Abstract
This work describes the project for an advanced undergraduate laboratory course in cell and molecular biology. One objective of the course is to teach students a variety of cellular and molecular techniques while conducting original research. A second objective is to provide instruction in science writing and data presentation by requiring comprehensive laboratory reports modeled on the primary literature. The project for the course focuses on a gene, MSH2, implicated in the most common form of inherited colorectal cancer. Msh2 is important for maintaining the fidelity of genetic material where it functions as an important component of the DNA mismatch repair machinery. The goal of the project has two parts. The first part is to create mapped missense mutation listed in the human databases in the cognate yeast MSH2 gene and to assay for defects in DNA mismatch repair. The second part of the course is directed towards understanding in what way are the variant proteins defective for mismatch repair. Protein levels are analyzed to determine if the missense alleles display decreased expression. Furthermore, the students establish whether the Msh2p variants are properly localized to the nucleus using indirect immunofluorescence and whether the altered proteins have lost their ability to interact with other subunits of the MMR complex by creating recombinant DNA molecules and employing the yeast 2-hybrid assay.
Keywords: undergraduate, laboratory course, yeast, mismatch repair, colorectal cancer, molecular biology
INTRODUCTION
Seminal reports from committees examining undergraduate education in the research sciences strongly support an inquiry-based approach to learning (National Research Council Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century, 2003; The Boyer Commission on Educating Undergraduates in the Research University, 1998). The course outlined in this paper is designed to use an inquiry-based approach to teach students a variety of current research laboratory methods in the context of a novel research project. Students gain practical skills and theoretical understanding while experiencing the progression and complexity of a real research project.
Future research biologists are predicted to need skills from many disciplines, including mathematics, physics, chemistry, engineering, and computer science (National Research Council Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century, 2003). The training of these multi-faceted future research scientists requires an integrated curriculum with courses that are accessible and interesting to a variety of students. The Advisory Committee to the National Science Foundation Directorate for Education and Human Resources (1996) and the authors of a report for Project Kaleidescope (Rothman and Narum, 1999) encourage endeavors to educate a variety of science students. The subject for the project outlined in this paper was selected to capture the interest of a broad spectrum of enrolled students. The topic is centered on a highly conserved, fundamental biological process with a direct impact on a significant human disease.
Background for the Research Project
In the United States in the year 2003, the American Cancer Society estimates that colorectal cancer (CRC) will be diagnosed in 147,500 individuals and will result in 57,100 deaths. Approximately 2–7% of these CRC cases are a consequence of the most commonly inherited form of the disease, hereditary non-polyposis colorectal cancer (HNPCC) (reviewed recently in Boland, 2000; Muller and Fishel, 2002; Peltomaki, 2001). As far back as 1913, HNPCC was recognized as having a genetic basis (Warthin, 1913). Eighty years later, an exciting convergence of basic science and medical research proved that HNPCC results from defects in DNA mismatch repair (MMR) genes (Aaltonen et al., 1993; Fishel et al., 1993; Ionov et al., 1993; Leach et al., 1993; Parsons et al., 1993; Peltomaki et al., 1993; Strand et al., 1993; Thibodeau et al., 1993).
DNA MMR is a highly conserved process (reviewed recently in Harfe and Jinks-Robertson, 2000; Marti et al., 2002). Early work in prokaryotes revealed that certain genes when inactivated led to a pronounced increase in the rate of spontaneous mutations (Cox et al., 1972; Nevers and Spatz, 1975; Siegel and Bryson, 1967). Identification of several of these genes including mutH, mutL, mutS, and uvdR/mutU established the importance of prokaryotic methyl-directed DNA mismatch repair in maintaining genomic integrity (Bauer et al., 1981; Glickman, 1979; Glickman and Radman, 1980; Jones and Wagner, 1981; Lu et al., 1983; Pukkila et al., 1983; Rydberg, 1978; Shanabruch et al., 1983).
In nature, DNA mismatches form primarily as a consequence of replication errors and chemical modification (reviewed recently by Aquilina and Bignami, 2001; Kunkel and Bebenek, 2000). Without an intact mismatch repair system, DNA begins to accumulate mutations, some of which will be deleterious to the cell or organism. In addition to the increased rate of single base-pair mutations, researchers observed that repetitive DNA elements (for example mono, di and tri-nucleotide repeats) are highly unstable in mismatch repair defective strains (Levinson and Gutman, 1987; Strand et al., 1993). Interestingly, these findings contributed to the establishment of the link between HNPCC and mismatch repair, given that a hallmark of HNPCC tumors is highly unstable repetitive DNA elements (Aaltonen et al., 1993; Ionov et al., 1993; Parsons et al., 1993; Thibodeau et al., 1993).
Closely related eukaryotic homologs of MutS (MutS Homolog or MSH genes) (Reenan and Kolodner, 1992) and MutL (MutL Homolog or MLH and some Post-Meiotic Segregation or PMS genes) (Horii et al., 1994; Lindblom et al., 1993; Nicolaides et al., 1994; Papadopoulos et al., 1994) are found in diverse organisms ranging from yeast to humans. While MutS functions as a homodimer to bind mispaired DNA in prokaryotes, the similar function is accomplished in eukaryotes by two distinct heterodimers known as MutS α, composed of Msh2 and Msh6, and MutSβ, composed of Msh2 and Msh3 (Figure 1). MutSα binds primarily to single-base pair mismatches and small insertion/deletion loops. MutS β binds to larger insertion/deletion loops. A heterodimer composed of Mlh1 and Pms1 (PMS2 in humans) functions as the primary MutL equivalent in DNA mismatch repair in eukaryotes. The MutH endonuclease confers stand specificity for repair in prokayotes, where the newly synthesized unmethylated strand is cleaved for repair. A strand specificity factor in eukaryotes is postulated to be Pol30/PCNA, although the precise mechanism of stand discrimination is not clear. Downstream events including strand displacement, excision and new synthesis are currently being investigated and include helicases (Mechanic et al., 2000; Yamaguchi et al., 1998), exonucleases (Burdett et al., 2001; Genschel et al., 2002), polymerases, and ligases (Tomkinson and Mackey, 1998).
Figure 1.
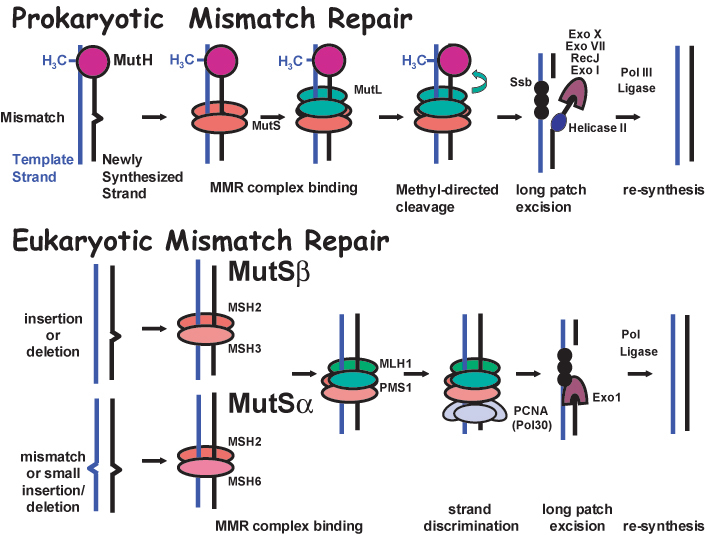
Schematic model for DNA mismatch repair. While MutS functions as a homodimer to bind mispaired DNA in prokaryotes, the similar function is accomplished in eukaryotes by two distinct heterodimers known as MutS β, composed of Msh2 and Msh6, and MutS α, composed of Msh2 and Msh3. MutSα binds primarily to single-base pair mismatches and small insertion/deletion loops. MutS β binds to larger insertion/deletion loops frequently generated at repetitive DNA elements as DNA polymerase slips during replication. A heterodimer composed of Mlh1 and Pms1 functions as the MutL dimer equivalent in DNA mismatch repair in eukaryotes. The MutH endonuclease confers stand specificity for repair in prokayotes, where the newly synthesized unmethylated strand is cleaved for repair. The strand specificity factor in eukaryotes is thought to be Pol30/PCNA. Downstream events including strand excision and new synthesis include the activities of helicases (Helicase II), exonucleases (ExoI, ExoX, ExoVII, RecJ and Exo1), single stranded binding proteins (SSb), DNA polymerases (Pol III and Polσ /ϵ) and ligases (Ligase).
After the establishment of the connection between HNPCC and DNA MMR, many germline mutations have been mapped in afflicted families. Most mutations reside in one two genes, MSH2 and MLH1. Individuals inheriting a defective mismatch repair locus have an 80–90% chance of manifesting cancer before the age of 50 (Lynch and Lynch, 2000). The high penetrance and early age of onset of the disease underscores the need for aggressive identification and screening of susceptible individuals.
The focus of this research project is MSH2. Since the discovery of the role hMSH2 in HNPCC many alterations in the gene have been found in afflicted families.1 Included among these changes is a collection of nearly 30 missense mutations. With missense mutations one must always consider that a single amino acid change does not necessarily result in a dysfunctional protein. Certain changes are functionally silent. In some cases, there are not sufficient family members to do a careful pedigree analysis to ensure that the mapped mutation is indeed pathogenic. It is part of the objective of the course to determine whether the mapped missense mutations found at hMSH2 loci result in a dysfunctional protein (pathogenic polymorphisms) or if the missense changes are likely to be benign polymorphisms in the human population. If missense mutations are to be used in genetic counseling, it is imperative to determine which changes result in a defective protein. We also use the mutations that have been found in clinical isolates to define important structure-function relationships in the Msh2 protein.
MATERIALS AND METHODS
Basic Molecular and Microbial Techniques
The mutagenesis procedures are provided in detail elsewhere (Muta-Gene Phagemid In Vitro Mutagenesis, Version 2, Bio-Rad Laboratories, Hercules, CA) (Ausubel et al., 1994). The mutagenesis components not commercially available include the mutagenic oligonucleotides (Princeton Syn/Seq Facility) and template DNA (see Figure 2). Transformation of CaCl2 and electro-competent bacterial cells and subsequent microbial manipulations are conducted according to Ausubel et al. (1994). Small-scale preparations of plasmid DNA are accomplished by the alkaline lysis method (Ausubel et al., 1994). Large-scale preparation of plasmid DNA is achieved by using anion exchange chromatography (Midi-prep procedure, Qiagen Inc., Valencia, CA). Polymerase chain reactions (PCR) are conducted using standard conditions (Ausubel et al., 1994). Plasmid DNA and PCR fragments are digested using the conditions specified by the manufacturer (New England Biolabs, Inc., Beverly, MA). Agarose gel reagents and electrophoresis parameters are detailed elsewhere (Ausubel et al., 1994). Missense mutations are sequenced at the nucleotide level using dideoxy nucleotide sequencing Sequenase Version 2 chain-termination sequencing reagents, [α-35S]dATP (Amersham Biosciences, Piscataway, NJ), MSH2 specific primers (Princeton Syn/Seq facility) and template DNA.
