Abstract
Although the importance of regulatory and functional sequence evolution in generating species differences has been studied to some extent, much less is known about the role of other types of genomic changes, such as fluctuation in gene copy number. Here, we apply analyses of gene function and expression of anthocyanin pigment pathway genes, as well as cosegregation analyses in backcross populations, to examine the genetic changes involved in the shift from blue to red flowers in Andean Iochroma (Solanaceae). We demonstrate that deletion of a gene coding for an anthocyanin pathway enzyme was necessary for the transition to red floral pigmentation. The downregulation of a second pathway gene was also necessary for the novel flower color, and this regulatory pattern parallels the genetic change in the two other red-flowered species in the sister family Convolvulaceae in which flower color change has been examined genetically. Finally, we document a shift in enzymatic function at a third locus, but the importance of this change in the transition to red flowers depends on the exact order with which the three changes occurred. This study shows that gene inactivation or loss can be involved in the origin of phenotypic differences between species, thereby restricting the possibility of reversion to the ancestral state. It also demonstrates that parallel evolution of red flowers in three different species occurs via a common developmental/regulatory change but by mutations in different genes.
Keywords: anthocyanins, flower color, gene loss, Iochroma, parallel evolution
Introduction
Identifying the number and kinds of genetic changes underlying species differences is crucial for addressing basic questions about the evolutionary process. Here, we document the genetic changes involved in flower color differences between two species in order to address two such questions. The first is whether evolutionary change constrains possible future evolutionary trajectories, including reversion to ancestral states (Gould 1970; Bull and Charnov 1985; Bridgham et al. 2009). One type of genetic change that is likely to constrain reversion is gene inactivation (pseudogenization) or gene deletion (Porter and Crandall 2003). Although comparative genomic studies indicate that pseudogenization and gene deletion are common occurrences (Hughes and Friedman 2004; Hahn et al. 2007), in very few cases is it known whether these events influence phenotypic differences between species and thus constrain subsequent phenotypic evolution (but see Quattrocchio et al. 1999).
The second evolutionary question we address is whether parallel phenotypic evolution is accomplished via parallel evolution at lower levels: biochemical, developmental, and genetic (Des Marais and Rausher 2010). We define parallelism at the genetic level as a similar phenotypic change in different lineages that results from similar genetic change in the same gene. Several cases of genetic parallelism have been reported, involving characters such as body armor and albinism (Colosimo et al. 2005; Protas et al. 2006; Gross et al. 2009). Such parallelism presumably arises either because alterations in only that gene can give rise to the phenotypic change or because alteration of that gene incurs substantially less deleterious pleiotropy than alterations in other genes that produce the same phenotypic effect. Investigations of other characters, however, have often revealed that different genes are involved in similar phenotypic change (Wittkopp et al. 2003; Yoon and Baum 2004; Steiner et al. 2009). Although seldom examined, one explanation for a lack of genetic parallelism is the absence of differences among genes in the degree of deleterious pleiotropy. In such cases, however, changes in different genes may result in similar developmental or biochemical changes, producing parallelism at these levels (Des Marais and Rausher 2010).
These issues have previously been investigated by characterizing the genetic changes involved in the evolutionary loss of floral anthocyanin pigments (Quattrocchio et al. 1999; Schwinn et al. 2006; Whittall et al. 2006; Hoballah et al. 2007; Streisfeld and Rausher 2009a). Two patterns are revealed by these studies. First, this phenotypic transition may often involve gene inactivation. In the white-flowered Petunia axillaris, gene inactivation has caused loss of floral pigmentation in five independent lineages (Quattrocchio et al. 1999; Hoballah et al. 2007) and similar inactivating mutations appear to be responsible for lack of pigmentation in several Antirrhinum species (Schwinn et al. 2006). Second, this phenotypic transition involves massive genetic parallelism: in all cases in which the gene responsible for the transition has been characterized, it is an R2R3 Myb transcription factor (Streisfeld and Rausher 2011).
By contrast, it is unknown whether changes in floral hue also exhibit these patterns of genetic parallelism at multiple levels. The genes involved in hue transitions have been determined for only two species, the red-flowered Ipomoea quamoclit and red-flowered populations of Phlox drummondii. In both cases, the transition is due exclusively to regulatory changes, and gene inactivation is not involved (Zufall and Rausher 2004; Des Marais and Rausher 2010; Hopkins and Rausher 2011). In an independent red-flowered Ipomoea lineage (represented by Ipomoea horsfalliae), a similar transition from blue to red flowers has been achieved through biochemical and developmental parallelism, but the genes involved have not yet been identified (Streisfeld and Rausher 2009b), precluding any conclusion about genetic parallelism.
Here, we examine another transition from blue to red flowers, one represented by Iochroma gesnerioides (Solanaceae). By analyzing segregating reciprocal backcross populations derived from a cross between I. gesnerioides and a related blue-flowered species, Iochroma cyaneum, we identify genes involved in the transition to red flowers. In doing so, we show that this transition involved gene inactivation, which likely constrains the possibility of reversion to blue flowers. In addition, we show that the transition from blue to red flowers in I. gesnerioides involves different genes from the transitions in other taxa but uses a similar developmental mechanism.
Materials and Methods
Study System
Iochroma and five small allied genera form a clade of 35–40 species, the Iochrominae sensu Olmstead et al. (2008), whose phylogeny has been elucidated in previous studies (Smith and Baum 2006). Blue flowers, colored by delphinidin pigments, are the ancestral state in this group, and at least six shifts to other flower colors are estimated to have occurred in this clade (Smith 2006; Smith and Baum 2007, supplementary fig. S1, Supplementary Material online). The shift to red flowers has occurred twice, once due to increases in carotenoid pigments (Smith SD, unpublished data) and once due to a shift from blue anthocyanins derived from the anthocyanidin delphinidin to red anthocyanins derived from pelargonidin. The pelargonidin-producing lineage is represented by two species, I. gesnerioides and Iochroma fuchsioides. Here, we focus on I. gesnerioides and use a closely related blue-flowered species, I. cyaneum to represent the blue ancestral state.
A series of crosses involving I. cyaneum and I. gesnerioides were performed to analyze the genetics of flower color segregation. The parental I. cyaneum individuals were derived from an accession cultivated at the Missouri Botanical Gardens by W. G. D'Arcy, and the I. gesnerioides individuals were grown from seed accession number 944750129 from the Solanaceae Germplasm collection in the Botanical Garden of Nijmegen in the Netherlands. Voucher specimens for each accession (Smith 265 and 266) have been deposited at the University of Wisconsin-Madison Herbarium. A single individual from each accession was crossed to create F1 individuals, which were subsequently crossed to each parent to produce two backcross populations. Backcross populations were used instead of F2 populations because F1 pollen is inviable. All populations were grown in the Duke University greenhouses in natural light.
Identification and Genetic Analysis of Candidate Loci
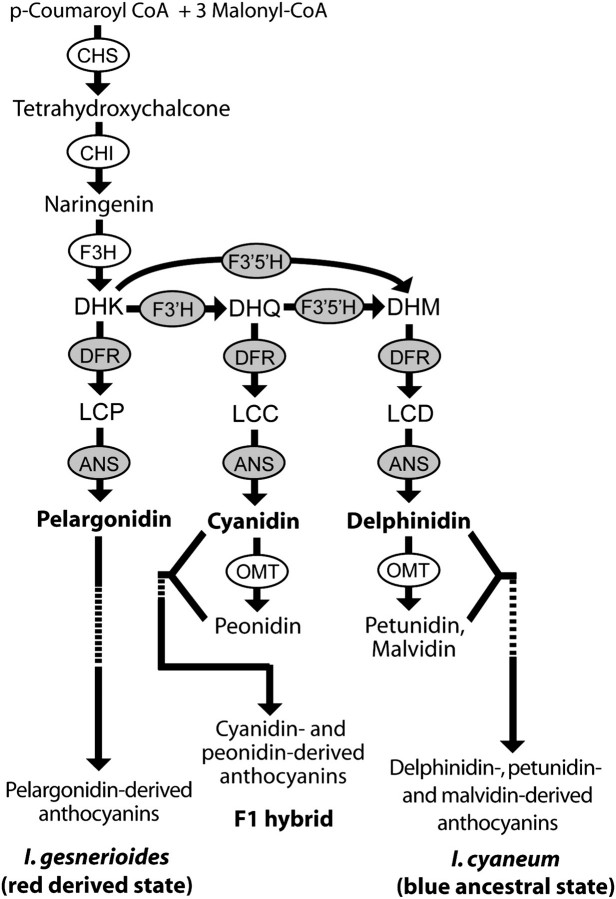
The well-conserved structure of the anthocyanin pathway provides a framework for identifying loci that could have contributed to the shift from delphinidin to pelargonidin production in I. gesnerioides. The production of delphinidin requires the action of two flavonoid hydroxylases (F3′H and F3′5′H), which convert dihydrokaempferol (DHK) into dihydromyricetin (DHM) and the subsequent action of dihydroflavonol-4-reductase (DFR) and anthocyanidin synthase (ANS) to convert the colorless DHM into the delphinidin pigments (fig. 1). A shift from delphinidin to pelargonidin production could be achieved by downregulation or inactivation of F3′H and F3′5′H, preventing conversion of DHK into DHM. Alternatively, the downstream enzymes DFR and ANS could have evolved increased specificity for the precursors of pelargonidin and/or decreased specificity for the precursors of cyanidin and delphinidin.
