19). For this reason, the binding of ligand 1 to the slipped sequence was also investigated by ITC using the 11-mer shown in Figure 3c. A 2 : 1 complex was observed with low-micromolar Kd values: Kd(1) = 3.5 µM and Kd(2) = 7.0 µM (Figure 3c).
Figure 3.
ITC binding studies of ligand 1 using (a) a DNA sequence containing a CCTG repeat, (b) an RNA sequence containing a CCUG repeat (RNA A) and (c) an RNA containing a slipped-CCUG motif (RNA B).
Figure 4.
Two possible stem–loop structures for CCUG repeats with two consecutive C–U mismatches (left) or alternating C–C and U–U mismatches in the slipped form (right).
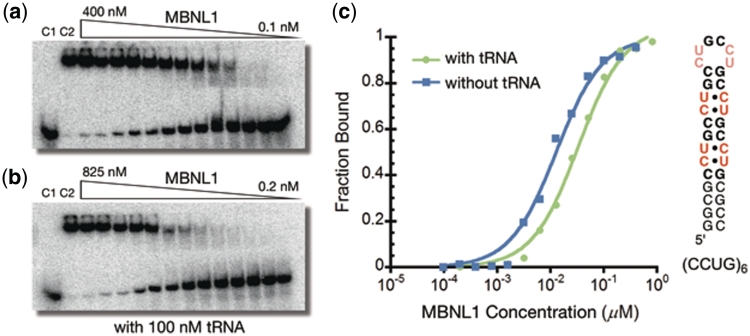
To determine whether ligand 1 can inhibit the MBNL1–CCUG interaction, an electrophoretic mobility shift assay was performed using a (CCUG)6 RNA. The Kd values for the complexes formed between (CCUG)6 and MBNL1 were determined in the absence and presence of 100 nM of competitor tRNA to be 15 ± 2 nM and 30 ± 4 nM, respectively (Figure 5). These values are in good agreement with literature values (19). No significant inhibition of the MBNL1–RNA interaction was observed with ligand 1 up to a concentration of 250 µM (see Supplementary Data).
Figure 5.
(a) Gel electrophoretic mobility shift assay of MBNL1 binding to (CCUG)6 RNA. (b) MBNL1 binding to (CCUG)6 in the presence of 100 nM tRNA. Control lane 1 (C1): RNA only. Control lane 2 (C2): RNA + MBNL1. (c) Plots of fractions of (CCUG)6 bound as a function of MBNL1 concentration in the presence (green) and absence (blue) of 100 nM tRNA.
Development of ligands 2 and 3
The high affinity of ligand 1 for U–U mismatches and moderate affinity for C–C mismatches led us to explore similar ligands that recognize C–U mismatches in CCUG repeats. To this end, a structurally similar analog 2 was synthesized and studied (Scheme 1). The ability of ligand 2 to bind CCUG and CCTG repeats was examined by ITC, however, these studies were hampered by poor water solubility. To achieve the high ligand concentration needed for ITC studies, ligand 2 required more than 10% (v/v) DMSO. With as little as 1% (v/v) DMSO, a 500 µM solution of ligand 3 could be prepared in a 300 mM NaCl solution (MOPS buffer; pH 7), presumably because it contains a water-solubilizing substituent. Because experiments with ligand 2 were performed in the presence of 10% DMSO, the melting temperature (Tm) of (CCUG)6 was measured in the presence and absence of 10% DMSO. The stability of the RNA was found to be identical within experimental error in the presence of 1–10% DMSO. In addition, the Tm of (CCUG)6 was concentration independent suggesting the formation of a hairpin not a duplex. The hydrolytic stability of (CCUG)6 RNA in the presence of ligands 2 and 3 was also evaluated. The (CCUG)6 RNA was incubated with ligands 2 and 3 for at least 4 h at room temperature and the RNA was evaluated by denaturing polyacrylamide gel electrophoresis (see Supplementary Data). No cleavage of the RNA was observed under these conditions. Therefore, the RNA is stable to hydrolytic cleavage in all of the binding and inhibition experiments reported herein.
Scheme 1.