Figure 2.
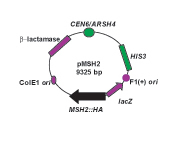
Schematic map of the MSH2 phagemid construct, pMSH2. The 9325 base pair (bp) pMSH2 construct depicted contains the promoter and open reading frame for the MSH2 gene inserted into the pRS413 shuttle vector (Sikorski and Hieter, 1989). The hemagglutinin (HA) epitope is repeated a total four times in the MSH2 gene (MSH2::HA) to allow for immunodetection of the protein product with α-HA antibodies. A single hemagglutinin (HA) epitope is located at an NcoI site within MSH2 (Alani et al., 1995) and a triple HA epitope also engineered by Eric Alani is located in the extreme C-terminal coding region. The construct is described as a phagemid since it is capable of replicating by a single-stranded phage origin F1 (+) or by a double-stranded plasmid origin of replication (ColE1 ori). This construct produces single-stranded DNA with the coding (+) strand of lacZ when replicated from the F1 (+) single-stranded origin of replication upon superinfection with the helper phage M13K07. The arrows represent the orientation of the relevant open reading frames. For selection in bacteria, the construct contains the β-lactamase gene, a gene conferring resistance to β-lactam antibiotics including ampicillin, carbinicillin, and penicillin. The plasmid is replicated in yeast via an autonomously replicating sequence (ARSH4) and maintained in yeast cells with a centromere (CEN6). The plasmid is selected for by growing his3 defective strains harboring the plasmid in media lacking histidine (–HIS). Single-stranded uracil containing pMSH2 template DNA was extracted and purified from the extruded phage particles (Ausubel et al., 1994) prior to the course for student use.
Preparation of Yeast Strains for the Mismatch Repair Functional Assays
Mutagenized plasmid, pMSH2 and pRS413 are used to transform AGY75 (Table 1), a reporter strain constructed prior to the course using the lithium acetate method (Burke et al., 2000). The selective medium is lacking histidine (–HIS) to select for transformants harboring the mutagenized plasmid, pMSH2, or pRS413. The medium also is missing tryptophan (–TRP) to select for the pSH44 dinucleotide reporter construct (described in Figure 4 and 5A) as well as leucine (–LEU) and threonine (–THR) to induce expression of the GT16.5–URA3 fusion transcript from the LEU2 inducible promoter. Finally, the medium lacks uracil (–URA) to ensure that no instabilities in the dinucleotide repeat had been incorporated before the functional assays are initiated. These selective plates are described as– HIS –TRP –LEU –THR –URA. All yeast manipulations, including media preparation, are conducted as described previously (Burke et al., 2000).
Table 1.
Yeast MSH2 alleles and their human equivalent analyzed
| Yeast msh2 allele designation | Human MSH2 equivalent |
|---|---|
| N123S | N127S |
| I141M | I145M |
| D164H | D167H |
| E194G | E198G |
| C195R | C199R |
| G317D | G322D |
| S318C | S323C |
| C345R | C333R |
| C345Y | C333Y |
| L402F | L390F |
| L457P | L440P |
| D524Y | D506Y |
| R542P | R524P |
| E580V | E562V |
| P640L | P622L |
| H658R | H639R |
| H658Y | H639Y |
| G693D | G674D |
| G693S | G674S |
| M707I | M688I |
| G711R | G692R |
| C716F | C697F |
| C716R | C697R |
Yeast allele designation is as follows: codon 123 changed from asparagine (N) to serine (S) is termed N123S. Please note that the human and yeast equivalent codons do not align exactly, for example human N127S is N123S in yeast.
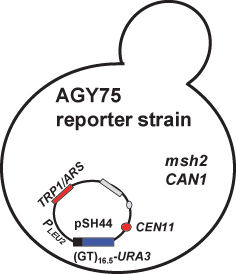
Figure 4.
Mismatch repair reporter strain: The genotype of the chromosome reporter strain (AGY75) is given in Table 1. Relevant features of strain are that the MSH2 gene is deleted (msh2Δ), it encodes a functional arginine permease gene (CAN1), and it conta ins a dinucleotide repeat instability reporter construct, pSH44, generously provided by Thomas Petes (University of North Carolina at Chapel Hill) (Henderson and Petes, 1992). pSH44 has a dinucleotide sequence, GT, repeated 16.5 times (GT16.5) in frame with the URA3 gene. Expression of the fusion product is driven by the LEU2 promoter (PLEU2), which is activated under conditions where leucine and threonine are limiting (–LEU –THR). The pSH44 construct is replicated in yeast via an autonomously replicating sequence (ARS) associated with TRP1 (TRP1/ARS). The plasmid is stably maintained in yeast by a centromere (CEN11) and is selected for by growing trp1 defectivestrains harboring the plasmid in media lacking tryptophan (–TRP).
Figure 5.
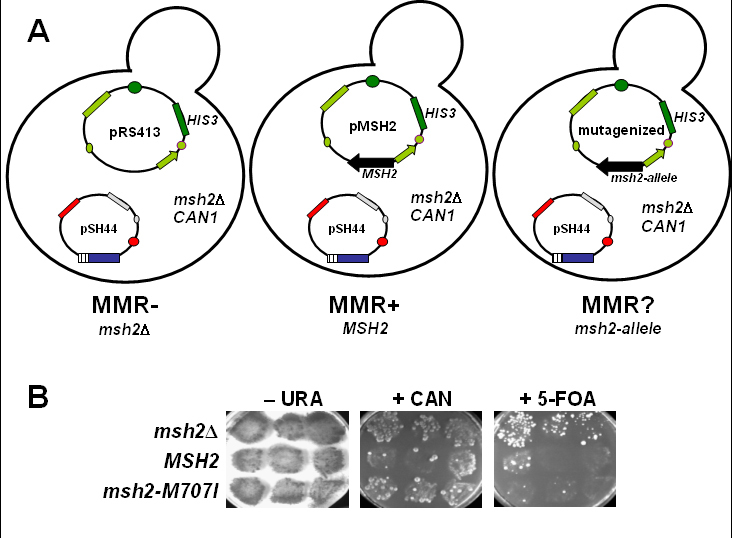
Mismatch repair assay: (A) Three strains for the MSH2 mismatch repair functional assays are generated by transformation of the AGY75 reporter strain detailed in Figure 3. Three plasmids are introduced to generate the strains. One strain will be lacking MSH2 (msh2Δ), a second will express the wild-type MSH2 gene (MSH2), and the third will express the MSH2 missense allele (msh2-allele). When the msh2Δ reporter strain contains the vector pRS413, the strain is completely defective for mismatch repair (MMR–) because no Msh2 protein is expressed. When the reporter strain contains pMSH2, the encoded MSH2 gene product is able to complement the defect of msh2Δ and therefore is mismatch repair proficient (MMR+). The mismatch repair phenotype of the reporter strain containing the plasmid expressing the msh2 allele (pMSH2-allele) is unknown (MMR?). To assess the function of the altered form of Msh2, the results are compared to the two controls to determine the MMR phenotype. (B) Qualitative functional mismatch repair student results. Yeast msh2Δ cells transformed with either pRS413 (msh2Δ), pMSH2 (MSH2), or pMSH2-M707I (msh2-M707I) were streaked in the indicated positions on– HIS –TRP –LEU –THR –URA plates and incubated at 30°C for 2 d. The patches were replica-plated onto –HIS –TRP– LEU –THR –URA (-URA), onto –HIS –TRP –LEU– THR –HIS –ARG + CAN (+ CAN), and onto –HIS –TRP– LEU –THR + 5-FOA (+ 5-FOA) plates. Significant growth on +CAN or +FOA plates indicates a mismatch repair defect. Similar growth for all patches on the –URA plate indicates that an equal number of cells were dispensed to all of the plates. (Panel B figure and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
Qualitative Mismatch Repair Assays
Three colonies from each of the transformations are transferred using sterile wooden sticks to a defined position approximately 20 mm in diameter on a fresh –HIS –TRP –LEU –THR –URA plate. After 2 d of growth at 30°C, yeast strains are replica-plated onto drug-containing media to assess the mismatch repair phenotype (described in detail below) and to –HIS –TRP medium as a control to ensure that the cells were properly delivered to all of the plates.
The drug selection for dinucleotide instability is synthetic medium lacking histidine, tryptophan, leucine, threonine and supplemented with 5-fluororotic acid monohydrate, 5-FOA (Toronto Research Chemicals Inc., North York, ON, Canada). The medium is designated –HIS –TRP –LEU –THR + 5-FOA to reflect the nutritional deficiencies and the presence of the drug 5-FOA. A second type of medium to score primarily for single base pair mismatch repair defects is supplemented with 60 μg/ml canavanine, CAN (Sigma-Alderich, Milwaukee, WI). The medium is also lacking histidine, tryptophan, arginine and is designated –HIS –TRP –ARG + CAN. Images of the plates are taken with a FOTO/Analyst Fotodyne Camera and FOTO/Analyst PC Image software (Fotodyne, Hartland, WI) and saved to a network drive so that the students are able to access their images from remote computers.
Quantitative Assays for Mismatch Repair
The same strains employed in the qualitative test above are used to inoculate liquid synthetic medium lacking histidine and tryptophan (–HIS– TRP). Cultures are grown overnight at 30°C with aeration. The approximate cell concentration is determined (Burke et al., 2000) and students either concentrate the culture or perform serial dilutions so that the appropriate number of cells is delivered to each plate.
Based on the qualitative assay, students are able to determine whether the msh2 allele has a mismatch repair phenotype that is similar to wild type (MMR+) or similar to msh2 null (MMR–). Based on this information, the cells with the msh2 missense allele are either processed like wild type or like msh2 null cells. The cultures must be treated differently because if the mutation rate is higher (as is the case for the msh2 null allele strain), fewer cells are plated on the drug-containing plates to allow for an accurate colony count. To select for Canr mutant ∼6 × 107 cells of the MMR + cells are plated on –HIS –TRP –ARG + CAN, by concentrating the cultures in a centrifuge for 5 min at 3000 rpm to allow for efficient plating volumes. Approximately 106 cells for each of the MMR–cells are plated on –HIS –TRP –ARG + CAN. To select for FOAr mutant∼1.4 × 106 MMR + and∼ 104 of the MMR–cells are plated onto –TRP– HIS –LEU –THR + FOA plates. To obtain a viable cell count for each culture ∼ 100 cells for each of the samples are plated onto– HIS –TRP plates. The plates are placed at 30°C for 2–5 d.