FIG. 1.
Anthocyanin biosynthesis pathway and production in the parental species, Iochroma gesnerioides and Iochroma cyaneum and the F1 hybrid. Pathway enzymes (circled) include flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), DFR, anthocyanidin synthase (ANS), and O-methyltransferase (OMT) (Holton and Cornish 1995). Anthocyanin precursors include DHK, DHQ, and DHM. The pathway produces three classes of anthocyanidins: 1) those derived from pelargonidin; 2) those derived from cyanidin (includes peonidin), and 3) those derived from delphinidin (includes petunidin and malvidin). Anthocyanidins are modified by the addition of sugars and other moieties to form anthocyanins. Candidate genes for the blue to red transition are shaded.
To distinguish among these possibilities, we tracked the genotypes at these four candidate loci (F3′h, F3′5′h, Dfr, Ans) in the segregating backcross populations. Large segments of each candidate gene were cloned from genomic DNA (gDNA) and cDNA from parental lines in order to design markers to genotype backcross individuals. gDNA was extracted from fresh leaf tissue using a 2× hexadecyltrimethylammonium bromide extraction (Doyle and Doyle 1987) and total RNA was extracted from 0.1 g of tissue with the Spectrum Total RNA extraction kit (Sigma, St Louis, MO) from corollas from floral buds (1–1.5 cm), flash frozen in liquid nitrogen. gDNA was removed from the extracted RNA with on-column DNase digestion (Qiagen, Valencia, CA), and first strand cDNA synthesis was performed with 1 μg of total RNA in a 20 μl reaction using SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). The four candidates, F3′h, F3′5′h, Dfr, and Ans, which appear as single copy genes in closely related Solanaceae (e.g., Solanum, Capsicum), were amplified from I. cyaneum and I. gesnerioides using previously published degenerate primers (De Jong et al. 2004; Whittall et al. 2006). Polymerase chain reaction (PCR) products were gel purified with the QIAquick gel extraction kit (Qiagen) and cloned using the pGEM-T easy vector system (Promega, Madison, WI) following the manufacturer's protocol. The candidate gene sequences from parental gDNA and cDNA were then used to design species-specific markers to genotype the backcross populations (supplementary table S1, Supplementary Material online).
Quantification of Pigment Composition
Pigment production in parents, F1 progeny, and backcross individuals was quantified using high performance liquid chromatography (HPLC). Three individuals from the parental accessions were sampled to characterize pigment production in the parental species. Anthocyanidins (pelargonidin, petunidin, cyanidin, malvidin, peonidin, and delphinidin) were extracted from 500 mg of floral tissue as outlined in Harborne (1984). Extracts were dried and re-eluted in 100 μl methanol with 1% HCl, and 50 μl was analyzed with a Shimadzu LC-10AT liquid chromatography system with a 150 × 4.6 mm Alltech Prevail reverse-phase C18 column (Alltech Associates, Deerfield, IL) at a flow rate of 1 ml/min. Anthocyanidins were separated by gradient elution at 30 °C using solvents A (HPLC-grad water, 0.1% trifluoroacetic acid) and C (1-propanol, 0.1% trifluoroacetic acid) with the following program: 15% C from 0 to 4 min; linear increase to 20% C from 4 to 10 min; 20% C from 10 to 14 min; linear increase to 22.5% C from 14 to 16 min; instantaneous increase to 27.5% C; 27.5% C from 16 to 18 min; instantaneous decrease to 15% C; 15%C from 18 to 21 min. Peaks were detected at 520 and 540 nm. Anthocyanidins were identified and quantified by comparison with standard solutions of delphinidin, petunidin, cyanidin, malvidin, peonidin, and pelargonidin (Polyphenols Laboratories, Sandnes, Norway). For each individual, the relative production of each of the three major classes of anthocyanidins (pelargonidin-derived, cyanidin-derived, delphinidin-derived, fig. 1) was calculated by the dividing the amount of each class (in micromoles) by the total amount of anthocyanidins produced. Two anthocyanidins (petunidin and malvidin) are delphinidin-derived, and one (peonidin) is cyanidin-derived (fig. 1).
Quantification of Anthocyanin Gene Expression
Expression levels of candidate genes in parents and backcross populations were examined using quantitative real-time PCR (qPCR). Three individuals from the parental accessions were used to characterize expression levels in the parental species. Floral bud cDNA was prepared as described above and diluted to 2.5 ng/μl for qPCR. Primers were designed in conserved coding regions of the candidate genes to amplify products between 100 and 250 bp (supplementary table S1, Supplementary Material online). Primers were also designed for the elongation factor (EF1-α) from Iochroma to serve as a reference gene. Specificity of the primers was determined by visualization of the products on a 2% agarose gel and by sequencing the products. Each 20 μl qPCR reaction contained 10 μl Dynamo SYBR green qPCR mix (Finnzymes, Espoo, Finland), 0.4 μl 1× 6-carboxy-X-rhodamine passive reference dye, 0.2 μM of each primer, and 2.5 ng of total cDNA. Reactions were run on an ABI Prism 7000 sequence detection system with the following conditions: 95 °C for 10 min, followed by 40 cycles of 95°C for 20 s, and 55°C for 1min. Two reactions were run for each sample and were repeated if the threshold (Ct) values for each reaction differed by greater than 10%. Relative expression was calculated following equation (1) in Pfaffl (2001), with cDNA from a single blue-flowered individual from the parental accession serving as reference sample for all runs. Efficiency for each gene was determined as described in Peirson et al. (2003).
Assay of DFR Function
Differences in DFR function between I. cyaneum and I. gesnerioides were examined using in vitro assays. Full-length coding regions were obtained using the GeneRacer Kit, directionally cloned into the pENTR-SD/D-TOPO vector, and recombined into the pDEST-14 overexpression vector (all products from Invitrogen). This construct was transformed into BL21 Star (DE3) Escherichia coli cells, and colonies were used to inoculate 5 ml overnight cultures in LB-Amp100. The overnight cultures were used to inoculate a 200 ml culture, which was grown at 37°C until reaching OD600 of 0.5, at which point protein expression was induced by adding 500 μl 200 mM isopropyl-beta-D-thiogalactopyranoside and growing for an additional 3 h at 28°C. After pelleting the bacteria, crude protein was extracted and assayed for the ability to convert 0.5 μM substrate following Des Marais and Rausher (2008). We examined three substrates, dihydrokaempferol (DHK), dihydroquercetin (DHQ), and dihydromyricetin (DHM), which are the precursors of pelargonidin, cyanidin, and delphinidin, respectively (fig. 1). The three substrates were obtained from TransMIT (Marburg, Germany), Sigma (St. Louis, MO), and Apin Chemicals (Abingdon, United Kingdom), respectively. Each assay was repeated with four independent clones (4 replicates) from each species, and the results were analyzed with a Shimadzu UV-2401PC spectrophotometer. Peak heights were converted into micromoles using standard curves for each product, and the relative efficiency of DFR from each species on each substrate was calculated as the micromoles of substrate converted divided by the total micromoles converted.