Synthesis of ligands 2 and 3.
Affinity and selectivity of ligand 3 for RNA
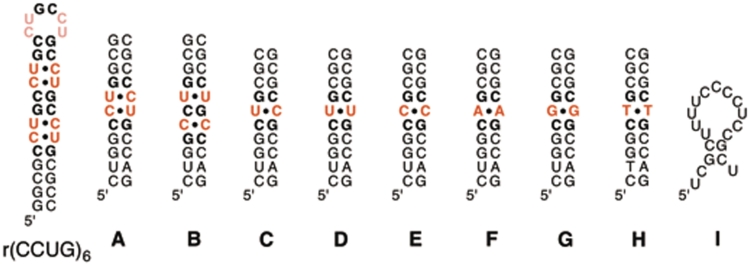
The affinity and selectivity of ligand 3 for binding to CCUG RNA was evaluated by measuring the affinity of ligand 3 for several oligonucleotides (Figure 6). The stabilities of these RNAs were evaluated by melting curves obtained by UV spectroscopy and are reported in Table 1. The duplexes containing C–U (RNA C), U–U (RNA D), C–C (RNA E), A–A (RNA F) and G–G (RNA G) were designed to evaluate the ability of ligand 3 to selectively recognize the mismatches that are most likely found in the poly(CCUG) RNA compared to related mismatched sequences. The binding of ligand 3 to the DNA analog of the DM1 sequence containing a T–T mismatch (DNA H) and the 18-nt fragment of the human cTNT (hcTNT) pre-mRNA, a natural target of MBNL1 (RNA I) was also evaluated. To investigate the non-specific binding of ligand 3 to nucleic acids in general, its affinity toward tRNA and herring sperm DNA (hsDNA) was measured (Table 1).
Figure 6.
Nucleic acids studied in ITC experiments (tRNA and herring sperm DNA are not shown). The structures shown represent the most stable structures predicted by m-fold. Note that RNA I is most likely single stranded under the conditions used to perform the binding or complex inhibition experiments.
Table 1.
Dissociation constants (Kd) and Tm values for all tested nucleic acids with ligand 3a
| Nucleic acids | Specific target | Tm (°C)b | Kd(1) | Kd(2) | Kd(3) | Selectivityc |
|---|---|---|---|---|---|---|
| (CCUG)6 | C–U pairsd | 61 | 5.2 ± 0.6 | 478 ± 85 | – | 1.4 |
| A | C–U pairs | 53 | 3.6 ± 0.8 | 258 ± 37 | – | 1.0 |
| B | C–C and U–U | 54 | 0.13 ± 0.04 | 10 ± 5 | 202 ± 14 | 0.04 |
| C | Single C–U pair | 62 | 85 ± 30 | 231 ± 55 | – | 24.0 |
| D | U–U | 63 | 25 ± 13 | 271 ± 83 | – | 6.9 |
| E | C–C | 62 | 0.77 ± 0.28 | 162 ± 8 | – | 0.2 |
| F | A–A | 62 | 118 ± 8 | 2270 ± 360 | – | 33.0 |
| G | G–G | 67 | 103 ± 26 | 7690 ± 280 | – | 29.0 |
| H | T–T | 51 | 1.4 ± 1.5 | 102 ± 25 | – | 0.4 |
| I | – | – | 334 ± 11 | – | – | 93.0 |
| tRNA | – | – | 2400 ± 2100 | – | – | 670.0 |
| hsDNA | – | – | 36 ± 7e | – | – | 10.0 |
aKd values are reported in µM. bConcentrations of duplexes are 10 µM; additional 1% (v/v) DMSO has negligible effect on the stability of the duplexes (ΔTm < 2°C). cSelectivity is the ratio of Kd(1) of the particular nucleic acid to that of RNA A. dm-fold predicts two 2 × 2 nt internal loops. eLigand 3 bound to about every 10 bp.