Steady-State Expression Levels of the Msh2p Variants by Immunoblotting
Proteins extracts (Burke et al., 2000) are resolved using SDS polyacrylamide gel electrophoresis (SDS-PAGE) (Ausubel et al., 1994). The resolving gel is a 7% acrylamide gel (37.5:1 acrylamide to bis-acrylamide ratio). Samples and markers are resolved in duplicate halves on the gel. One half is subjected to for Coomassie Brilliant Blue R (Sigma–Alderich) staining and the duplicate is processed for immunoblotting (Ausubel et al., 1994). Immunodetection of Msh2p expressing the hemagglutinin epitope (HA) is conducted according to the ECL method (Amersham Biosciences). The primary antibody used is mouse 12CA5 monoclonal antibody specific for the HA epitope (Princeton Monoclonal Facility). The blot is incubated at room temperature in the presence of the primary antibody for 1 hour. After the specified washes, the secondary antibody, α-mouse-horse radish peroxidase (HRP)-conjugated secondary antibody (Amersham Biosciences) is applied for 45 min. Both antibodies are used at a 1:2500. After the final washes, the ECL detection reagents are applied and the blot is processed as recommended by the manufacturer.
Localization of the Msh2p Variants Using Indirect Immunofluorescence
The procedure for indirect immunofluorescence of HA epitope tagged Msh2 (Msh2::HA) is based on previous protocols (Pringle et al., 1989). Cultures of exponentially growing yeast strains (the same strains used in the functional assays) are fixed in ∼4% formaldehyde for 30 min at 30°C, washed and digested for 1 hour with Zymolyase 100T (Seikagaku Corporation, Japan) as recommended. The primary antibody for detection of Msh2::HA is 12CA5 diluted 1:400. In addition, control wells, are prepared with a mixture of 12CA5 and with rat monoclonal α-tubulin (YL1/2, Serotec, Raleigh, NC) each diluted 1:400. The primary antibody incubation is for 30 min at room temperature. Wells are washed and the secondary antibody mixture is applied for 45 min at room temperature in a moist chamber. The secondary antibody mixture is a combination of goatα -mouse IgG Alexa Fluor 488 diluted 1:200 and donkey α-rat IgG Alexa Fluor 594 diluted 1:400 (both from Molecular Probes, Eugene, OR). Cells are washed extensively, stained with freshly diluted 4',6'-diamidino-2-phenylindole, DAPI (1 μg/ml in PBS) for 5 min at room temperature, washed twice and prepared for microscopy as recommended (Pringle et al., 1989). Cells are examined with a Nikon Eclipse E600 (Nikon, Melville, NY) containing a DIC H Plan Fluor 100X oil immersion objective lens (NA = 1.3). Fluorescence was visualized using Nikon filter sets: UV-2A (for DAPI stained nuclei and mitochondria), B-2A (for Msh2 localization), and G-2A (for tubulin localization). Images are recorded using a Nikon DXM1200 digital camera and ACT-1 software, version 2 (Nikon) and stored as electronic files onto network drives.
Recombinant DNA Cloning
The entire sample containing digested pMSH2-allele construct and 1 μg of digested pGBD-MSH2 are resolved by agarose gel electrophoresis. The appropriate bands are excised and extracted using the Bio101 Gene Clean Spin Extraction Kit (Qbiogene, Carlsbad, CA). The yield of extracted insert and vector DNA is determined by comparing a portion of the sample to a concentration curve of digested pGBD-MSH2. The pmol/μl of 5' termini for each fragment are determined and ligation reactions are prepared such that∼ 3- to 5-fold molar excess of 5' termini of insert DNA compared to vector is present in the sample. A control tube is prepared with pGBD-MSH2 vector fragment alone with no insert fragment. In addition, ligase controls include a singly cut vector with and without T4 DNA ligase. Ligation reaction reactions are conducted according to the specifications of the enzyme manufacturer (New England Biolabs). Products of the ligation reaction and the control samples are used to transform bacterial cells by electroporation (Ausubel et al., 1994). A control tube in which no DNA has been added is also prepared to control for sterility. A final uncut vector control is prepared to compare with the singly cut vector with and without ligase. Screening for recombinant molecules employs basic molecular techniques described above, including the alkaline lysis extraction procedure, PCR, restriction endonuclease digestions, and analytical agarose gel electrophoresis. The control DNA for these diagnostic tests is pGBD-MSH2. In addition, a second diagnostic restriction endonuclease digestion is conducted to verify that the digestion sites used in the procedure are still encoded in the recombinant plasmids.
Yeast 2-hybrid Assays
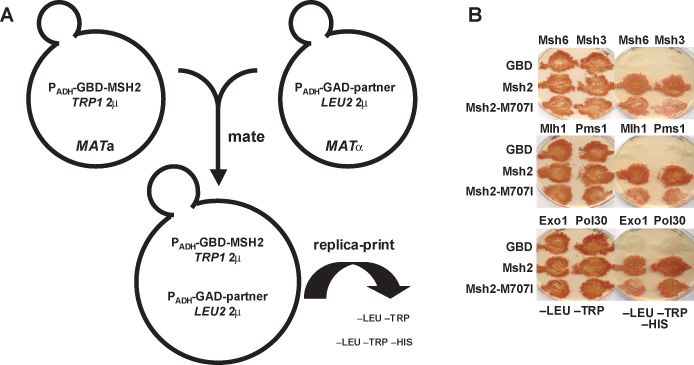
pGBD-C2, pGBD-MSH2, and the recombinant plasmid containing the fusion between GBD and the msh2 allele (pGBD-MSH2-allele) are introduced into PJ69-4A, a MATa yeast 2-hybrid reporter strain (James et al., 1996) using the lithium acetate transformation method (Burke et al., 2000). Cells are plated onto medium lacking tryptophan (–TRP) to select for the transformants. Before the course, the GAD fusion constructs (Figure 9A) were introduced into PJ69-4α, a MATα yeast 2-hybrid reporter strain (James et al., 1996) and distributed. The students set up crosses to form diploid yeast strains on YEPD agar plates. The plates were incubated for 24 h at 30°C and replicaprinted to selective plates lacking leucine and tryptophan (–LEU– TRP) or lacking leucine, tryptophan, and histidine (–LEU– TRP –HIS). The plates are incubated for 2–5 d at 30°C and are photographed as described above for the qualitative mismatch repair assays.
Figure 9.
2-hybrid assay to analyze subunit interaction. (A) Schematic illustration of the basic 2-hybrid constructs. The 8992 bp pGBD-MSH2 construct contains a modification of the alcohol dehydrogenase promoter (PADH) allowing for constitutive expression of an in-frame fusion between MSH2, including the hemagglutinin epitope and the GAL4 DNA Binding Domain (GBD). The vector is pGBD-C2 described previously (James et al., 1996). These plasmids are replicated in yeast via the high-copy 2 μ origin of replication and are selected for in trp1 defective yeast strains using medium lacking tryptophan (–TRP). The GAD constructs also contain the PADH promoter allowing for constitutive expression of an in-frame fusion between the open reading frames for the functional partners of Msh2p and the GAL4 Activating Domain (GAD). The functional partners include MSH6, MSH3, MLH1, PMS1, EXO1 and POL30. The vector is pGAD-C2 (James et al., 1996). The fusion constructs were made by PCR amplification of the partner open reading frames (partner) from yeast genomic DNA and the in frame fusion constructs were formed by in vivo recombination techniques (Oldenburg et al., 1997). The GAD plasmids are replicated in yeast via the high-copy 2 μ origin of replication and are selected for by growing leu2 defective yeast strains containing the constructs in medium lacking leucine (–LEU). Each haploid yeast strain contains a plasmid that will express a hybrid protein. After mating the haploid cells and the resulting diploids will carry both constructs necessary for assaying the 2-hybrid protein-protein interaction. (B) Student results from the 2-hybrid experiments to assay MMR subunit formation. MATa cells were transformed with pGBD-C2, pGBD-MSH2, and pGBD-msh2-M707I and mated on YEPD with individual MATÿstrains harboring pGAD-MSH6, pGAD-MSH3, pGAD-MLH1, pGAD-PMS1, pGAD-EXO1, and pGAD-POL30. Cells were sequentially replica plated onto –LEU –TRP –HIS, and –LEU –TRP and incubated at 30ÿC for 4 days. –LEU –TRP selects for diploids harboring both pGAD and pGBD constructs. Growth on –LEU –TRP –HIS indicates a 2-hybrid interaction. GBD fusions are listed to the left of each plate; GAD fusions are listed above each plate. GBD indicates cells transformed with the pGBD-C2 vector control. (Panel B figure and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
RESULTS
Overview of the Basic Project
Students work in pairs to study a MSH2 missense mutation that is listed as pathogenic in the human databases (see Table 1 for a listing of alleles generated). The missense mutation is introduced in the cognate yeast MSH2 coding sequence so that the MSH2 gene harboring the mutation is distinguishable from the wild type by restriction endonuclease digestion. Students definitively prove that the mutation has been engineered by nucleotide sequence analysis of the mutated region. Ultimately, the mutant alleles are analyzed using in vivo DNA mismatch repair assays. The remainder of the course is focused on discerning in what way are the altered Msh2 proteins malfunctioning in mismatch repair. Students examine steady-state protein levels by immunoblotting to determine if the altered amino acid might affect the stability of the protein. Additionally, they determine whether the Msh2p variants are properly localized to the nucleus using indirect immunofluorescence. Finally, the students test whether the altered proteins have lost their ability to interact with other subunits of the mismatch repair complex using the yeast 2-hybrid assay.
Mutagenesis Strategy
Using site-directed mutagenesis the students create human missense mutations designated as pathogenic in the cognate yeast MSH2 gene. To introduce the changes the students employ a mutagenesis technique known as the dut ung method (Kunkel, 1985). The technique uses single-stranded uracil-containing template DNA, a mutagenic oligonucleotide to prime DNA synthesis, deoxynucleotides and T7 DNA polymerase to synthesize the mutagenized DNA strand, and T4 DNA ligase to create a covalently closed double stranded DNA plasmid. After the in vitro synthesis, the synthesized plasmids are introduced into a bacterial strain that will preferentially repair the parental uracil-containing strand, resulting in a high yield of transformants containing the mutagenized plasmid. Most of the components for the mutagenesis are commercially available. The unique reagents include the template DNA and the mutagenic oligonucleotides. The MSH2 containing template DNA (pMSH2) was constructed prior to the course and is described in detail in Figure 2 and the mutagenic oligonucleotides are designed as detailed in the following section.
Designing Mutagenic Oligonucleotides
The students begin the project by designing mutagenic oligonucleotides. The procedure relies on Accelrys SeqWeb Version 2, a Web-based sequence analysis program, to aid in the design. Each pair is assigned a missense allele found in the human databases. The databases consulted include the International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC), and the Human Gene Mutation Database (in association with Celera). The cognate amino acid is found in yeast gene by comparing the human and yeast sequences. A yeast codon preference table (ftp://genome-ftp.stanford.edu/pub/codon/ysc.g63.cod) is consulted when planning the mutagenesis to ensure that the mutated protein will not have decreased expression as a consequence of rare codon usage at the altered amino acid site. In addition, the students engineer a mutation that will either create or destroy a restriction endonuclease site within the coding region of the gene. Oligonucleotides are designed to be 23 nucleotides in length with 10 nucleotides flanking the codon to be altered. Finally, the proper strand must be specified so that the mutagenic oligonucleotide will anneal to the single-stranded template DNA using the information given in Figure 2. Before ending the computer session, the students determine the restriction endonuclease digestion pattern that would be predicted for the unmutagenized and mutagenized plasmids.