Southern Blotting
Initial cloning of amplicons of F3′5′h suggested the presence of multiple copies in Iochroma, and this possibility was examined using Southern blotting. For this experiment, 20 μg of total gDNA from each parental species was digested with EcoRI and HindIII for 10 h at 37°C. Digested DNA was precipitated with 0.1 volume 3 M sodium acetate (pH 5.2) and 1 volume isopropanol. After pelleting and washing with 70% ethanol, the DNA was re-eluted in Tris–ethylenediaminetetraacetic acid buffer and run for 15 h on an 0.8% agarose gel at 15 V with 30 ng of digoxigenin (DIG)-labeled ladder (Marker VII, Roche, Indianapolis, IA). Fragments were transferred to a positively charged nylon membrane (Roche), which was probed with a DIG-labeled DNA probe following manufacturer's protocols. The probe comprised 592 bases from the last exon of F3′5′h, which was cloned using the pGEM-T easy vector system (Promega) and amplified from the vector using Expand High Fidelity polymerase and PCR DIG labeling mix (Roche). The resulting DIG-labeled PCR product was visualized on an agarose gel to ensure incorporation of DIG nucleotides and purified before use as a probe. After hybridization, stringency washes and detection following manufacturer's protocols, the CSPD substrate was applied to the membrane, and luminescing bands were visualized by exposing the membrane to X-ray film.
Statistical Analyses
We tested the effects of candidate genes on pigment production with univariate and multivariate analyses of variance (MANOVAs and ANOVAs). These analyses were performed with PROC GLM in SAS for Windows version 9.2 (SAS Institute, Inc., Cary, NC). In all cases, the response variables were the relative proportions of each major class of pigments (delphinidins, cyanidins, pelargonidins), and interactions between genes were incorporated into all models when relevant.
Results
Differential Gene Expression and Gene Deletion in the Red I. gesnerioides
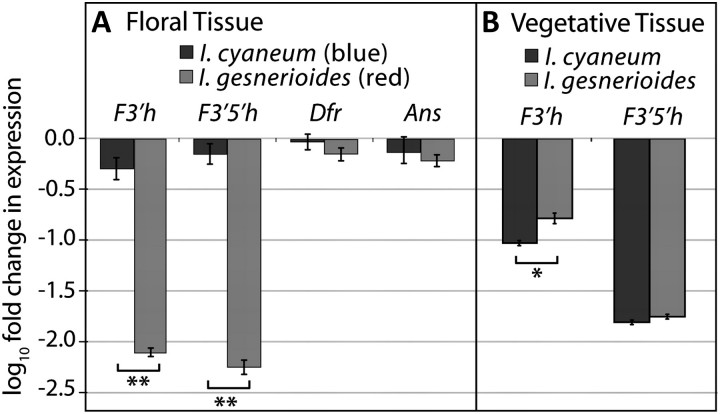
To assess differences in anthocyanin pathway functioning between I. gesnerioides and I. cyaneum, we first measured expression levels of pathways genes in each species. Comparison of expression levels showed that two genes, F3′h and F3′5′h, were strongly downregulated in red I. gesnerioides flowers relative to blue I. cyaneum flowers (Student's t-test, both P < 0.001, fig. 2). F3′h was upregulated in leaves of I. gesnerioides compared with leaves of I. cyaneum (Student's t-test, P < 0.05, fig. 2), consistent with the presence of cyanidin-derived anthocyanins in I. gesnerioides leaves (supplementary fig. S2, Supplementary Material online). The restriction of F3′h downregulation to flowers suggests that expression has been decoupled in flowers and vegetative tissue, allowing for a flower-specific loss of expression. F3′5′h also appeared strongly downregulated in flowers of I. gesnerioides (fig. 2). However, additional experiments revealed that this apparent downregulation actually reflects deletion of the single functional copy in I. gesnerioides.
FIG. 2.
Differences in anthocyanin gene expression between Iochroma gesnerioides and Iochroma cyaneum. (A) F3′h, F3′5′h, Dfr, and Ans expression levels (±1 standard error) in floral tissue of three I. gesnerioides individuals and two I. cyaneum individuals relative to the single I. cyaneum parent from the crosses. Values of −1 and −2 correspond, respectively, to 10 and 100 times fewer copies of transcript relative to the I. cyaneum parental individual. **P < 0.001. (B) F3′h and F3′5′h expression in vegetative tissue of three I. gesnerioides individuals and two I. cyaneum individuals relative to expression in the floral tissue of the single I. cyaneum parent from the crosses. *P < 0.05.
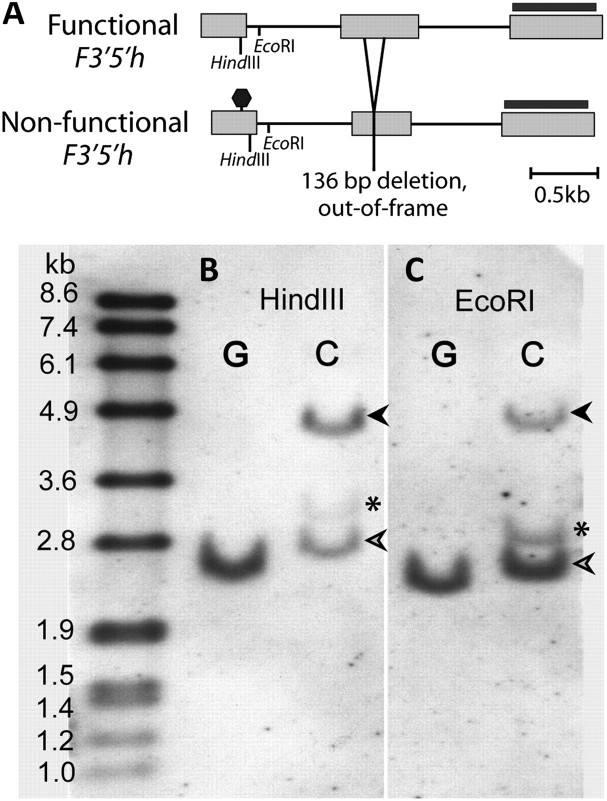
In I. cyaneum, two copies of F3′5′h are detected by PCR from gDNA. This gene is represented by a single copy in most Solanaceae, including those most closely related to Iochroma (Holton et al. 1993; Jung et al. 2005; Olsen et al. 2010). Given that polyploidy is not known in Iochroma (Smith et al. 2008), these two copies presumably represent a lineage-specific gene duplication. One of the two F3′5′h copies is clearly nonfunctional due to a premature stop codon in the first exon and a 136 bp out-of-frame deletion in the second exon. The other copy, which is expressed in I. cyaneum floral cDNA, presumably represents the functional gene (fig. 3). By contrast, the functional copy was not detectable by PCR from I. gesnerioides floral cDNA or gDNA, and only the nonfunctional copy was detectable by PCR from gDNA. Moreover, Southern blotting confirmed that I. gesnerioides has only one copy of F3′5′h, presumably the nonfunctional copy that is easily amplified from gDNA (fig. 3). The absence of a functional F3′5′h is consistent with the production of cyanidin-derived, rather than delphinidin-derived, anthocyanins in the leaves of I. gesnerioides (supplementary fig. S2, Supplementary Material online).
FIG. 3.
Deletion of the functional F3′5′h copy in Iochroma gesnerioides. (A) Schematic diagram of functional and nonfunctional F3′5h copies showing the premature stop (octagon) and the 136 bp deletion in the nonfunctional copy, as well as the enzymes and probe (black bar) used in B and C. I. gesnerioides (“G”) and Iochroma cyaneum (“C”) gDNA were digested with HindIII (B) and EcoRI (C) and probed with the third exon of F3′5′h to reveal both copies. Open arrows indicate the nonfunctional copy present in both species; dark arrows show the presumed functional copy, absent in I. gesnerioides. The band marked with an asterisk is either a third copy of F3′5′h in I. cyaneum or represents nonspecific binding but is also absent in I. gesnerioides.