ITC was used to measure the affinity of ligand 3 for the duplexes (Table 1). For (CCUG)6 RNA, ligand 3 bound with low micromolar affinity Kd(1) = 5.2 ± 0.6 µM and a significantly weaker Kd(2) = 478 ± 85 µM. Ligand 3 binds to RNA A, which contains a single CCUG site, i.e. two proximal U–C mismatches (RNA A), similarly, exhibiting a low micromolar affinity Kd(1) = 3.6 ± 0.8 µM and Kd(2) = 258 ± 37 µM. The affinity for the slipped-CCUG structure (RNA B) was significantly higher, with first and second Kd values of 0.13 ± 0.04 µM and 10 ± 5 µM, respectively. For comparison purposes, the Kd values for the duplexes containing a single C–U (RNA C, Kd = 85 ± 30 µM), U–U (RNA D, Kd = 25 ± 13 µM) and C–C mismatches (RNA E, Kd = 0.77 ± 0.28 µM) were measured.
Completing the picture, the binding of ligand 3 was measured to duplexes F and G containing A–A and G–G mismatches, respectively, and to DNA H with a CTG site relevant in DM1 (20). The purine–purine mismatches were bound weakly by ligand 3 (Kd > 100 µM) whereas the CTG sequence (DNA H), was bound with a low micromolar affinity (Kd = 1.4 ± 1.5 µM). Ligand 3 was found to bind weakly (Kd = 334 ± 11 µM) to the 18-nt fragment of hcTNT pre-mRNA (RNA I) and tRNA (Kd = 2400 ± 2100 µM). Finally, ligand 3 bound herring sperm DNA with Kd = 36 ± 7 µM.
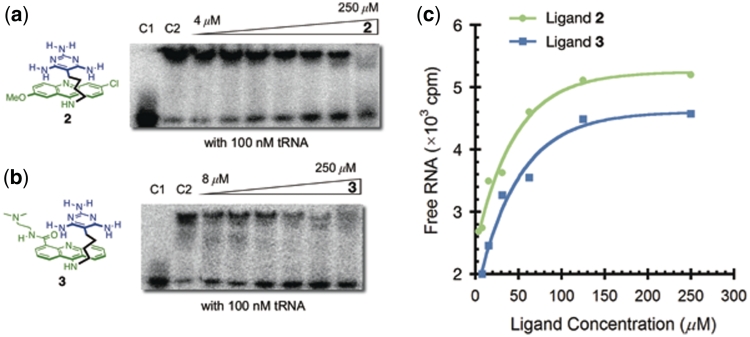
Inhibition of the MBNL1–CCUG complex by ligands 2 and 3
Despite the limited solubility of ligand 2 under the ITC conditions, its ability to inhibit MBNL1 binding to RNA could be determined because of the lower concentrations required for gel electrophoretic mobility shift assays. Both ligands 2 and 3 were found to significantly destabilize the MBNL1–CCUG complex. The IC50 of ligands 2 and 3 were determined by incubating varying concentrations of the ligands with a constant concentration of MBNL1–CCUG complex and evaluating the results using gel shift assays. The Ki of these ligands was determined using the equation Ki = IC50 × (Kd/[protein]total), in which Kd is the dissociation constant of the MBNL1–RNA complex and the concentration of MBNL1 is in large excess of the Kd value. The comparison of Ki values, rather than the IC50 values, under different conditions is important in order to compare the effects of the conditions on the behavior of the ligands independently of the effects of the varying conditions on the Kd of the MBNL1–CCUG complex. Importantly, ligands 2 and 3 do not bind to MBNL1 as measured by ITC.
To evaluate the specificity of ligands 2 and 3 for CCUG RNA, the inhibition experiments were performed in the presence of competitor RNA sequences. For ligand 2, the Ki value is increased only slightly to 3.4 ± 0.9 µM in the presence of 100 nM tRNA (Figure 7a and c). Evaluation of ligand 3 provided IC50 values of 52 ± 8 µM and 35 ± 3 µM in the absence and presence of 100 nM tRNA, respectively, with a Ki of 2.2 ± 0.3 µM and 2.8 ± 0.3 µM, respectively (Figure 7b and c). The selectivity of ligands 2 and 3 for the target sequences (CCUG)6 relative to (CUG)12 (i.e. DM2 versus DM1) and their relative abilities to inhibit MBNL1 binding to the same sequences were also evaluated. Ligand 2 inhibited binding to (CCUG)6 and (CUG)12 with similar Ki values of 2.4 and 4.8 µM, respectively, whereas ligand 3 exhibited a ∼7-fold higher inhibition of the MBNL1–CCUG interaction compared to the MBNL1–CUG interaction. The ability of ligand 3 to destabilize complexes formed between MBNL1 and cTNT (RNA I, Figure 6) was also evaluated. cTNT is the 18-nt fragment (19) of the hcTNT pre-mRNA, which is a natural target of MBNL1. Ligand 3 only weakly inhibited the MBNL1–cTNT interaction. Table 2 summarizes the inhibition studies of ligands 2 and 3.