Site-Directed Mutagenesis
Over several laboratory sessions, the mutagenic oligonucleotide is phosphorylated and used to prime synthesis of the complementary strand of the uracil-containing DNA template in an in vitro mutagenesis reaction. Each student pair prepares three samples. One contains all of the listed components, a second lacks the mutagenic oligonucleotide (no primer control), and a third lacks the T4 DNA ligase (the no ligase control).
One tenth of the reaction products and controls are resolved by agarose gel electrophoresis in the presence of ethidium bromide to determine whether the expected double-stranded covalently closed products are generated before introducing the molecules into bacteria.The students are provided with double-stranded and single-stranded pMSH2 controls for comparison purposes (see Figure 3A for an example of a student's results).
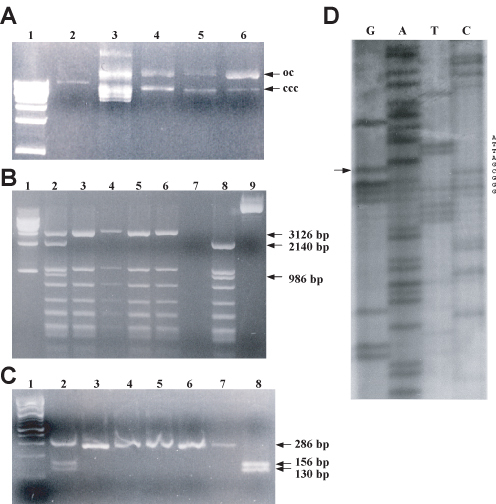
Figure 3.
Student results for creating and confirming a site-directed missense mutation in the MSH2 gene. (A) Electrophoresis of site-directed mutagenesis products. Electrophoresis was performed using a 1% agarose gel containing 0.5 μg/ml ethidium bromide. The lanes were loaded as follows: 1 kb ladder (lane 1); double-stranded pMSH2 DNA (lane 2); single-stranded uracil-containing pMSH2 DNA (lane 3); single-stranded uracil-containing DNA incubated with phosphorylated mutagenic oligonucleotide, ATP, dNTPs, T7 DNA polymerase, and T4 DNA ligase (lane 4); `no primer' control (lane 5); and `no ligase control' (lane 6). Bands were visualized by UV transillumination. oc, open circular (nicked) DNA; ccc, covalently closed circular DNA. ccc DNA migrates faster than oc DNA through an agarose gel due to ethidium bromide-induced supercoiling. (B) Confirmation of missense mutation M707I by digestion and electrophoresis of whole plasmid DNA. Unmutagenized pMSH2 (lane 8) and alkaline lysis purified DNA from six E. coli transformants (lanes 2–7) produced by transformation with in vitro synthesized plasmid DNA were digested with Hae III at 37°C for 1 h. Digestion products were analyzed by electrophoresis with a 1.5% agarose containing 0.25 μg/ml ethidium bromide and visualized by UV transillumation. Lane 1 contains the 1 kb ladder and lane 9 contains undigested pMSH2. The missense mutation eliminates a HaeIII site. The relevant fragment sizes are indicated next to the image of the gel. (C) Confirmation of missense mutation by digestion and electrophoresis of PCR products. Unmutagenized pMSH2 (lane 8) and alkaline-lysis purified DNA from the six E. coli transformants (lanes 2–7) described above were subjected to 35 cycles of PCR amplification (see Materials and Methods) and were subsequently digested with HaeIII at 37°C for 1 h. Digestion products were analyzed by electrophoresis on a 3% agarose gel with 0.25 μg/ml ethidium bromide and visualized by UV transillumation. Lane 1 contains the 100-bp ladder. The relevant fragment sizes are indicated next to the image of the gel. (D) Dideoxynucleotide sequencing of the missense mutation. Plasmid DNA confirmed above to be likely to contain the missense mutation was digested for 15 min at 37°C with PvuII in the presence of 0.5 pmol/μl of primer to relieve supercoiling. After denaturing at 100°C for 3.5 min, ice-cold samples were incubated with [α-35S]dATP-containing labeling mixture at room temperature for 3.5 min and incubated with the appropriate termination mixture for 5 min. DNA was denatured and run on a 6% polyacrylamide sequencing gel. Arrow indicates the engineered mutation representing codon 707 in the yeast MSH2 gene. The sequence shown corresponds to the non-coding strand, with 5' at the bottom of the gel. The relevant nucleotides are listed to the right of the image. (Figures and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
The synthesized molecules from the mutagenesis are used to transform bacterial cells by means of the calcium chloride method. The bacterial strain produces a functional uracil N-glycosylase that allows for selection against the parental, unmutagenized strands. Students transform the bacteria with the complete reaction mixture, and with two controls. One control lacks input DNA (the negative, or sterility control, which should give no transformants), and the second contains double-stranded pMSH2 DNA (a positive control, which should give a high yield of transformants).
Confirming the Mutation by Restriction Endonuclease Digestion of Plasmid DNA and PCR Amplified Fragments
Six transformants are chosen to recover the plasmid DNA for restriction endonuclease screening to confirm the presence of the mutation. Digestions are performed on the whole plasmid and on polymerase chain reaction (PCR) amplified fragments of the mutagenized regions. Many of the relevant restriction endonucleases cleave the pMSH2 plasmid into multiple fragments (some as many as 73 fragments), making the diagnosis difficult. Thus, analysis of the amplified region of interest proves to be a highly effective way for the students to identify the mutagenized plasmids.
Students select two primers from a listing of available primers to amplify the mutagenized region. The goal is to amplify a fragment that upon restriction endonuclease digestion will give a banding pattern distinct from that of unmutagenized DNA. The students are directed to choose primers keeping in mind considerations such as the annealing positions and the orientation of the 3'-OH end of the primers with respect to the altered codon.
The purified plasmid DNA and the unmutagenized plasmid DNA control (pMSH2) as well as the PCR products are digested with diagnostic restriction endonucleases. Students are required to find the correct buffer and temperature conditions for each diagnostic digestion and prepare an agarose gel of a suitable percentage, depending on the desired resolving power. Students identify the mutagenized plasmids by analyzing the digestion patterns and comparing them to the predicted patterns and to the unmutagenized DNA control lanes (see Figure 3B and 3C for examples of student gels). Typically, greater than 50% of the samples analyzed encode the desired mutation.
Dideoxy Nucleotide Sequence Analysis of the Mutagenized Region
As a final confirmation that the mutagenesis was precisely as engineered, the students perform dideoxy nucleotide sequencing reactions of the relevant region using highly purified plasmid DNA. Students must select an appropriate primer from the listing of MSH2 primers stocks to prime the synthesis reaction. The students take into consideration the orientation of the mutation with respect to the position of the primer's 3'-OH end. The primer is chosen so that it will anneal ∼100 nucleotides away from the altered codon to allow for optimal resolution of the sequence in the mutagenized region (see Figure 3D for a student result).
Qualitative and Quantitative DNA Mismatch Repair Assays
After confirmation of the mutagenesis at the nucleotide level, the mutagenized plasmid (containing the msh2 allele of interest), the pMSH2 unmutagenized plasmid (containing wild-type MSH2), and the pRS413 vector control (no MSH2) are used to transform the msh2Δ yeast reporter strain AGY75 (Table 2, Figure 4, and Figure 5A). As a control for sterile technique, the students prepare a transformation sample with no plasmid DNA.
Table 2.
Yeast strains used in this study
| Strain | Genotype | Plasmid | Plasmid markers | Source |
|---|---|---|---|---|
| AGY75* | MAT a ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2::LEU2 RAD5 | pSH44 | PLEU2-(GT)16.5-URA3 TRP1/ARS CEN | This study |
| PJ69-4A | MAT a trp1-901 leu2-3,112 ura3-52 his3 ?200 gal4 Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | P. James, University of Wisconsin (James et al., 1996) | ||
| PJ69-4α | MATα trp1-901 leu2-3,112 ura3-52 his3 Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | P. James, University of Wisconsin (James et al., 1996) |
AGY75 strain is isogenic with W303 except that it is wild-type for RAD5 and CAN1. In addition, the MSH2 gene has been deleted
Using the created yeast strains, qualitative and quantitative mismatch repair assays are performed. Two types of drug-containing medium are used to assay DNA mismatch repair. The drugs select for yeast cells that have incorporated mutations into their DNA at an elevated rate. In these assays, an increased number of survivors on the drug-containing medium reflects a pronounced defect in DNA mismatch repair. The selection for dinucleotide instability (typically seen as a consequence of MutSβ loss of function) is synthetic medium supplemented with 5-fluororotic acid monohydrate, 5-FOA, whichprovides a potent selection for yeast cells that are ura3 defective (Boeke et al., 1987). In the assay, the loss of Ura3p is a consequence of a frameshift in the reporter construct (Figure 4) dinucleotide repeat region that is fused in frame with URA3 (Henderson and Petes, 1992). Dinucleotide tracts expand or contract at an elevated rate as a consequence of replication errors (reviewed in Kunkel and Bebenek, 2000). If not repaired by the MMR machinery, the reading frame for the URA3 gene will be disrupted and the cell will no longer produce a functional Ura3p gene product. Mismatch repair defective cells display an elevated rate of resistance to 5-FOA (FOAr) using this reporter, which is indicative of dinucleotide instability (Henderson and Petes, 1992). The second type of medium to score primarily for single base pair mismatch repair defects (typically seen as a consequence of MutSα loss of function) is supplemented with canavanine, a toxic analog of arginine (Rosenthal, 1977). Both canavanine and arginine are taken into the cells via the arginine permease (Grenson et al., 1966) encoded at the CAN1 locus (Broach et al., 1979). If the CAN1 gene is defective, then the yeast become resistant to canavanine, Canr (Whelan et al., 1979). Mismatch repair defective strains show an elevated rate of canavanine resistance (Marsischky et al., 1996).
Students examine the results of growth on the drug plates and non-selective plates and compare the msh2-allele strain to the wild-type strain and the msh2 null strain. The wild type strain typically shows very little growth on the drug containing plates compared to the msh2 null strain, which characteristically shows pronounced growth. By comparison with the controls, the students make a qualitative judgment as to whether the msh2 allele results in defects in DNA mismatch repair (see Figure 5B for an example of student results).
Using the same strains and drug selection media, the students perform quantitative assays to determine the rate of mutation in the yeast strains described above. In order to calculate the rate, the students determine the number of Canr or FOAr mutants in the culture and the total number of viable cells. From this information the students determine the rate as an expression of the number of mutations (derived from the mutant count) per cell division (derived from the number of viable cells plated). The median number of colonies per plate from each drug-containing medium (either FOA or CAN) is used to calculate the mean number of mutations that must have occurred (Lea and Coulson, 1949). For each strain, the number of colonies on the nonselective plates is averaged and the appropriate dilution factor is applied to give the number of viable cells. For microbial growth, it is an accepted approximation that the number of cells in a population is equivalent to the number of cell divisions. Having gathered the required data and after making the appropriate conversions, the rate of mutation is expressed in terms of the number of mutations per cell division.