Color Variation in the Backcross to Blue Caused by Alteration of DFR Function
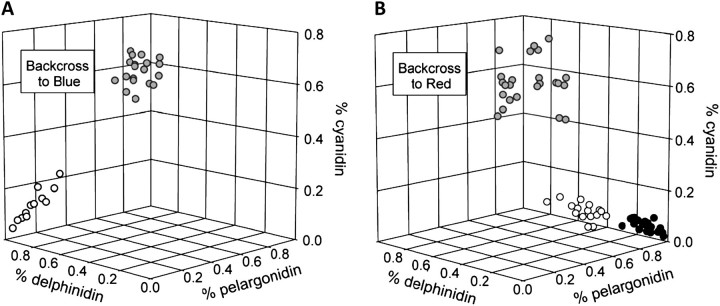
In order to determine whether these changes at F3′h and F3′5′h, as well as potential differences at the other candidate loci, Dfr and Ans, affect floral color, we examined the progeny of F1 individuals backcrossed to both parents. In contrast to the red and blue parents, the F1 individuals produced purple flowers containing primarily cyanidin-derived anthocyanins (fig. 1, supplementary table S2, Supplementary Material online). In the backcross to the blue I. cyaneum parent, two discrete color classes segregated: blue, similar to I. cyaneum flowers in appearance and pigment composition and purple, similar to those of F1 individuals (fig. 4). The ratio of the two classes, 15:20, did not differ significantly from 1:1 (χ2 = 0.714, P = 0.40), suggesting that the phenotypic difference is controlled by variation at a single locus. A species-specific marker in Dfr cosegregated perfectly with phenotype, whereas the other candidate loci did not (supplementary table S3, Supplementary Material online). In addition, a MANOVA of the effects of markers in F3′h, Dfr, and Ans on pigment composition indicated that Dfr was highly significant, whereas F3′h and Ans were not significant (table 1). F3′5′h was not included in this analysis because genotypes in this backcross were not distinguishable. Thus, the backcross to the blue parent implicates Dfr as contributing to the flower color difference between the two species.
FIG. 4.
Floral pigment phenotypes in backcross populations. (A) The backcross to blue population had two distinct phenotypic classes: blue flowers with mostly delphinidin (open circles) and purple flowers with mostly cyanidin (gray circles). Blue flowers are homozygous for blue alleles at Dfr and purple flowers are heterozygous. (B) The backcross to red population had three distinct phenotypic classes: purple flowers with mostly cyanidin (gray circles), pink flowers with intermediate composition (open circles), and red flowers with 80% or more pelargonidin (black circles). Phenotypic classes in this backcross are determined largely by F3′5′h genotype and F3′h expression (fig. 6).
Table 1.
Effects of Candidate Genes on Pigment Production in Backcross Populations Based on Multivariate and Univariate Analyses of Variance.
| MANOVA | df | Univariate Analyses |
||||||
| Delphinidin |
Cyanidin |
Pelargonidin |
||||||
| Wilks' λ | MS | F | MS | F | MS | F | ||
| (A) Analysis of backcross to blue population (N = 33)a | ||||||||
| F3′h | 0.84 | 1 | 6.60 × 10−3 | 1.63 | 5.77 × 10−4 | 0.17 | 3.01 × 10−3 | 2.02 |
| Dfr | 0.03*** | 1 | 2.92 | 722.97*** | 2.23 | 642.24*** | 0.05 | 30.71*** |
| Ans | 0.86 | 1 | 0.01 | 2.99 | 0.01 | 3.86 | 2.37 × 10−5 | 0.02 |
| F3′h × Dfr | 0.95 | 1 | 3.13 × 10−3 | 0.78 | 3.61 × 10−3 | 1.04 | 3.14 × 10−6 | 0.00 |
| F3′h × Ans | 0.98 | 1 | 1.21 × 10−3 | 0.30 | 1.98 × 10−4 | 0.06 | 4.71 × 10−4 | 0.31 |
| Dfr × Ans | 0.81 | 1 | 7.96 × 10−3 | 1.97 | 1.56 × 10−4 | 0.05 | 5.73 × 10−3 | 3.83 |
| F3′h × Dfr × Ans | 0.86 | 1 | 0.01 | 3.06 | 0.01 | 3.20 | 2.37 × 10−5 | 0.02 |
| Error | 25 | 4.05 × 10−3 | 3.48 × 10−3 | 1.50 × 10−3 | ||||
| MANOVA | df | Univariate Analyses | ||||||
| Delphinidin | Cyanidin | Pelargonidin | ||||||
| Wilks' λ | MS | F | MS | F | MS | F | ||
| (B) Analysis of backcross to red population (N = 37)b | ||||||||
| F3′5′h | 0.39*** | 1 | 0.10 | 31.80*** | 0.02 | 1.23 | 0.19 | 16.88** |
| Dfr | 0.81 | 1 | 4.82 × 10−3 | 1.55 | 3.53 × 10−4 | 0.03 | 6.84 × 10−3 | 0.60 |
| T | 0.13*** | 1 | 1.27 × 10−3 | 0.41 | 2.13 | 159.83*** | 2.04 | 178.69*** |
| F3′5′h × Dfr | 0.68* | 1 | 0.02 | 6.36* | 0.03 | 2.41 | 0.10 | 8.61** |
| Dfr × T | 0.95 | 1 | 4.39 × 10−5 | 0.01 | 1.76 × 10−3 | 0.13 | 1.81 × 10−3 | 0.16 |
| F35′h × T | 0.73* | 1 | 0.02 | 6.30* | 2.22 × 10−3 | 0.17 | 0.04 | 3.20 |
| F3′5′h × Dfr × T | 0.67* | 1 | 0.03 | 9.14** | 0.01 | 0.75 | 0.07 | 6.27* |
| Error | 29 | 0.03 | 0.01 | 0.01 | ||||
| Delphinidin | Cyanidin | Pelargonidin | ||||||
| MS | F | MS | F | MS | F | |||
| (C) Specific contrasts between genotypic classes in the backcross to red (df = 1 for all) | ||||||||
| FRFRDBDRTRTR versus FRFRDRDRTRTR | 4.15 × 10−4 | 0.13 | 1.05 × 10−3 | 0.08 | 2.51 × 10−3 | 0.22 | ||
| FRFRDRDRTBTR versus FRFRDRDRTRTR | 1.25 × 10−3 | 0.40 | 0.46 | 38.40*** | 0.43 | 37.39*** | ||
| FBFRDRDRTRTR versus FRFRDRDRTRTR | 0.06 | 19.87*** | 0.02 | 1.64 | 0.15 | 13.59** | ||
NOTE.—N = number of individuals, df = degrees of freedom, MS = mean square (based on type III sums of squares). The pigment classes, delphinidin, cyanidin, and pelargonidin are the relative production of delphinidin and its derivatives, cyanidin and its derivatives, and pelargonidin, respectively.
F3′5h genotype was not included because the two possible genotypes (one or two functional copies) cannot be discriminated.
The four-way interaction term was not included in the model because there are insufficient degrees of freedom.
***P < 0.0001, **P < 0.01, *P < 0.05.
The effect of Dfr on pigment production could be due to either an expression difference or a functional difference. Because DFR and F3′H compete for the precursor DHK (fig. 1), an increase in Dfr expression could possibly increase the relative production of pelargonidin-derived anthocyanins. However, qPCR revealed no detectable difference in expression level between the parental species (fig. 2). Also, Dfr expression level had no effect on pigment production in either backcross population (supplementary fig. S3, Supplementary Material online). By contrast, in vitro assays of DFR function showed that DFR from the red I. gesnerioides acts more than twice as efficiently on DHK, the precursor of red pelargonidin pigments and less than one-third as efficiently on DHM, the precursor of blue delphinidin pigments, as DFR from I. cyaneum (fig. 5). Thus, the DFR of each species is specialized on the substrate corresponding to the type of anthocyanins it produces. Sequence comparison revealed 12 amino acid differences between the species that could contribute to this effect (supplementary fig. S4, Supplementary Material online), and one of these changes (146 V to I) lies within a region known to affect substrate specificity (Johnson et al. 2001; Petit et al. 2007).