Figure 7.
(a) Gel electrophoretic mobility shift assay of ligand 2 with (CCUG)6 RNA in the presence of 100 nM tRNA. Control lane 1 (C1): RNA only. Control lane 2 (C2): RNA + MBNL1. (b) Gel electrophoretic mobility shift assay of ligand 3 with (CCUG)6 RNA in the presence of 100 nM tRNA. Control lane 1 (C1): RNA only. Control lane 2 (C2): RNA + MBNL1. (c) Plots illustrating inhibition of MBNL1–(CCUG)6 complex with ligand 2 (green) and 3 (blue).
Table 2.
Summary of inhibition studies of various RNAs with ligands 2 and 3
| RNAa | tRNA (nM) | Kd (nM) | [MBNL1] (nM) |
2 |
3 |
||
|---|---|---|---|---|---|---|---|
| IC50 (µM) | Ki (µM) | IC50 (µM) | Ki (µM) | ||||
| (CCUG)6 | – | 15 ± 2 | 350 | 59 ± 5 | 2.5 ± 0.2 | 52 ± 8 | 2.2 ± 0.3 |
| 100 | 30 ± 4 | 350 | 40 ± 11 | 3.4 ± 0.9 | 35 ± 3 | 3.0 ± 0.3 | |
| (CUG)12 | – | 150 ± 20 | 1000 | 37 ± 11 | 5.5 ± 1.4 | 118 ± 40 | 18 ± 5 |
| cTNTb(I) | – | 1.0 ± 0.2 | 15 | 101 ± 26 | 6.7 ± 1.6 | >250 | >17 |
aRNA concentrations were 0.1 nM. b18-nt fragment of the cTNT pre-mRNA (RNA I in Figure 6).
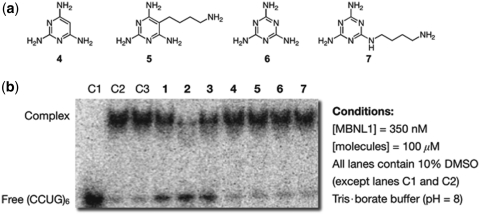
Finally, the ability of simple triaminopyrimidine (ligands 4 and 5) and triaminotriazine (ligands 6 and 7) recognition wedges to inhibit MBNL1 binding was examined (Figure 8). Consistent with our previous results for DM1, wedges without tethered intercalators did not destabilize the MBNL1–CCUG complexes (Figure 8).
Figure 8.
(a) Chemical structures of wedge molecules. (b) Gel electrophoretic mobility shift assay showing that only π-stacked intercalators inhibit the MBNL1–CCUG complex. Control lane 1 (C1): RNA only. Control lane 2 (C2): RNA + MBNL1. Control lane 3 (C3): RNA + MBNL1 with 10% DMSO. Except when otherwise noted, all lanes contain 350 nM MBNL1, 100 µM ligands and 10% DMSO.
DISCUSSION
The multisystemic clinical features of DM2 resemble those of DM1, although patients with DM2 do not suffer from the severe congenital form of disorder that occurs in DM1 (21). The prevalence of DM2 can be as high as in DM1 (1 in 8000) depending on the ethnicity of the population (22). As noted in the introduction, the phenotype of the disease can be reversed by inhibiting the interaction of MBNL1 and CUG repeats (9,10). For example, Berglund and coworkers (10) showed that pentamidine (IC50 = 58 µM) was able to rescue the missplicing of the IR and cTNT in HeLa cells. Based on these studies, it is likely that small molecule ligands with similar IC50 values that are selective inhibitors of the MBNL1–CCUG interaction will serve as good leads for development of a therapeutic agent to treat DM2. Therefore, small molecule ligands structurally similar to ligand 1 (IC50 = 43 µM for MBNL1–CUG) were explored.