Each pair determines the rates for the MSH2 wild-type, the msh2 missense allele and msh2 null control strain. Class data are pooled to gather more accurate mutation rates for each strain and to illustrate the importance of multiple trials to obtain meaningful data. Four pairs typically analyze each allele so that a total of 12 trials for each msh2 allele strains and 75–90 trials for each of the control strains are performed yearly. Students are asked to compare the calculated mutation rates with those derived from the combined data.
By the end of the first half of the semester, the students have engineered a mutation and established whether the change effects DNA mismatch repair. In a typical year, at least one of the human-derived allele has a wild-type phenotype in yeast. The students are encouraged to carefully scrutinize the original publications that designated the wild type-like alleles as pathogenic and to include their findings in the first laboratory report.
Determination of the Steady-State Levels of Msh2 and Msh2 Variant Proteins
To begin to understand why certain missense mutations in MSH2 result in mismatch repair defects, the steady-state expression levels of the Msh2 and Msh2-variant proteins are determined. This assay determines whether the amino acid change results in dramatically altered stability of the Msh2 protein. The same three strains used in the mismatch repair assays are used in this analysis. The immunodetection of wild type Msh2p and the variant Msh2p are performed using antibodies that bind the hemagglutinin epitope (HA) encoded in the MSH2 gene (see Figure 2). The no epitope control sample is protein extracts from the msh2 null strain harboring the pRS413 vector. Student results from an immunoblot and Coomassie staining can be seen in Figure 6. Each year, many students observe a decrease in overall expression of the variant Msh2 proteins. Portions of the lectures and informal discussions are devoted to understanding the possible models for decreased expression. Students are encouraged to devise a strategy for distinguishing between the possibilities.
Figure 6.
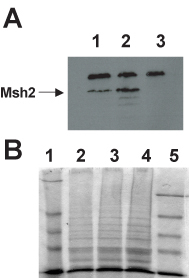
Student results determination of steady state levels of Msh2 and the altered Msh2 protein. (A) Immunoblot analysis of Msh2 levels. Protein extracts containing hemagglutinin (HA) tagged versions of the wild type Msh2 protein and the Msh2-M707I protein along with a no epitope control extract were fractionated by SDS-PAGE (7% acrylamide separating gel) and transferred to a nitrocellulose membrane. Immunodetection was performed with mouse α-HA monoclonal primary antibody 12CA5 and α-mouse IgG HRP-conjugated secondary antibody. Images were formed on X-OMAT AR-5 Kodak film for 5 min. Lane 1, Msh2-M707I protein extracts; lane 2, wild-type Msh2p protein extracts; lane 3, no epitope protein extracts. The indicated band at 111 kD represents the Msh2-HA fusion proteins (Msh2p). Sizes were determined by comparison with prestained molecular weight standards (not shown). The identity of the high molecular weight (∼175 kD) cross-reacting band seen in all lanes was not determined. The lower molecular weight bands in lane 2 are likely to be Msh2p degradation products. (B) Coomassie staining of electrophoresed yeast protein extracts. Protein extracts were separated by SDS-PAGE as described above. A duplicate half of the gel was stained with Coomassie Blue R250 Stain, destained overnight, and dried under vacuum at 80ÿC for 1 h. The lanes are as follows: lane 1, NEB prestained protein markers; lane 2, Msh2-M707I protein extracts; lane 3, wild-type Msh2p protein extracts; lane 4, no epitope protein extracts; and lane 5, Invitrogen SeeBlue prestained standards. (Figure and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
Localization of Msh2 and Msh2 Variant Protein Using Indirect Immunofluorescence
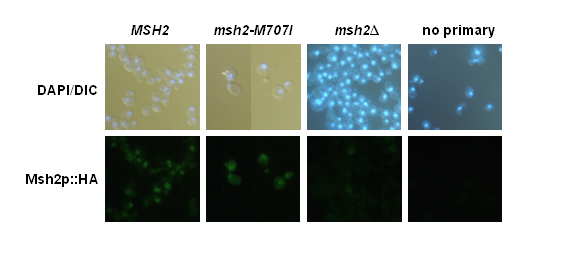
Students also test the hypothesis that the defect in mismatch repair phenotype conferred by some msh2 alleles might be attributable to failure of the variant Msh2 protein to localize to the nucleus. Indirect immunofluorescence is employed to localize the wild-type and mutated forms of the Msh2 protein. The same three yeast strains used in the previous analyses are used for the localization studies. The strains include the wildtype (expressing Msh2p containing the HA epitope), the msh2 missense mutant strain (expressing the variant Msh2p containing the HA epitope), and the msh2 null strain (that does not express Msh2 or the HA epitope).
Two important negative controls for the experiment include a sample of wild-type, antigen expressing cells with no primary antibody added and a sample with msh2 null cells that do not express the antigen (no epitope control), but are treated similarly to the experimental cells. These controls emphasize the importance of including a no epitope control if possible, since the background level of fluorescence observed in the no epitope control is typically much higher than the no primary antibody control. The positive control for the experiment consists of cells expressing the wild-type Msh2 protein. In addition, a double labeling of Msh2 and tubulin (see Materials and Methods) is performed as an internal control to prove that the cells were adequately prepared for immunofluorescence. All cells should show tubulin staining since tubulin is essential for viability. In addition, all cells are treated with the DNA specific dye, DAPI, to indicate nuclear position.
Students observe the overall cellular morphology with differential interference contrast (DIC) optics and visualize the Msh2 or tubulin localization and the cellular DNA using a microscope equipped with epi-fluorescence capabilities. After examining and photographing the control and experimental samples, the students are able to determine whether the variant Msh2p co-localizes with the nuclear DNA. Students are introduced to the practical and theoretical aspects of fluorescence microscopy while obtaining a tangible result. Results from a student immunofluorescence experiment may be seen in Figure 7. This laboratory session has a high rate of success in that the controls work well. One caveat is that the variant proteins that display decreased overall expression are sometimes difficult to detect. For these exceptions, the staff provides prepared slides where the allele has been over expressed, so that the localization may be observed. Many alleles are properly expressed and the variant protein is localized to the nucleus. The final experiment detailed below often proves helpful in understanding the defect for certain alleles.
Figure 7.
Student results from indirect immunofluorescence of Msh2p. Yeast msh2Δ cells transformed with pRS413 (msh2Δ), pMSH2 (MSH2), or pmsh2-M707I (msh2-M707I) are shown in the figure. Fixed and digested cells were incubated with mouse α-HA monoclonal primary antibody (12CA5), with 12CA5 + rat monoclonalα -tubulin (YL1/2), or with BSA-PBS (no primary) and subsequently incubated with a secondary antibody mixture of goat α-mouse IgG Alexa Fluor 488 (green emission) and α-rat IgG Alexa Fluor 594 (red emission, not shown). Nuclei were visualized by DAPI staining. Secondary antibody staining was visualized by fluorescence microscopy, and DAPI staining was visualized by simultaneous DIC and fluorescence microscopy (DAPI/DIC). (Figure and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
Formation of a Recombinant DNA Molecule to Test MMR Subunit Interaction
The purpose of the final experiment is to determine whether the changed amino acids in the variant Msh2 proteins effect interactions with other members of the MMR complex. The students employ the yeast 2-hybrid assay to monitor the subunit interaction (see James et al., 1996 for a full explanation of the technique). Prior to the course, the basic components of the 2-hybrid MMR interaction were constructed (Table 3 and Figure 9A). Students design and create a recombinant DNA molecule that allows for expression of the variant Msh2 protein in the yeast 2-hybrid assay.
Table 3.
Plasmids used in analysis
| Plasmid | Relevant markers | Source |
|---|---|---|
| pMSH2 | MSH2::HA HIS3 CEN6 ARSH4 ampr | This work |
| pRS413 | HIS3 CEN6 ARSH4 ampr | Sikorski and Hieter, 1989 |
| pGBD-C2 | GBD TRP1 2μ ampr | James et al., 1996 |
| pGAD-C2 | GAD LEU2 2μ ampr | James et al., 1996 |
| pGBD-MSH2 | GBD-MSH2 TRP1 2μ ampr | This work |
| pGAD-MSH6 | GAD-MSH6 LEU2 2μ ampr | This work |
| pGAD-MSH3 | GAD-MSH3 LEU2 2μ ampr | This work |
| pGAD-MLH1 | GAD-MLH1 LEU2 2μ ampr | This work |
| pGAD-PMS1 | GAD-PMS1 LEU2 2μ ampr | This work |
| pGAD-EXO1 | GAD-EXO1 LEU2 2μ ampr | This work |
| pGAD-POL30 | GAD-POL30 LEU2 2μ ampr | This work |
The SeqWeb program described above is employed to devise an efficient strategy for constructing the recombinant DNA molecule. A construct prepared before the class, pGBD-MSH2, serves as the “vector backbone” (Figure 9A) and a fragment of DNA containing the msh2 missense mutation represents the“ insert.” Students select two unique restriction enzymes that cut within the MSH2 coding region on the pGBD-MSH2 construct. Upon double digestion pGBD-MSH2 should be cleaved at sites within the MSH2 coding sequence that flank the mutation and generate non-ompatible, cohesive-ends. Using noncompatible cohesive ends improves the efficiency of obtaining the desired construct and also ensures that the reading frame is in tact upon ligation. Each enzyme cleavage site must be unique in pGBD-MSH2 to preserve the vector backbone, but does not need to be in the pMSH2-derivative construct carrying the mutation. The students generate a restriction map with unique cut sites in pGBD-MSH2. When selecting the enzymes, consideration is given to buffer compatibilities of the selected enzymes and how well the fragments can be resolved by agarose gel electrophoresis. Provided the recombinant DNA cloning strategy is reasonable, the students proceed with their own design.
The students digest the DNA, resolve the fragments with agarose gel electrophoresis and purify the desired bands from agarose slices. The students determine the yield of insert and vector DNA and ligation reactions and controls are prepared. Controls include the pGBD-MSH2 vector fragment alone with no insert fragment, and a singly cut vector with and without T4 DNA ligase to measure for the efficiency of the ligation. The ligated molecules are introduced into bacterial cells using electroporation. An uncut plasmid DNA control is included to measure the efficiency of the electroporation and to serve as a comparison for how efficiently the ligase worked to generate covalently closed plasmids. If the ligation is 100% efficient, then the cut plasmid with ligase should give the same number of transformants as the uncut control; whereas the cut control with no ligase should result in considerably fewer transformants. Standard molecular procedures screen for the recombinant molecules. For an example of student results see Figure 8.
Figure 8.