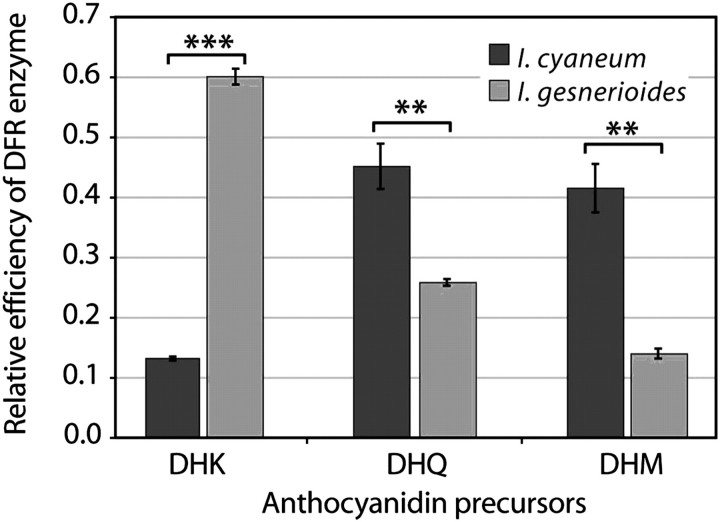
FIG. 5.
Relative efficiency of DFR enzyme from Iochroma cyaneum and Iochroma gesnerioides on precursors of anthocyanidins. DFR activity was assessed on DHK, DHQ, and DHM, the precursors for pelargonidin, cyanidin, and delphinidin, respectively (fig. 1). Relative efficiency is calculated as micromoles substrate converted/ total micromoles converted. Error bars indicate ±1 standard deviation across four replicates. The species differ significantly in relative efficiency on each substrate (MANOVA: I. cyaneum vs. I. gesnerioides, Wilks' λ = 0.005, P < 0.0001; univariate ANOVAs, F = 1128.00, 25.42, 45.60; and P = 0.0001***, 0.002**, 0.0005** for DHK, DHQ, and DHM, respectively).
F3′h and F3′5′h Control Pigment Production in the Backcross to Red
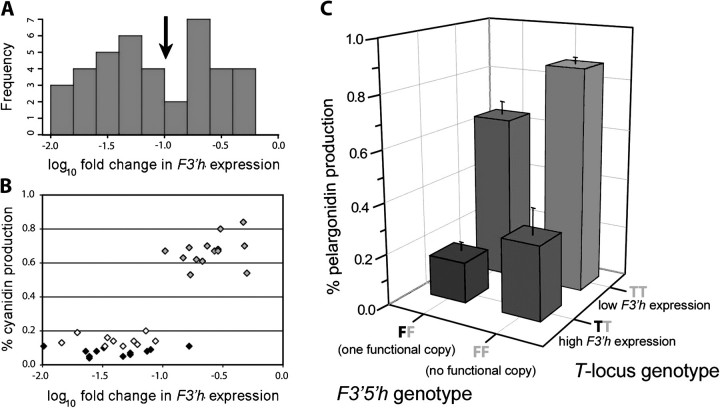
In the backcross to the red-flowered parent, individuals with flowers indistinguishable in visual appearance and in pigment composition to those of both I. gesnerioides (N = 20) and F1 individuals (N = 25) were produced (fig. 4, supplementary table S2, Supplementary Material online). In addition, individuals intermediate in appearance and pigment composition (N = 16) were also produced (fig. 4). A preliminary MANOVA using markers in all four genes indicated that only F3′5′h significantly affected pigment composition (supplementary table S4, Supplementary Material online). However, further analyses suggested that although F3′h genotype had no effect on pigment composition (supplementary tables S3 and S4, Supplementary Material online), F3′h expression had a strong effect on the relative production of cyanidin and pelargonidin. Several lines of evidence suggest that this effect may be due to a single trans-acting locus with two alleles. First, the frequency histogram of F3′h expression levels in the backcross to the red parent appears to be bimodal, with the boundary between the two groups corresponding to log change in expression of −1.0 (fig. 6A). The reality of these two groups is supported by the observation that, with one exception, the individuals they represent differ substantially in percent cyanidin production (Fig. 6B). Moreover, the numbers of individuals in these two groups occur in roughly a 1:1 ratio (supplementary tables S1 and S3, Supplementary Material online), as expected in a backcross population. Finally, if this variation were due to nongenetic causes, we would expect some individuals within I. gesnerioides to exhibit purple flowers with high expression of F3′h and high levels of cyanidin-based anthocyanidins, which we do not see. For the following analysis, we designate the inferred locus controlling F3′h expression the T-locus. The absence of an effect of the marker allele at the F3′h locus on either pigment composition or its own expression indicates that T is a trans-acting regulator.
FIG. 6.
Effects of F3′h expression and F3′5′h deletion in the backcross to red (BCR) population. (A) Relative expression of F3′h across 37 BCR individuals. Expression levels are shown relative to the blue Iochroma cyaneum parent; a value of −1.0 corresponds to 10-fold lower expression of F3′h relative to I. cyaneum. The arrow denotes the cutoff between low expression and high expression corresponding to low and high cyanidin classes in B. (B) F3′h expression versus cyanidin production in 37 BCR individuals. All individuals except one with expression greater than −1.0 produce mostly cyanidin (gray symbols). All individuals with expression lower than −1.0 produce mostly pelargonidin; these are divided into two groups: pink (50–80% pelargonidin, open symbols) and red (>80% pelargonidin, black symbols). (C) Combined effects of the F3′5′h and the inferred T-locus controlling F3′h expression in BCR. The cutoff between high and low F3′h expression follows (A). The average pelargonidin production for each genotypic combination is shown ±1 standard deviation. Black letters (T or F) denote a blue I. cyaneum allele and gray, a red I. gesnerioides allele. BCR individuals that lack a functional copy of F3′5′h (i.e., are homozygous for the red null allele at that locus) and have low F3′h expression (i.e., inferred to be homozygous for red alleles at the T-locus) recover the red parental phenotype (>80% pelargonidin production). Each of the four genotypic combinations (e.g., homozygous at both F3′5′h and T) includes individuals that are homozygous and heterozygous at Dfr.
Based on these observations and the preliminary MANOVA (supplementary table S4, Supplementary Material online), we performed a MANOVA that included three factors potentially affecting pigment composition: Dfr genotype, F3′5′h genotype, and T-locus genotype, where assignment of the latter was determined by whether F3′h expression was above or below −1 on a log10 scale (arrow in fig. 6A). In this analysis, the main effects of F3′5′h and T were highly significant (P < 0.0001; table 1). As expected from the topology of the anthocyanin pathway (fig. 1), an additional “red” allele at F3′5′h decreases the relative amount of delphinidin produced while increasing the relative amount of pelargonidin and an additional red allele at T decreases cyanidin while increasing pelargonidin (table 1). There were minor but significant epistatic effects involving the F3′5′h × Dfr, F3′5′h × T, and F3′5′h x T × Dfr interactions (table 1). In addition, as determined by specific contrasts (table 1), homozygosity for the derived red allele at both F3′5′h (FR) and T (TR) is necessary for producing the pigment composition of I. gesnerioides flowers (greater than 80% pelargonidin production; fig. 6C).
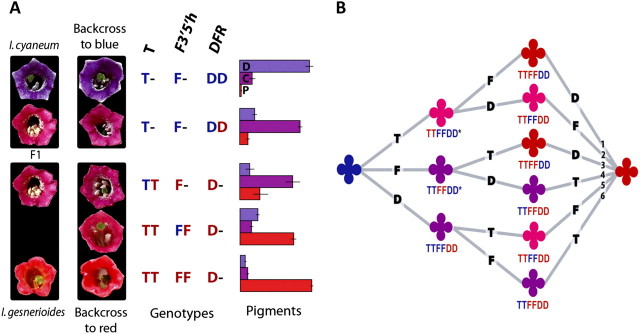
In the parental species, the F1 hybrid and two sets of backcrosses, a total of four discrete phenotypes were observed. These four phenotypes correspond to four classes of pigment composition and four classes of genotypes (fig. 7 and supplementary text, Supplementary Material online). Although there is slight variation in pigment composition within classes, this variation is small compared with variation between classes, giving rise to the visual discreteness of the phenotypes (fig. 7). Moreover, the cumulative effect of substitutions of the red alleles at these three loci account for almost 100% of the difference in floral pigment composition between the parental species (fig. 7 and supplementary text, Supplementary Material online). Effects of any other unidentified loci on pigment composition are thus expected to be relatively minor.