In comparison to CUG repeats, targeting CCUG repeats represents a particular challenge because neither an NMR nor X-ray structure of RNA containing CCUG repeats is available. The CCUG repeat structure is predicted by m-fold to be comprised of two C–G base pairs and two C–U mismatches (23), although alternative structures cannot be ruled out (24). For example, Warf and Berglund (19) have examined an alternative, slipped CCUG structure containing alternating C–C and U–U mismatches separated by C–G base steps (Figure 4). Although a less stable form, it was found that MBNL1 bound this slipped structure with ∼1.5-fold higher affinity than to the structure containing two C–U mismatches.
The triaminotriazine unit in ligand 1 was designed to form a base triplet with a U–U mismatch. Whether the designed complex forms is not known, but ligand 1 binds r(CUG)4 and r(CUG)2 sequences with Kd = 0.43 µM and Kd = 2.1 µM, respectively, whereas the values for r(CAG)2 and r(CGG)2 sequences were Kd > 300 µM (12). Ligand 1 does bind with moderate affinity (Kd = 14 µM) to r(CCG)2 sequences, which contain a C–C mismatch.
The high affinity of ligand 1 for U–U mismatches and moderate affinity for C–C mismatches led us to explore the use of ligand 1 to recognize CCTG and CCUG. Previously, it was found that ligand 1 bound the CTG sequence with a 1:1 stoichiometry (12). In sequences containing CTG repeats, there was a definite negative cooperativity for adjacent sites. In contrast, ligand 1 appears by ITC fitting, CD titration studies and Job analysis to bind the CCTG sequence in 1 : 3 stoichiometry. Although care must be taken not to overinterpret the Kd values obtained from ITC fitting, the tight binding reflected in the Kd(2) value may indicate a conformational change with formation of a 1 : 1 complex. Thus, the positive cooperativity could result from the first ligand inducing formation of two C-bulges and a central T–T mismatch, the latter site providing the same tight binding observed at CTG sites, whereas the C-bulge recognition would be comparatively weaker (25). This C-bulge and T–T mismatch structure, which is suggested to be a potential drug target, is recently observed by NMR spectroscopy for CCTG repeats (26).
The strong binding of ligand 1 for CCTG-containing DNA did not translate into strong binding for an RNA oligonucleotide containing a CCUG repeat (Ligand 1 binds CTG and CUG with similar affinity; see ref. 12). Furthermore, no inhibition of MBNL1 binding to (CCUG)6 was observed. Although the detailed structure of the CCUG repeat is currently unknown, it has been proposed to adopt an A-form helical conformation, similar to that of the CUG repeat (6,27). At this time it is not known why ligand 1 is unable to recognize strongly the CCUG repeat. Nonetheless, the fact that ligand 1 is capable of strong and selective CUG binding in RNA and is able to inhibit binding of MBNL1 suggested that it should be possible to discover and develop ligands exhibiting similar behavior with CCUG sequences. This was the impetus for exploring ligands 2 and 3.
To improve the ability of ligand 1 to bind to CCUG RNA, structurally similar ligands such as 2 were pursued. Ligand 2 is identical to 1, but with a triaminopyrimidine moiety that forms a C-linked Janus-wedge. It was an appealing recognition unit to explore because 5-(4-aminobutyl)-2,4,6-triaminopyrimidine (17) was a known compound, allowing 2 to be easily synthesized by the same route as used for ligand 1. Unfortunately and somewhat surprisingly, ligand 2 exhibited very limited water solubility compared to the structurally similar ligand 1. The limited solubility of 2 prevented it from being studied by ITC with <10% DMSO. Ligand 3 was prepared with a more water-soluble intercalator, N-[2-(dimethylamino)ethyl]acridine-4-carboxamide (DACA) in place of the 9-amino-6-chloro-2-methoxyacridine unit and this allowed the triaminopyrimidine motif to be examined.