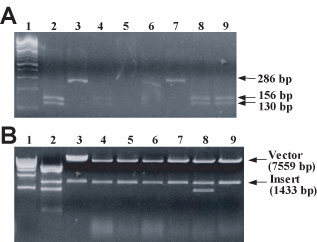
(A) Gel electrophoresis of HaeIII of digested PCR-amplified alkaline-lysis miniprep DNA. A 286-bp fragment of pGBD-MSH2 and pGBD-msh2-M707I DNA produced by directional cloning and isolated from MV1190 transformants by alkaline lysis was amplified by PCR, as described in Materials and Methods. Amplified products were digested with HaeIII, and digest products were fractionated on a 3% agarose + 0.25 μg/ml EtBr gel. Lane 1, 100-bp ladder; lane 2, pMSH2; lane 3, pmsh2-M707I (previously verified by dideoxynucleotide sequencing); lanes 4–9, plasmid DNA from transformants 1–6. Relevant fragment sizes are indicated in the figure. (B) Gel electrophoresis of StyI/KpnI-digested alkaline-lysis miniprep DNA. 1 μg each of pMSH2 (lane 2), pGBD-MSH2 (lane 3), and 6 alkaline lysis miniprep DNA samples (lanes 4–9) were digested with StyI and KpnI, and digest products were fractionated on a 1% agarose + 0.25 μg/ml EtBr gel. DNA was visualized by UV illumination. StyI cleaves once within pMSH2 and once within pGBD-MSH2; KpnI cleaves three times within pMSH2 and once within pGBD-MSH2. Lane 1, 1-kb ladder. Vector (pGBD-MSH2 lacking the smaller StyI-KpnI fragment) and insert (fragment containing the mutagenized region) DNA sizes are indicated in the figure. (Figure and legend courtesy of Ruth Tennen, Princeton University. Reproduced with permission.)
The recombinant plasmid DNA as well as pGBD-C2 vector DNA and pGBD-MSH2 are used to transform a MATa yeast 2-hybrid strain (Table 1, Figure 9A). In addition, a no-plasmid DNA sterility control is also prepared. Prior to the course, the GAD fusion constructs (Figure 9A) were introduced into a MATα yeast 2-hybrid reporter strain. Thus, a panel of haploid yeast strains is available, each carrying a construct that will express a relevant hybrid protein. The MATa strains harbor the GBD fusions or GBD-alone control, while the MATα strains contain the GAD-partner fusions.
The students set up crosses to form diploid yeast strains. Ultimately, each diploid expresses both of the hybrid proteins necessary to examine the MMR subunit interactions (Figure 9A). Control diploid strains are formed to illustrate no interaction (cells expressing GBD alone in combination with each of the GAD-Partner fusions) and to illustrate a positive interaction (GBD-Msh2 in combination with each of the GAD-Partner fusions). In the yeast reporter strain, the HIS3 gene is fused downstream of the GAL1 promoter, thus the 2-hybrid interaction may be assayed using medium lacking histidine (–HIS). Two other reporter genes, ADE2 and lacZ are encoded in the 2-hybrid reporter strain, but the expression of these genes is not sufficient to accurately assess the interactions of the MMR components. Growth on –HIS indicates that the hybrid proteins are interacting. Thus, the determination of whether the mutated amino acids interfere with the ability of Msh2p to bind its functional partners can be assayed using a simple growth assay. Results from a typical 2-hybrid experiment can be seen in Figure 9B. Many of the missense mutations appear to prevent mismatch repair complex formation. Students are encouraged to examine the crystal structure of the closely related MutS protein (Lamers et al., 2000; Obmolova et al., 2000) to identify the amino acid of interest and to develop a hypothesis for why subunit formation might be effected.
Instruction in Writing in the Scientific Style
Students enrolled in the course are required to execute all of the experiments detailed above and to write two 25-page papers in the format of a scientific publication. This method of student evaluation was chosen since most assessments of scientific achievement are dependent upon written documents.
Although the students are writing up an ongoing project, each laboratory report has a different emphasis. The first report has an important implication for human health, thus the background material should include the relevant medical literature. The first report contains the following sections: Designing a mutagenic oligonucleotide; Creating a msh2 missense mutation by site-directed mutagenesis; Confirmation of the missense mutation by restriction enzyme analysis and dideoxy sequencing; and Mutation frequency assays of msh2 missense alleles. In the first report, the students have the satisfaction of discussing a concrete result with a medically significant conclusion. The second report investigates how the variant Msh2 proteins are defective and thus the background should emphasize the molecular mechanism of DNA mismatch repair. The second report specifically includes: Immunoblot analysis of altered Msh2 proteins; Cloning msh2 alleles into the yeast 2-hybrid system; Interactions with members of the mismatch repair machinery; and Msh2p localization studies. The second report has many results and allows for integration of several pieces of data to form a coherent model for a particular MSH2 allele.
As with most research articles, laboratory reports include the following sections: Title Page, Abstract, Introduction, Materials and Methods, Results (including data presentation), Discussion, and References. High standards are imposed in terms of scholarship, accuracy, scientific writing style, figure quality, citation professionalism, proposals for future work, and critical analysis. A page limit is imposed to encourage the students to write succinctly and to reflect reality since most scientific documents, including manuscripts and grant proposals, have page constraints.
Guidance for writing the reports is provided through several avenues. Seven pages of the manual are devoted to providing advice and suggestions for writing the laboratory reports. In addition to typical office hours, review sessions are held and a discussion board is posted on the course web site to answer specific technical questions asked electronically.
When evaluating the written work we strive to provide sufficient feedback on the document and to maintain grading equity. A rubric has been devised to aid teaching assistants in evaluating the reports (supplement supplied). The rubric has been useful for supplying students with concrete feedback as to their level of achievement in a variety of areas and for providing a more quantitative form of evaluation to aid in establishing equity across the grading groups. The rubric designed for this course allows for distinguishing among very accomplished Princeton molecular biology students. Educators might need to shift the emphasis depending on the typical student. Overall, the rubric has been well received by the students and the teaching assistants.
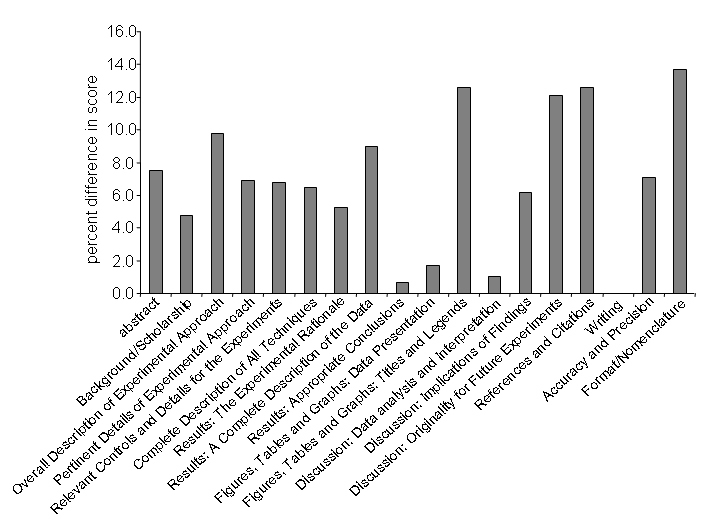
The quantitative assessment also allows for an analysis of specific areas of improvement in student writing. For the two years during which the rubric listed in the supplement was used, students showed an overall improvement of 5.4 ± 0.4 percent between the first and second report. The areas of improvement are in almost all assessment areas (see Figure 10); however, the students showed reproducible improvement in sections that are unique to scientific documents. The data is consistent with the observation that the students enter the class with basic writing and analytical skills and therefore do not show pronounced improvement in these areas. In addition, the data suggests that the feedback given after the first report aids the students specifically in the areas of writing in the scientific style.
Figure 10.
Improvement in Laboratory Report Scores. The chart shows the percent difference in scores comparing the first and second laboratory reports. Each category corresponds to the assessment areas found in the grading rubric. Data are averages from classes taught in 2002 and 2003.
Course Assessment
Princeton University's Office of the Registrar provides an assessment service for all courses. The assessment includes numerical scores in response to specific categories and written evaluations. All evaluations are anonymous and conducted under the supervision of a student volunteer rather than the course director. Based on the numerical and written evaluations, the course described in this paper has been well received by the students (see Tables 4 and 5). The breakdown of the numerical score for 2003 is shown in Table 4. Similar marks were given for the 5 years the course has been taught using the format described in this paper (not shown). The scores from a year prior to when the project was implemented (1997) and the University average are included for comparison purposes (Table 4). There was a significant improvement in the approval rating in all areas surveyed in the year that the course described in the paper was first introduced. Of particular interest is the score regarding the student's assessment of the overall quality of the laboratories the year that the project was implemented (Table 5 compare 1997 scores to those following).
Table 4.
Evaluations for 2003 compared with 1997 ratings prior to the described course and the university average
| 2003 course evaluation | n | 2003 avga | 1997 avgb | University avgc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unacceptable | <> | Excellent | |||||||||||
| 1 | 2 | 3 | 4 | 5 | |||||||||
| LECTURES | |||||||||||||
| Stimulation of independent thinking | 9% | 27% | 64% | 56 | 4.6 | 3.6 | 4.2 | ||||||
| Clear presentation of subject matter | 11% | 89% | 56 | 4.9 | 3.6 | 4.2 | |||||||
| Overall quality of lectures | 21% | 79% | 56 | 4.8 | 3.6 | 4.2 | |||||||
| LABORATORIES | |||||||||||||
| Usefulness to lectures and readings | 2% | 98% | 55 | 5.0 | 4.3 | 3.8 | |||||||
| Instructor's organization and guidance | 5% | 95% | 55 | 4.9 | 4.1 | 4.0 | |||||||
| Overall quality of laboratories | 100% | 55 | 5.0 | 4.2 | 3.9 | ||||||||
| READINGS: | |||||||||||||
| Stimulation of independent thinking | 33% | 67% | 12 | 4.7 | 3.7 | 4.1 | |||||||
| Clear presentation of subject matter | 17% | 83% | 12 | 4.8 | 3.8 | 4.0 | |||||||
| Overall quality of readings | 27% | 73% | 11 | 4.7 | 3.7 | 4.1 | |||||||
| PAPERS, REPORTS: | |||||||||||||
| Guidance in preparation of written work | 2% | 16% | 24% | 58% | 55 | 4.4 | 4.0 | 3.9 | |||||
| Comments in response to written work | 21% | 36% | 43% | 53 | 4.2 | 3.7 | 4.0 | ||||||
| Overall value of papers, reports | 2% | 4% | 37% | 57% | 54 | 4.5 | 4.0 | 4.0 | |||||
| GENERAL: | |||||||||||||
| Contribution to knowledge of subject | 2% | 21% | 77% | 56 | 4.8 | 4.3 | 4.4 | ||||||
| Contribution to critical evaluation | 2% | 29% | 70% | 56 | 4.7 | 4.3 | 4.3 | ||||||
| Contribution to interest in subject | 5% | 32% | 63% | 56 | 4.6 | 4.1 | 4.2 | ||||||
| Overall quality of the course | 13% | 88% | 56 | 4.9 | 4.2 | 4.3 | |||||||
Course enrollment = 57. Evaluations returned = 56. Response = 98%
Course enrollment = 54. Evaluations returned = 50. Response = 93%
Total enrollment = 17,235. Evaluations returned = 12,007. Response =70%
Table 5.