FIG. 7.
Floral phenotypes, backcross genotypes, and pigment variation. (A) Flowers of the parents, the F1 hybrid, and the backcrosses are shown. The blue backcross has blue and purple phenotypes and the red backcross has purple, pink, and red phenotypes. Genotypes are given for each backcross phenotype at the three associated loci: the T-locus controlling F′3h expression, F3′5′h, and Dfr. Blue alleles at each locus are colored blue and red are red. T-locus genotypes were inferred from F3′h expression levels (fig. 6). Bar graphs show pigment composition for each genotype; D, C, and P are the mean proportions of each class of pigment (delphinidin, cyanidin, and pelargonidin) ±1 standard deviation. For analyses underlying this figure, see supplementary text, Supplementary Material Online. (B) Possible orders of changes to Dfr, F3′5′h, and T in the transition from blue to red flowers. Phenotypes follow the same four groupings observed in the backcrosses (A and fig. 4), blue, purple, pink, and red. The six possible paths are numbered for reference in the text. The two starred phenotypes are inferred from both the crosses and the structure of the pathway (fig. 1), whereas the remaining phenotypes can be inferred from the crosses alone.
Discussion
Multiple Genetic Changes Were Necessary for Evolving Red Flowers
Analyses of genotypic and phenotypic variation in the backcross populations demonstrated that changes of major effect in three genes are responsible for almost all of the difference in floral pigmentation between I. cyaneum and I. gesnerioides. First, we found a major shift in substrate specificity of the enzyme DFR, with the copy from red-flowered I. gesnerioides showing preference for the precursor of red pigments and that of blue-flowered I. cyaneum showing preference for the precursors of blue and purple pigments. Second, we documented a deletional inactivation of the gene for the branching enzyme F3′5′H, responsible for producing blue pigment precursors, in the red I. gesnerioides. Finally, we showed that a change in an unidentified transcription factor that causes a significant downregulation of F3′h in I. gesnerioides also contributes to greater production of red pigments. We infer the existence of this transcription factor, as opposed to a cis-regulatory change because F3′h itself does not cosegregate with either pigment composition or its own expression level. Although it is possible that more than one trans-acting element contributes to this effect, the bimodality of F3′h expression and the phenotypic discreteness of the high and low expression classes suggests that a single factor is likely.
Of the three documented changes, at least two of them, the loss of F3′5′h and the downregulation of F3′h, were required for the evolution of red flowers in I. gesnerioides. As observed in the backcross to red, only individuals homozygous for red null alleles at F3′5′h and red alleles at the inferred T-locus recover the red parental phenotype with high pelargonidin production (fig. 7). The loss of F3′5′h alone results in only a small increase in pelargonidin (table 1) because F3′H will continue to convert DHK, the precursor of red pelargonidin pigments, into DHQ, the precursor of purple cyanidin pigments (fig. 1). Although the loss of F3′h expression, through the change at the inferred trans-acting T-locus, results in a greater increase in pelargonidin (table 1), the phenotype remains pink because it does not produce as much pelargonidin as the red flowers of I. gesnerioides (fig. 7). This difference occurs because F3′5′h converts some DHK to DHQ, resulting in the production of some cyanidin and delphinidin. In combination, however, the fixation of both changes results in the red parental phenotype because the side branches of the pathway that produce purple cyanidin and blue delphinidin pigments (fig. 1) are nearly inactivated, leaving only red pelargonidin pigments.
It is less clear whether the functional shift in Dfr contributed to the blue-to-red transition because its role depends on the order of the genetic changes. The backcross to red suggests that being homozygous for the red allele at Dfr would at most convert a blue flower into a purple flower with flux mainly through the cyanidin branch. Thus, if substitution(s) in Dfr occurred first in the ancestrally blue lineage, a novel purple phenotype would have been produced, but changes in the other two genes would also have been necessary to further reduce cyanidin and delphinidin production (fig. 7, paths 5 and 6). On the other hand, if changes to F3′5′h and T occurred first (fig. 7, paths 1 and 3), the substitutions in Dfr may not have been necessary: with F3′5′H absent and greatly reduced levels of F3′h, little cyanidin or delphinidin could be produced, allowing production of only derivatives of pelargonidin. Under this scenario, the altered substrate specificity of Dfr would presumably reflect selection to improve enzyme efficiency on the pelargonidin precursor DHK.
Determining the order of molecular changes involving Dfr, F3′h, and F3′5′h could be aided by studying other species of Iochroma more closely related to I. gesnerioides. The two species studied here, both easily grown and popular as ornamentals, are not each other's closest relatives. I. gesnerioides and its red sister species I. fuchsioides are most closely related two blue-flowered species from the cloud forests of Ecuador, Iochroma calycinum and Iochroma baumii (supplementary fig. S1, Supplementary Material online). Because the downregulation of F3′h and the inactivation of F3′5′h both appear necessary for the production of red flowers, we would expect these changes to also be present in I. fuchsioides but absent in the closely related blue species. However, examination of F3′5′h in I. fuchsioides might reveal only partial degeneration of the functional copy, which would suggest a stepwise degradation beginning with inactivating mutations and followed by the gene deletion. Similarly, comparison of Dfr sequence in related taxa could elucidate the order of changes involved in the functional shift. Amino acid changes shared between the two red species but absent in their blue relatives would be strong candidates for the evolution of preference for the red pigment precursors. Given that our studies were based on a single accession of I. gesnerioides, examining variation at these loci within and among populations may also be relevant. For example, changes in Dfr that are fixed across all red-flowered populations are more likely to be of functional importance in the shift to red than those that are continuing to segregate.
Gene Losses in the Evolution of Species Differences
Comparative studies of full genome sequences have documented an enormous flux in genomic content along the tree of life, with many genes and in some cases entire gene families being gained and lost or pseudogenized in different lineages (Hughes and Friedman 2004; Demuth et al. 2006; Hahn et al. 2007). In some cases, these historical expansions and contractions have been related to major life history or phenotypic transitions (Moran and Mira 2001; McBride and Arguello 2007; Demere et al. 2008; Preston et al. 2011), such as the loss of enamel loci accompanying the loss of teeth in the bird lineage (Sire et al. 2008). However, without traditional genetic or transgenic studies, it remains unclear whether the genomic differences observed in present day taxa were responsible for the initial phenotypic transition or whether these differences reflect subsequent evolution. To date, only one study has definitively linked a change in gene copy number, in this case a gene gain, with a phenotypic difference between species (Hanikenne et al. 2008), and, to our knowledge, there are no similar cases involving gene loss. There are also few examples of phenotypic differences between species attributable to pseudogenization, and one of the best examples involves a shift from pigmented to white flowers in P. axillaris (Quattrocchio et al. 1999).
In Iochroma, gene inactivation was clearly necessary for the phenotypic transition to red coloration because individuals carrying a functional copy of F3′5′H do not produce the red phenotype, regardless of the genetic composition at other loci. Although we have shown that this gene is deleted in I. gesnerioides, we cannot distinguish between the possibility that deletion caused inactivation and the possibility that deletion occurred after initial inactivating mutations, such as those observed in spontaneous red-flowered mutants (Snowden and Napoli 1998; Chen et al. 2007). In either case, however, gene inactivation appears to have contributed to the evolution of red flowers. Compared with other possible mechanisms for accomplishing this transition (e.g., regulatory changes), gene inactivation might be considered a priori an unlikely candidate because it could result in decreased fitness due to pleiotropic effects. Nevertheless, gene inactivation, including deletion, is known to contribute to phenotypic variation within species (Grant et al. 1998; Werner et al. 2005) and domesticates (Dixon et al. 1996; Cai et al. 1997), and potential adverse pleiotropic effects of gene inactivation may be reduced if there are additional copies of the gene or if the gene affects only the phenotype of interest.