Ligand 3 bound to (CCUG)6 and RNA A with similar affinity and stoichiometry. Specifically, both sequences fit a binding model with a single, reasonably tight binding site (3–5 µM) and a second, significantly weaker binding site (250–480 µM). The tighter binding is presumably associated with one of the two CCUG sites in (CCUG)6 and the single CCUG site in RNA A (Supplementary Data). Potential weaker sites in (CCUG)6 include the remaining CCUG site, (CCUG)2 loop region and GC stem regions. In fact, titration with RNA A, lacking the second CCUG site and loop, give an ITC curve with better curvature. This suggests the residual heat at the end of the titration for (CCUG)6 likely arises from a general, non-specific binding of the weaker sites. Indeed, the affinity is similar to that observed for ligand 3 binding to the single-stranded hcTNT pre-mRNA (RNA I). Overall the data are consistent with ligand 3 binding to only one of the two C–U mismatches and a single CCUG site when two sites are neighboring. The origin of this latter effect is unclear.
Similar to the preference exhibited by MBNL1 (19), ligand 3 binds with ∼40-fold higher affinity to the slipped-CCUG structure (RNA B) than to the duplex containing adjacent C–U mismatches (RNA A). These results are in good agreement with the Kd values for the duplexes containing the single U–U (RNA D) and C–C mismatches (RNA E), suggesting that ligand 3 binds strongly to the C–C mismatch in the slipped structure and to the U–U with a reduced affinity.
Surprisingly, ligand 3 does not bind strongly (Kd = 85 ± 30 µM) to RNA C, which contains a single C–U step with two adjacent CG steps. Ligand 3 also binds weakly (Kd > 100 µM) to the purine–purine mismatches in duplexes F and G. The binding of ligand 3 to DNA containing a CTG sequence (DNA H), which is one of the targets of DM1 (20), occurs with low micromolar affinity (Kd = 1.4 ± 1.5 µM). The difference in binding affinity of ligand 3 toward analogous RNA and DNA duplexes (RNA D, U–U; Kd = 25 µM) and (DNA H, T–T; Kd = 1.4 µM) may result from structural differences between B-form DNA and A-form RNA.
Importantly, ligand 3 not only binds CCUG, it inhibits MBNL1 binding to CCUG with low micromolar Kd values. Together with the ITC results, these data suggest that ligand 3 is a good lead ligand for targeting either the CCUG or CCUG-slipped repeat structures implicated as the causative agent of DM2. Finding agents that complex CCUG and inhibit MBNL1 binding is a challenge, but finding agents that are highly selective is perhaps even more difficult and likely more important given the difficulty in developing highly selective agents targeted to nucleic acids. The selectivity profile of ligand 3 has been examined in several different contexts and can be found in Tables 1 and 2. For example, it is notable that ligand 3 binds (CCUG)6 >600-fold tighter than it does tRNA. Thus, even in the presence of tRNA, ligand 3 is capable of disrupting the MBNL1–CCUG interaction with minimally diminished Ki values. It is also significant that ligand 3 does not inhibit the interaction between MBNL1 and its natural target, the 18-nt fragment of hcTNT pre-mRNA (IC50 > 250 µM).
Developing a systematic structure–activity relationship is beyond the scope of the current study, but the data from the gel electrophoretic mobility shift assays using simple wedge recognition compounds 4–7 indicate that the acridine unit is critically important. These results agree with the previously proposed π-stacked intercalator model wherein the acridine is responsible for affinity whereas the selectivity is dictated by the nature of the wedge motif. In this regard, it is intriguing that the small structural difference between triaminotriazine and triaminopyrimidine units leads to the preferential binding to CUG for the former and to CCUG for the latter. It is likely that the basicity difference between triaminotriazine (pKa ∼ 5) (28) and triaminopyrimidine (pKa ∼ 6.7) (29) plays an important role and this is an aspect that merits additional study.