Breakdown of responses to “Overall quality of laboratories”
| 1 | 2 | 3 | 4 | 5 | |||
|---|---|---|---|---|---|---|---|
| Year | n | Unacceptable (%) | Poor (%) | Fair (%) | Good (%) | Excellent (%) | Score |
| 1997 prior to course | 50 | 0 | 0 | 16 | 50 | 34 | 4.2 |
| 1999 first year | 68 | 0 | 0 | 0 | 22 | 78 | 4.8 |
| 2000 second year | 50 | 0 | 0 | 0 | 6 | 94 | 4.9 |
| 2001 third year | 42 | 0 | 0 | 2 | 12 | 86 | 4.8 |
| 2002 fourth year | 46 | 0 | 0 | 2 | 7 | 91 | 4.9 |
| 2003 fifth year | 56 | 0 | 0 | 0 | 0 | 100 | 5.0 |
Many of the written evaluations indicated the student interest in the project. Below are several examples of student comments regarding the project.
I was excited by the application of such knowledge. Core lab is an excellent example: the class is dynamic, involving students in a larger investigation of colon cancer while teaching us key techniques that we used for our theses and beyond.
—Daniel Stover Class of 2003 http://www.molbio.princeton.edu/undergrad_stud_perspct.php
You add to the body of scientific knowledge while learning techniques from sequencing to cloning to replica-plating. The overall idea is really cool: you are part of a long-term, large-scale study of colon cancer in a yeast model. So, while you are helping understand a major human killer, you are learning manytechniques that you will use down the line for your thesis.... The lab reports are very helpful, because it's like a mini-thesis you have all the same sections, learn how to make figures and diagram results. All in all, this class is really fun.
—Anonymous
Semester Taken: Spring 2002. Taken from the Princeton University Student Course Guide, a student-run course guide at Princeton University.
[Y]ou learn applicable lab techniques, safety measures, experiment design approaches, and formal write up approaches.... This is a great way of determining whether you want to pursue a career in lab sciences and it is also a great way to apply what you have learned in many of your other classes toward producing a final project write up that you can be proud of.
—Anonymous
Semester Taken: Spring 2002. Taken from the Princeton University Student Course Guide, a student-run course guide at Princeton University.
A survey of Molecular Biology seniors conducted in the spring of 2002, provides an indication of the broader utility of the course. In response to a senior thesis survey question “Which classes helped you the most in preparing you for your independent work?” 85% of the 26 respondents cited MOL350, the course described in this paper. In that survey, specific comments were solicited about the course described in this paper, since it is part of the Princeton molecular biology core curriculum. In response to the following question: “Now that it has been one or more years since you have taken the core requirements, what are your views on these courses?” Student testimony about MOL350 (also known as core lab) in response to the above question was as follows: 4 had no comment, 1 was neutral (“alright”), 1 was negative (“too formulaic”), and 25 were positive. 41% of the positive comments specifically mentioned the value of the course with respect to conducting research laboratory work and writing a molecular biology thesis.
In addition, the course confers a technical benefit beyond completing a degree at Princeton University. A recent example is from Jeffrey Kim, Class of 2003, who started working after graduation as a research technician at Rockefeller University.
I started work there (Rockefeller University) immediately after school ended and things have been going really well.... The preparation in Core Lab junior year was invaluable as a resource now because I didn't need any additional training to do all of the major lab techniques and I was able to get started on my independent work as soon as I got there. The people in lab were impressed with the experience that I got from your class....
DISCUSSION
Summary
This paper outlines an advanced laboratory course in cell and molecular biology. The students enrolled in the course gain experience with current laboratory techniques and scientific writing while conducting original research. The topic was selected to appeal to a wide range of students and the format of the project was designed to provide a practical and relevant training for future work in a molecular biology research laboratory setting.
Project Format: Advantages of Using Model Organisms
We use the facile model organism Saccharomyces cerevisiae, to study eukaryotic DNA mismatch repair. Because mismatch repair is a highly conserved process, we are able to ask medically relevant questions using a safe and noncontroversial model organism. Other practical considerations include the vast research resources available that have been generated by the yeast scientific community over the years. Furthermore, yeast research is inexpensive when compared to many model organisms or tissue culture systems. Yeast cells are easily manipulated, rapidly growing, efficiently stored for long periods of time, genetically tractable, readily transformed, and non-hazardous. Finally, most molecular biology laboratories employ either bacteria or yeast to manipulate cloned DNA, therefore becoming proficient in microbial work is a useful skill. Employing bacteria for the molecular manipulations and yeast for the mismatch repair functional assays, we are able to cover many topic areas and use a variety of techniques in the time allocated for the course.
Project Format: Techniques
The format of the project ensures exposure to commonly used research laboratory methods, including site-directed mutagenesis, the polymerase chain reaction, restriction enzyme analysis, agarose gel electrophoresis, DNA sequencing, immunoblotting, microbial manipulations, fluorescence microscopy, recombinant DNA technology, and in vivo reporter assays. The procedures for the project are provided in a loose-leaf manual. Protocols are derived from a variety of sources and the heterogeneity of styles has been preserved with the objective of introducing the students to an assortment of protocol formats. Original procedures have been modified slightly to eliminate the most confusing ambiguities.
Project Format: Time Frame
Each student works in the laboratory twice a week (Mon and Wed or Tue and Thu) for 3 h each session for a total of 12 wk (see the supplied Timeline). Thus, over the semester the students spend ∼72 h in the laboratory. In addition, they are often required to come in during“ off-hours” to perform minor tasks to keep the project progressing.
To ensure execution of all of procedures, it is necessary to have the intermediates as well as the final constructs and strains available when a particular aspect of the experiment fails. The “back-ups” allow the students to experience the entire project and ensure that a substantial amount of data is available to describe and analyze in their laboratory reports.
If an educator is considering implementing this course, but class time is limited, an option is to implement only the first half of the course. Biophysics majors currently exercise this option at Princeton. Many of the most basic molecular techniques are covered in the first half of the course. If resources are limited, the dideoxy nucleotide sequencing laboratory could be eliminated. In addition, many teaching laboratories are not equipped with fluorescent microscopes and thus this module could be eliminated. Prior to purchasing the microscope, gene dosage experiments were conducted rather than the immunofluorescence.
Supervision and Encouragement of Independence
The course employs a technician, a lecturer, a postdoctoral instructor, and four graduate student teaching assistants to prepare reagents and to teach and supervise 50–75 undergraduate students, most of whom are juniors in the Department of Molecular Biology. Each laboratory session has ∼30 students, or 15 pairs, divided into two laboratory rooms. Each room has one teaching assistant and either the lecturer or the postdoctoral instructor present. Thus, the ratio of instructional staff to students is ∼2:15.
The teaching staff provides instruction and support, but whenever possible encourages self-sufficiency. For example, students prepare reagents and maintain stock solutions. In addition, they routinely calculate the appropriate dilutions from a stock to the final solution. Students are required to plan as well as execute their experiments. For example, they design a mutagenesis, calculate diagnostic restriction digestion patterns, choose appropriate primers for PCR and sequencing reactions, and devise a recombinant DNA cloning strategy.
Student Motivation and Learning
An ongoing challenge to educators is to understand what motivates their students to learn and retain the material being presented. Many recent reports on how students learn have supported the approach outlined in this paper (for example see The National Research Council's, Bio2010, Transforming Undergraduate Education for Future Research Biologists, 2003).
The course has been designed to expose student to relevant and useful techniques and to let the students experience the excitement of original research in an interesting and applicable field. The students are able to apply what they have learned to their senior thesis independent research projects conducted in the various molecular biology research laboratories at Princeton. In addition, many of the Princeton molecular biology students go on to use the technical skills in their future careers, some 40% of which will earn a Ph.D.
The topic for this course was chosen to capture the interest of a wide array of enrolled students, including physics, engineering, chemistry, and biology majors. The Advisory Committee to the National Science Foundation Directorate for Education and Human Resources (1996) and the authors of a report for Project Kaleidescope (Rothman and Narum, 1999) support this effort to appeal to a variety of science students. The diverse students enrolled in this course express a high degree of enthusiasm for the topic. Because DNA mismatch repair is a fundamental biological process with an understandable relevance to human health, the subject matter engages those interested in basic science, medicine, or both.
The students are also excited about participating in a novel research project and express in written evaluations that they find it preferable to performing a set of experiments where the answer is known. The inquiry-based approach to learning described in this paper is supported by the National Research Council's recent recommendations for undergraduate education for future research biologists (National Research Council Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century, 2003). Other influential educational reports recommend making research-based learning for undergraduates the standard (The Boyer Commission on Educating Undergraduates in the Research University, 1998). In summary, in an effort to motivate the students to learn and retain the material, a great deal of energy is directed toward making this course exciting, real, and relevant for future scientists.
Acknowledgments
We would like to express our gratitude to Barbara Devlin for her dedication and technical assistance and to Ruth Tennen for providing the figures and legends to illustrate the student results. We appreciate the input and advice of Mark Rose on the course and the manuscript. We would like to thank Lorraine Symington, Eric Alani, Thomas Petes, and Rodney Rothstein for generously supplying strains and plasmids. Finally we thank the 296 hard-working Molecular Biology undergraduates and 19 teaching assistants at Princeton University who have participated in the course over the past five years. The MOL350 course was funded by the following sources: Princeton University, The Department of Molecular Biology at Princeton University, and The Howard Hughes Medical Institute.
Footnotes
Abbreviations used: CRC, colorectal cancer; HNPCC, hereditary non-polyposis colorectal cancer; MMR, mismatch repair; PCR, polymerase chain reaction; DIC, differential interference contrast; DAPI, 4',6'-diamidino-2-phenylindole; WT, wild type; Gal, galactose; Glc, glucose; YEPD, rich media containing yeast extract/peptone/dextrose; YEP-GAL, rich media containing yeast extract/peptone/galactose; 5-FOA, 5-fluoro-ortic acid; Luria-Bertani medium (LB); CAN, canavanine.
Current address: Department of Molecular Genetics, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97201-3098.
Compiled by the Human Gene Mutation Database, Copyright © University of Wales College of Medicine 2003)
References
- Aaltonen, L.A., Peltomaki, P., Leach, F.S., Sistonen, P., Pylkkanen, L., Mecklin, J.P., Jarvinen, H., Powell, S.M., Jen, J., Hamilton, S.R., and others (1993). Clues to the pathogenesis of familial colorectal cancer. Science 260,812 -816. [DOI] [PubMed] [Google Scholar]
- Advisory Committee to the National Science Foundation Directorate for Education and Human Resources (1996). Shaping the Future: New Expectations for Undergraduate Education in Science, Mathematics, Engineering, and Technology. A Report on the Review of Undergraduate Education. Washington DC: National Science Foundation.