In the case of F3′5′h, however, only a single functional copy is present in Iochroma, suggesting that its inactivation incurred relatively little deleterious pleiotropy. This inference is consistent with known properties of this enzyme. Although F3′5′H can carry out 3′ hydroxylation of DHK to produce DHQ (fig. 1), this reaction is performed more effectively by F3′H, which continues to function in this role in I. gesnerioides vegetative tissue. Indeed, deletion of F3′5′h has no detectable effect on floral cyanidin production in the presence of a functional F3′h (fig. 7 and supplementary text, Supplementary Material online). Furthermore, the other flavonoid products requiring F3′5′H activity (e.g., pentahydroxyflavanone and myricetin) are uncommon in Solanaceae relative to the products requiring F3H and F3′H activity (e.g., kaempferol and quercetin) (Eich 2008). These considerations suggest that the deletion of F3′5′h may have had minimal negative consequences.
Gene inactivation appears to have contributed to the evolution of unpigmented flowers in P. axillaris (Quattrocchio et al. 1999) and possibly in other white flowered species (Schwinn et al. 2006). Our results indicated that it also contributes to the evolution of red flowers from blue-flowered ancestors. The involvement of gene knockouts in floral color evolution is not unexpected. As has been shown extensively with spontaneous mutants, knockouts of pathway genes can accomplish changes in both the amount and the type of floral pigment (Streisfeld and Rausher 2011). However, not all of these gene knockouts are equally represented in flower color divergence between species. In white-flowered species, knockouts appear to be differentially restricted to genes that incur relatively little adverse pleiotropy (Streisfeld and Rausher 2011). Our results suggest that this may also be true for transitions to red flowers.
Gene inactivation is expected to often constrain future evolutionary paths. For I. gesnerioides, inactivation of F3′5′h, along with the changes in F3′h expression and DFR function, means that the re-evolution of blue flowers by renewed production of delphinidin-derived anthocyanins is highly unlikely. As shown in the backcross to red, the addition of a “blue” Dfr allele that prefers the delphinidin precursor DHM over the pelargonidin precursor has no detectable effect on pigment production in the red background where there is little to no F3′H or F3′5′H activity. Increasing F3′h expression by adding a blue allele at the T-locus in a red background would cause a significant shift away from red flowers by decreasing pelargonidin and increasing cyanidin. However, a complete reversal to blue flowers with delphinidin pigments would additionally require the presence of a functional F3′5′H. Although F3′5′H belongs to the larger CYP75 family of P450 genes, its sequences are roughly 50–60% diverged at the amino acid level from the most closely related member, F3′H (Nelson et al. 2004; Shih et al. 2006), suggesting its function could not easily be replaced by recruiting another gene family member. Alternately, a functional F3′5′H could be restored through introgression, but the allopatric distribution of I. gesnerioides makes this a remote possibility. Thus, the deletion of F3′5′h may effectively preclude reversion to a blue-flowered delphinidin–producing state. If a similar inactivation is common in other red-flowered species, its occurrence may partly explain the observation that transitions from blue to red often appear directional or even irreversible in phylogenetic studies (Whittall and Hodges 2007; Wilson et al. 2007).
Parallel Genetic Mechanisms of Flower Color Evolution
Evolutionary transitions from an ancestral state of blue flowers to a derived state of red flowers have occurred repeatedly in angiosperms by shifting from cyanidin- or delphinidin-based anthocyanins to pelargonidin-based anthocyanins (Whittall and Hodges 2007; Wilson et al. 2007; Streisfeld and Rausher 2009b). These examples, along with spontaneous mutations that have been examined indicate that this shift can be accomplished by eliminating expression or activity of the flavonoid hydroxylases (F3′h and F3′5′h) (Streisfeld and Rausher 2011). Our results, coupled with those of studies in Ipomoea, suggest that mutations that reduce F3′h expression, rather than eliminating its function, are preferentially fixed in these parallel flower color transitions. As in I. gesnerioides, F3′h has been significantly downregulated in two other independent origins of red flowers with pelargonidin pigments (Ip. quamoclit and Ip. horsfalliae) and in all three cases, the downregulation occurs only in flowers, not in vegetative tissue (Streisfeld and Rausher 2009b; Des Marais and Rausher 2010).
This pattern of parallel evolution at the developmental/regulatory level contrasts with the limited information on spontaneous mutations causing red flowers with pelargonidin production. Although the sample size is small, all red-flowered mutants known from horticultural accessions or segregating in natural populations involving the flavonoid hydroxylases involve loss-of-function mutations rather than regulatory mutations. This pattern suggests that regulatory mutants affecting these genes are much rarer than knockout mutations, and that the involvement of F3′h regulatory mutations in red-flowered Iochroma and Ipomoea species reflects preferential fixation of regulatory mutants (P < 0.005, Fisher Exact Test). We suspect that this pattern arises because flower-specific downregulation has fewer deleterious pleiotropic effects than a loss of F3′h function throughout the plant. One possible reason for this deleterious pleiotropy is that derivatives of DHQ (produced by F3′h) provide significantly higher tolerance to UVB radiation compared with derivatives of DHK (Ryan et al. 2001, 2002). In addition, the rarity of pelargonidin derivatives relative to cyanidin derivatives in plant foliage (Price and Sturgess 1938) argues that cyanidin derivatives in foliage are generally advantageous and that purifying selection acts on F3′h to preserve function in vegetative tissues.
Parallelism may occur at multiple levels. In the case of transitions from blue to red flowers, it appears to occur at the levels of phenotype, biosynthetic pathway, and developmental/regulatory mechanism but not at the level of specific genes involved. In I. gesnerioides, F3′h downregulation appears to be caused by changes in a transcription factor, whereas in I. quamoclit it is caused by a cis-regulatory change in F3′h itself. This pattern suggests that it may not matter whether the regulatory mutations are in cis-elements of the downregulated gene or in floral-specific transcription factors so long as the developmental effect (decreased floral F3′h expression) is the same. Our results thus support the suggestion by Yoon and Baum (2004) that although the type of developmental/regulatory change that contributes to a new phenotype is often constrained, there is less differential constraint among the different types of mutation that can produce the favored developmental/regulatory change.
This pattern of phenotypic and biochemical parallelism arising from a similar developmental mechanism has emerged from other studies of pigment evolution, although its generality is likely to rely largely on the genetic architecture of pigmentation in different systems. For example, variation in abdominal pigmentation across Drosophila species is closely tied to expression of the yellow gene, and this regulatory change is due to cis-acting factors in some species but a combination of cis- and trans-factors in others (Wittkopp et al. 2002). By contrast, studies of the evolution of vertebrate pigmentation suggest not only similar developmental mechanisms but also shared genetic and even mutational bases for the same phenotypes (Manceau et al. 2010). Changes in patterns of pigmentation have repeatedly involved shifts in agouti expression (Steiner et al. 2007; Linnen et al. 2009), whereas differences in the amount of pigmentation have often evolved via changes in the Mc1r coding sequence, in some cases involving the fixation of the same derived amino acids in distantly related lineages (Rompler et al. 2006; Steiner et al. 2009; Rosenblum et al. 2010). This extension of parallelism to the genetic and even the mutational level may reflect a narrow range of genetic mechanisms that can produce the derived phenotype or reduced pleiotropy associated with the particular substitutions. We consider the latter factor as the likely cause for the repeated involvement of MYB transcription factors in evolutionary losses of floral anthocyanin pigmentation (Quattrocchio et al. 1999; Schwinn et al. 2006; Whittall et al. 2006; Hoballah et al. 2007; Streisfeld and Rausher 2009a). An alternative possibility is that the targeting of candidate genes in evolutionary genetic studies results in ascertainment bias, giving a biased perception of the relative contribution of particular genes, like Mc1r, in phenotypic evolution (Manceau et al. 2010). Studies that combine genome-wide approaches (crossing studies, quantitative trait locus mapping) with analysis of candidate genes will be needed to fully address the extent to which parallelism at the phenotypic and biochemical level arise from parallel developmental and genetic mechanisms.