Finally, the broadest finding from the current study is that ligands 2 and 3 exhibit among the highest documented selectivity for CCUG and, thus, represent an excellent starting point for the development more selective and potent inhibitors of MBNL1 binding. Because of the presence of multiple repeating units in the toxic RNAs that cause DM1 and DM2, an obvious approach involves oligomerization of these or analogous ligands (11,14). This is an approach that has already proven effective. For example, Disney reported that the oligomerization of a DNA dye (Hoechst 33258) and kanamycin A, ligands that exhibit only moderate selectivity for the CUG and CCUG sequence, respectively, result in oligomers of better affinity and selectivity (11,14,15). The use of intercalators presents an inherent limitation on selectivity because an unstacked intercalator can non-selectively complex a wide range of duplex DNAs and RNAs. Developing new ligands where the recognition wedge remains stacked or in close proximity to the intercalator, or where the intercalator is replaced by a different unit that provides affinity, may obviate this potential source of poor selectivity. These and other approaches to optimize this class of RNA binding agents are under active investigation and will be reported in due course.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (RO1AR058361). C.H.W. is a Croucher Foundation Scholar and an Illinois Department of Chemistry Ullyot Fellow. Y.F. is a recipient of a Department of Chemistry Fellowship. Funding for open access charge: National Institutes of Health (R01AR058361).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Harper PS. Myotonic Dystrophy. 3rd edn. London: W.B. Saunders; 2001. [Google Scholar]
- 2.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion J-P, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′-end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 3.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LPW. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 4.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 5.Ranum LPW, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 6.Mooers BHM, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl Acad. Sci. USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper TA. A reversal of misfortune for myotonic dystrophy? New England J. Med. 2006;355:1825–1827. doi: 10.1056/NEJMcibr064708. [DOI] [PubMed] [Google Scholar]
- 8.Mulders SAM, van den Broek WJAA, Wheeler TM, Croes HJE, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl Acad. Sci. USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA−MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J. Am. Chem. Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem. Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. The role of flexibility in the rational design of modularly assembled ligands targeting the RNAs that cause the myotonic dystrophies. ChemBioChem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mascal M, Hansen J, Fallon PS, Blake AJ, Heywood BR, Moore MH, Turkenburg JP. From molecular ribbons to a molecular fabric. Chem. Eur. J. 1999;5:381–384. [Google Scholar]
- 18.Wu M, Wu W, Gao X, Lin X, Xie Z. Synthesis of a novel fluorescent probe based on acridine skeleton used for sensitive determination of DNA. Talanta. 2008;75:995–1001. doi: 10.1016/j.talanta.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 21.Day JW, Ricker K, Jacobsen JF, Rasmussen LJ, Dick KA, Kress W, Schneider C, Koch MC, Beilman GJ, Harrison AR, et al. Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology. 2003;60:657–664. doi: 10.1212/01.wnl.0000054481.84978.f9. [DOI] [PubMed] [Google Scholar]
- 22.Udd B, Meola G, Krahe R, Thornton C, Ranum LPW, Bassez G, Kress W, Schoser B, Moxley R. 140th ENMC International workshop: myotonic dystrophy DM2/PROMM and other myotonic dystrophies with guidelines on management. Neuromuscul. Disord. 2006;16:403–413. doi: 10.1016/j.nmd.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 24.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong HC, Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. Molecular recognition of a thymine bulge by a high affinity, deazaguanine-based hydrogen-bonding ligand. Chem. Commun. 2009:668–670. doi: 10.1039/b817733n. [DOI] [PubMed] [Google Scholar]
- 26.Lam SL, Wu F, Yang H, Chi LM. The origin of genetic instability in CCTG repeats. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr185. doi:10.1093/nar/gkr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U–U wobble’: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt RC, Schmitt RG. Ultraviolet absorption spectra of derivatives of symmetric triazine—II: oxo-triazines and their acyclic analogs. Spectrochim. Acta. 1958;12:127–138. [Google Scholar]
- 29.Roth B, Strelitz JZ. Protonation of 2,4-diaminopyrimidines. I. Dissociation constants and substituent effects. J. Org. Chem. 1969;34:821–836. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.