- Alani, E., Chi, N.W., and Kolodner, R. (1995). The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions.Genes Dev. 9,234 -247. [DOI] [PubMed] [Google Scholar]
- Aquilina, G., and Bignami, M. (2001). Mismatch repair in correction of replication errors and processing of DNA damage. J. Cell Physiol. 187,145 -154. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Scidman, J.G., Smith, R.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. New York: Wiley.
- Bauer, J., Krammer, G., and Knippers, R. (1981). Asymmetric repair of bacteriophage T7 heteroduplex DNA. Mol. Gen. Genet. 181,541 -547. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., Trueheart, J., Natsoulis, G., and Fink, G.R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154,164 -175. [DOI] [PubMed] [Google Scholar]
- Boland, C.R. (2000). Molecular genetics of hereditary non-polyposis colorectal cancer. Ann. N.Y. Acad. Sci. 910, 50-59; discussion 59-61. [DOI] [PubMed] [Google Scholar]
- Boyer Commission on Educating Undergraduates in the Research University (1998). Reinventing Undergraduate Education: A Blueprint for America's Research Universities. Princeton, NJ: Carnegie Foundation for the Advancement of Teaching, pp.15 -17.
- Broach, J.R., Strathern, J.N., and Hicks, J.B. (1979). Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene 8,121 -133. [DOI] [PubMed] [Google Scholar]
- Burdett, V., Baitinger, C., Viswanathan, M., Lovett, S.T., and Modrich, P. (2001). In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 98,6765 -6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., and Stearns, T. (2000).Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual . Plainview, NY: Cold Spring Harbor Laboratory Press.
- Cox, E.C., Degnen, G.E., and Scheppe, M.L. (1972). Mutator gene studies in Escherichia coli: the mutS gene.Genetics 72,551 -567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel, R., Lescoe, M.K., Rao, M.R., Copeland, N.G., Jenkins, N.A., Garber, J., Kane, M., and Kolodner, R. (1993). The human mutator gene homolog MSH2 and its association with hereditary non-polyposis colon cancer. Cell 75,1027 -1038. [DOI] [PubMed] [Google Scholar]
- Genschel, J., Bazemore, L.R., and Modrich, P. (2002). Human exonuclease I is required for 5' and 3' mismatch repair. J. Biol. Chem. 277,13302 -13311. [DOI] [PubMed] [Google Scholar]
- Glickman, B.W. (1979). Spontaneous mutagenesis in Escherichia coli strains lacking 6-methyladenine residues in their DNA: an altered mutational spectrum in dammutants. Mutat Res. 61,153 -162. [DOI] [PubMed] [Google Scholar]
- Glickman, B.W., and Radman, M. (1980). Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 77,1063 -1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke, E., and Manney, T.R. (1979). Expression of radiation-induced mutations at the arginine permease (CAN1) locus in Saccharomyces cerevisiae. Genetics 91,53 -66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson, M., Mousset, M., Wiame, J.M., and Bechet, J. (1966). Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 127,325 -338. [DOI] [PubMed] [Google Scholar]
- Harfe, B.D., and Jinks-Robertson, S. (2000). DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34,359 -399. [DOI] [PubMed] [Google Scholar]
- Henderson, S.T., and Petes, T.D. (1992). Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell Biol. 12,2749 -2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii, A., Han, H.J., Sasaki, S., Shimada, M., and Nakamura, Y. (1994). Cloning, characterization and chromosomal assignment of the human genes homologous to yeast PMS1, a member of mismatch repair genes.Biochem. Biophys. Res. Comm. 204,1257 -1264. [DOI] [PubMed] [Google Scholar]
- Ionov, Y., Peinado, M.A., Malkhosyan, S., Shibata, D., and Perucho, M. (1993). Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis.Nature 363,558 -561. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144,1425 -1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M., and Wagner, R. (1981). N-Methyl-N'-nitro-N-nitrosoguanidine sensitivity of E. coli mutants deficient in DNA methylation and mismatch repair. Mol. Gen. Genet. 184,562 -563. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 82,488 -492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T.A., and Bebenek, K. (2000). DNA replication fidelity. Annu Rev Biochem. 69,497 -529. [DOI] [PubMed] [Google Scholar]
- Lamers, M.H., Perrakis, A., Enzlin, J.H., Winterwerp, H.H., de Wind, N., and Sixma, T.K. (2000). The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch.Nature 407,711 -717. [DOI] [PubMed] [Google Scholar]
- Lea, D.E., and Coulson, C.A. (1949). The distribution of the numbers of mutants in bacterial populations. J. Genet. 49,264 -285. [DOI] [PubMed] [Google Scholar]
- Leach, F.S., Nicolaides, N.C., Papadopoulos, N., Liu, B., Jen, J., Parsons, R., Peltomaki, P., Sistonen, P., Aaltonen, L.A., Nystrom-Lahti, M., and others (1993). Mutations of a mutS homolog in hereditary non-polyposis colorectal cancer. Cell 75,1215 -1225. [DOI] [PubMed] [Google Scholar]
- Levinson, G., and Gutman, G.A. (1987). High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K–12. Nucleic Acids Res. 15,5323 -5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom, A., Tannergard, P., Werelius, B., and Nordenskjold, M. (1993). Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat. Genet. 5,279 -282. [DOI] [PubMed] [Google Scholar]
- Lu, A.L., Clark, S., and Modrich, P. (1983). Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl. Acad. Sci. U.S.A. 80,4639 -4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, H.T., and Lynch, J. (2000). Lynch syndrome: genetics, natural history, genetic counseling, and prevention. J. Clin. Oncol. 18,19S -31S. [PubMed] [Google Scholar]
- Marsischky, G.T., Filosi, N., Kane, M.F., and Kolodner, R. (1996). Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10,407 -420. [DOI] [PubMed] [Google Scholar]
- Marti, T.M., Kunz, C., and Fleck, O. (2002). DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol. 191,28 -41. [DOI] [PubMed] [Google Scholar]
- Mechanic, L.E., Frankel, B.A., and Matson, S.W. (2000). Escherichia coli MutL loads DNA helicase II onto DNA. J. Biol. Chem. 275,38337 -38346. [DOI] [PubMed] [Google Scholar]
- Muller, A., and Fishel, R. (2002). Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC). Cancer Invest. 20,102 -109. [DOI] [PubMed] [Google Scholar]
- National Research Council Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century (2003). Bio 2010: Transforming Undergraduate Education for Future Research Biologists. Washington, DC: National Academies Press.
- Nevers, P., and Spatz, H.C. (1975). Escherichia coli mutants uvr D and uvr E deficient in gene conversion of lambda-heteroduplexes. Mol. Gen. Genet. 139,233 -243. [DOI] [PubMed] [Google Scholar]
- Nicolaides, N.C., Papadopoulos, N., Liu, B., Wei, Y.F., Carter, K.C., Ruben, S.M., Rosen, C.A., Haseltine, W.A., Fleischmann, R.D., Fraser, C.M., and others (1994). Mutations of two PMS homologues in hereditary non-polyposis colon cancer. Nature 371,75 -80. [DOI] [PubMed] [Google Scholar]
- Obmolova, G., Ban, C., Hsieh, P., and Yang, W. (2000). Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407,703 -710. [DOI] [PubMed] [Google Scholar]
- Oldenburg, K.R., Vo, K.T., Michaelis, S., and Paddon, C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25,451 -452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos, N., Nicolaides, N.C., Wei, Y.F., Ruben, S.M., Carter, K.C., Rosen, C.A., Haseltine, W.A., Fleischmann, R.D., Fraser, C.M., Adams, M.D., and others (1994). Mutation of a mutL homolog in hereditary colon cancer. Science 263,1625 -1629. [DOI] [PubMed] [Google Scholar]
- Parsons, R., Li, G.M., Longley, M.J., Fang, W.H., Papadopoulos, N., Jen, J., de la Chapelle, A., Kinzler, K.W., Vogelstein, B, and Modrich, P. (1993). Hypermutability and mismatch repair deficiency in RER+tumor cells. Cell 75,1227 -1236. [DOI] [PubMed] [Google Scholar]
- Peltomaki, P. (2001). Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 10,735 -740. [DOI] [PubMed] [Google Scholar]
- Peltomaki, P., Aaltonen, L.A., Sistonen, P., Pylkkanen, L., Mecklin, J.P., Jarvinen, H., Green, J.S., Jass, J.R., Weber, J.L., Leach, F.S., and others (1993). Genetic mapping of a locus predisposing to human colorectal cancer. Science 260,810 -812. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., Preston, R.A., Adams, A.E., Stearns, T., Drubin, D.G., Haarer, B.K., and Jones, E.W. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol. 31,357 -435. [DOI] [PubMed] [Google Scholar]
- Pukkila, P.J., Peterson, J., Herman, G., Modrich, P., and Meselson, M. (1983). Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli.Genetics 104,571 -582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan, R.A., and Kolodner, R.D. (1992). Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins.Genetics 132,963 -973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, G.A. (1977). The biological effects and mode of action of L-canavanine, a structural analogue of L-arginine. Q. Rev. Biol. 52,155 -178. [DOI] [PubMed] [Google Scholar]
- Rothman, F. G., and Narum, J. L. (1999). Then, Now and In the Next Decade: A Commentary on Strengthening Undergraduate Science, Mathematics, Engineering and Technology Education. Washington, DC: Project Kaleidoscope.
- Rydberg, B. (1978). Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 52,11 -24. [DOI] [PubMed] [Google Scholar]
- Shanabruch, W.G., Rein, R.P., Behlau, I., and Walker, G.C. (1983). Mutagenesis, by methylating and ethylating agents, in mutH, mutL, mutS, and uvrD mutants of Salmonella typhimurium LT2.J. Bacteriol. 153,33 -44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, E.C., and Bryson, V.(1967). Mutator gene of Escherichia coli B. J. Bacteriol. 94,38 -47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P.1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122,19 -27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, M., Prolla, T.A., Liskay, R.M., and Petes, T.D. (1993). Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365,274 -276. [DOI] [PubMed] [Google Scholar]
- Thibodeau, S.N., Bren, G., and Schaid, D. 1993. Microsatellite instability in cancer of the proximal colon.Science 260,816 -819. [DOI] [PubMed] [Google Scholar]
- Tomkinson, A.E., and Mackey, Z.B. 1998. Structure and function of mammalian DNA ligases. Mutat. Res. 407, 1-9. [DOI] [PubMed] [Google Scholar]
- Warthin, A.S. (1913). Heredity with reference to carcinoma. Arch. Intern. Med. 12,546 -555. [Google Scholar]
- Whelan, W.L., Gocke, E., and Manney, T.R. (1979). The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics 91,35 -51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M., Dao, V., and Modrich, P. (1998). MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J. Biol. Chem. 273,9197 -9201. [DOI] [PubMed] [Google Scholar]