Supplementary Material
Supplementary text, tables S1–S4, and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank members of the Rausher and Dong laboratories for technical advice, the Duke greenhouses for support, and the Nijmegen Botanical Gardens and L. Bohs for material of I. gesnerioides and I. cyaneum, respectively. We also appreciate the helpful comments of three anonymous reviewers and the associate editor. This work was supported by Duke University's Center for Evolutionary Genomics, by a National Institutes of Health NRSA fellowship (F32GM080082) to S.D.S., and by National Science Foundation Grants DEB-0448889 and DEB-0841521 to M.D.R.
References
- Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL. On irreversible evolution. Evolution. 1985;39:1149–1155. doi: 10.1111/j.1558-5646.1985.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Cai DG, Kleine M, Kifle S, et al. (13 co-authors) Positional cloning of a gene for nematode resistance in sugar beet. Science. 1997;275:832–834. doi: 10.1126/science.275.5301.832. [DOI] [PubMed] [Google Scholar]
- Chen S, Matsubara K, Kokubun H, Kodama H, Watanabe H, Marchesi E, Ando T. Reconstructing historical events that occurred in the petunia Hf1 gene, which governs anthocyanin biosynthesis, and effects of artificial selection by breeding. Breed Sci. 2007;57:203–211. [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- De Jong WS, Eannetta NT, De Jong DM, Bodis M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet. 2004;108:423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- Demere TA, Mcgowen MR, Berta A, Gatesy J. Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst Biol. 2008;57:15–37. doi: 10.1080/10635150701884632. [DOI] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLos One. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution. 2010;64:2044–2054. doi: 10.1111/j.1558-5646.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Eich E. Secondary metabolites. Berlin: Springer; 2008. Solanaceae and Convolvulaceae. [Google Scholar]
- Gould SJ. Dollo on Dollo's Law: irreversibility and the status of evolutionary laws. J Hist Biol. 1970;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- Grant MR, McDowell JM, Sharpe AG, Zabala MDT, Lydiate DJ, Dangl JL. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLos Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLos Genet. 2007;3:2135–2146. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Kramer U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- Harborne JB. Phytochemical methods. London: Chapman and Hall; 1984. [Google Scholar]
- Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell'Olivo A, Arnold M, Kuhlemeier C. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Stevenson TW, Cornish EC. Cloning and expression of cytochrome-P450 genes-controlling flower color. Nature. 1993;366:276–279. doi: 10.1038/366276a0. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature. 2011;469:411–414. doi: 10.1038/nature09641. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Differential loss of ancestral gene families as a source of genomic divergence in animals. Proc R Soc B Biol Sci. 2004;271:S107–S109. doi: 10.1098/rsbl.2003.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ET, Ryu S, Yi HK, Shin B, Cheong H, Choi G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001;25:325–333. doi: 10.1046/j.1365-313x.2001.00962.x. [DOI] [PubMed] [Google Scholar]
- Jung CS, Griffiths HM, De Jong DM, Cheng SP, Bodis M, De Jong WS. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theor Appl Genet. 2005;110:269–275. doi: 10.1007/s00122-004-1829-z. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos Trans R Soc B Biol Sci. 2010;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:research0054. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 2004;135:756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM. A molecular phylogeny of the Solanaceae. Taxon. 2008;57:1159–1181. [Google Scholar]
- Olsen KM, Hehn A, Jugde H, Slimestad R, Larbat R, Bourgaud F, Lillo C. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010;10:21. doi: 10.1186/1471-2229-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:E73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit P, Granier T, d'Estaintot BL, Manigand C, Bathany K, Schmitter JM, Lauvergeat V, Hamdi S, Gallois B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J Mol Biol. 2007;368:1345–1357. doi: 10.1016/j.jmb.2007.02.088. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:E45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ML, Crandall KA. Lost along the way: the significance of evolution in reverse. Trends Ecol Evol. 2003;18:541–547. [Google Scholar]
- Preston JC, Martinez CC, Hileman LC. Gradual disintegration of the floral symmetry gene network is implicated in the evolution of a wind-pollination syndrome. Proc Natl Acad Sci U S A. 2011;108:2343–2348. doi: 10.1073/pnas.1011361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JR, Sturgess VC. A survey of anthocyanins VI. Biochem J. 1938;32:1658–1660. doi: 10.1042/bj0321658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompler H, Rohland N, Lalueza-Fox C, Willerslev E, Kuznetsova T, Rabeder G, Bertranpetit J, Schoneberg T, Hofreiter M. Nuclear gene indicates coat-color polymorphism in mammoths. Science. 2006;313:62. doi: 10.1126/science.1128994. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Rompler H, Schoneberg T, Hoekstra HE. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc Natl Acad Sci U S A. 2010;107:2113–2117. doi: 10.1073/pnas.0911042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KG, Swinny EE, Markham KR, Winefield C. Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry. 2002;59:23–32. doi: 10.1016/s0031-9422(01)00404-6. [DOI] [PubMed] [Google Scholar]
- Ryan KG, Swinny EE, Winefield C, Markham KR. Flavonoids and UV photoprotection in Arabidopsis mutants. Z Naturforsch Sect C J Biosci. 2001;56:745–754. doi: 10.1515/znc-2001-9-1013. [DOI] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Chu IK, Yip WK, Lo C. Differential expression of two flavonoid 3′-hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 2006;47:1412–1419. doi: 10.1093/pcp/pcl003. [DOI] [PubMed] [Google Scholar]
- Sire JY, Delgado SC, Girondot M. Hen's teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD. Floral diversification and pollination biology of the Andean clade Iochrominae (Solanaceae) Madison (WI): Department of Botany, University of Wisconsin; 2006. [Google Scholar]
- Smith SD, Baum DA. Phylogenetics of the florally diverse Andean clade Iochrominae (Solanaceae) Am J Bot. 2006;93:1140–1153. doi: 10.3732/ajb.93.8.1140. [DOI] [PubMed] [Google Scholar]
- Smith SD, Baum DA. Systematics of Iochrominae (Solanaceae): patterns in floral diversity and interspecific crossability. Acta Hortic. 2007;745:241–254. [Google Scholar]
- Smith SD, Kolberg VJ, Baum DA. Morphological and cytological evidence for homoploid hybridization in Iochroma. Madroño. 2008;55:280–284. [Google Scholar]
- Snowden KC, Napoli CA. Psl: a novel Spm-like transposable element from Petunia hybrida. Plant J. 1998;14:43–54. doi: 10.1046/j.1365-313x.1998.00098.x. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Rompler H, Boettger LM, Schoneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol Biol Evol. 2009;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLos Biol. 2007;5:1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. Altered trans-regulatory control of gene expression in multiple anthocyanin genes contributes to adaptive flower color evolution in Mimulus aurantiacus. Mol Biol Evol. 2009a;26:433–444. doi: 10.1093/molbev/msn268. [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. New Phytol. 2009b;183:751–763. doi: 10.1111/j.1469-8137.2009.02929.x. [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. Population genetics, pleiotropy and the preferential fixation of mutations during adaptive evolution. Evolution. 2011;65:629–642. doi: 10.1111/j.1558-5646.2010.01165.x. [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci U S A. 2005;102:2460–2465. doi: 10.1073/pnas.0409474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall JB, Hodges SA. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447:706–709. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
- Whittall JB, Voelckel C, Kliebenstein DJ, Hodges SA. Convergence, constraint and the role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Mol Ecol. 2006;15:4645–4657. doi: 10.1111/j.1365-294X.2006.03114.x. [DOI] [PubMed] [Google Scholar]
- Wilson P, Wolfe AD, Armbruster WS, Thomson JD. Constrained lability in floral evolution: counting convergent origins of hummingbird pollination in Penstemon and Keckiella. New Phytol. 2007;176:883–890. doi: 10.1111/j.1469-8137.2007.02219.x. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc Natl Acad Sci U S A. 2003;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Baum DA. Transgenic study of parallelism in plant morphological evolution. Proc Natl Acad Sci U S A. 2004;101:6524–6529. doi: 10.1073/pnas.0401824101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, Rausher MD. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature. 2004;428:847–850. doi: 10.1038/nature02489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.