Dear Editor,
Gene targeting in mouse embryonic stem (ES) cells has revolutionized the field of mouse genetics and allowed for the analysis of diverse aspects of gene function in vivo. For more than two decades, researchers have been actively searching for ES cells from large animals such as pigs and cattle. Unfortunately, to date, no ES cell lines from large animals have passed the crucial test of germ line contribution. The sole method of gene targeting to date in these species remains somatic cell nuclear transfer (SCNT) combined with DNA homologous recombination (HR). Due to the limited proliferation competency and extremely low frequency of HR in somatic cells (less than 10−6), this process is highly inefficient and only a few successful examples have been achieved, even though enrichment strategies such as positive-negative marker selection, promoter-trap and adeno-associated viral delivery were previously used 1, 2, 3. The low efficiency of gene targeting in cultured somatic cells is the main barrier for gene targeting in large animals. Recently, zinc-finger nuclease (ZFN) technology has emerged as a powerful tool for genome editing. The success of ZFN technology for gene targeting in fruit flies, zebra fish, rodents as well as human cell lines encouraged us to establish a high-efficiency gene-targeting platform in large animals such as pigs 4, 5, 6, 7, 8.
In zebrafish and rats, methods of direct embryo injection with ZFN-encoding mRNA or plasmids have been employed to generate knockout (KO) mutations 5, 6. This method generates unpredictable deletion/insertion mutations in loci, which commonly result in a mosaic pattern in the first generation of offspring. Further breeding, which is often straightforward in rodents but tedious in large animals, is then necessary to obtain a heritable mutation. In the current study, we combined ZFN technology with SCNT to produce KO pigs with a defined mutation in peroxisome proliferator-activated receptor-gamma (Ppar-γ) (Supplementary information, Data S1). Thiazolidinediones (TZDs), selective ligands of PPAR-γ, are insulin sensitizers clinically used for the treatment of type 2 diabetes 9. Although TZD-mediated activation of PPAR-γ seems to have beneficial effects on cardiovascular diseases (CVD) in mouse models, the therapeutic efficacy of TZDs has been severely compromised because of the increased risk of adverse cardiovascular events in patients 9. As pigs are often considered an ideal animal model for studying human CVD, the Ppar-γ KO pig model will provide an invaluable tool for studying the role of PPAR-γ in CVD.
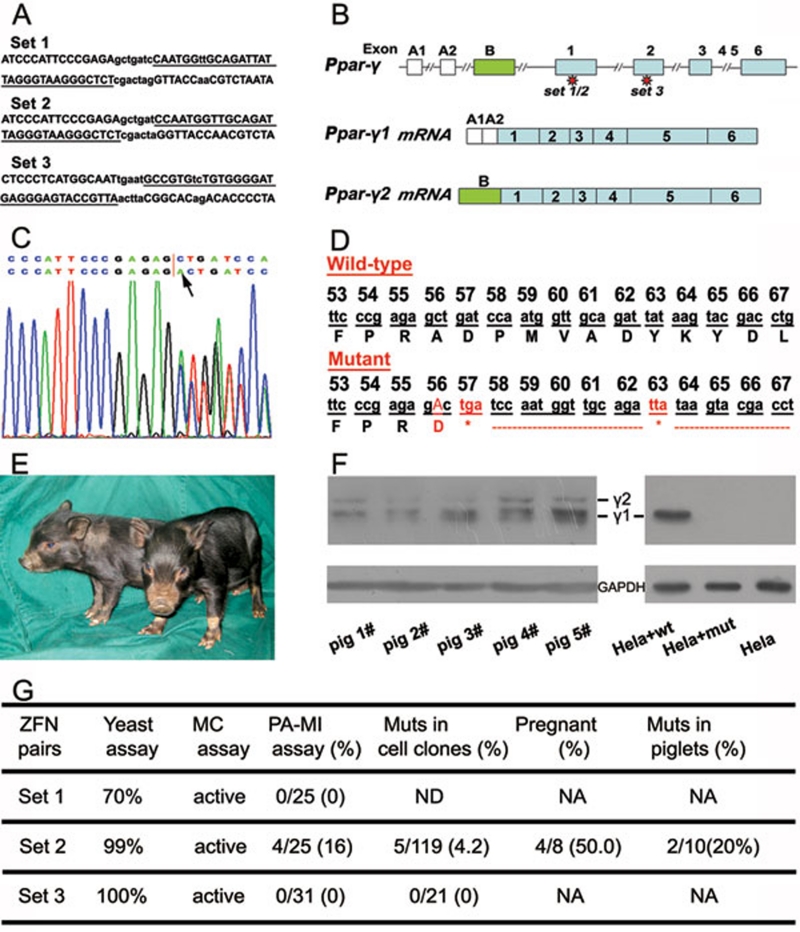
Three pairs of ZFNs with confirmed activity in a yeast MEL-1 assay were obtained commercially from Sigma-Aldrich (Figure 1A). In this study, ZFN set #3 showed the highest activity in the yeast assay. Furthermore, using a method described by Porteus 10, we found that ZFN set #3 was active in a cultured mammalian cell line-based assay (Figure 1G). Therefore, similar to the previously reported method in ZFN-KO rats 6, we performed direct injection of mRNAs encoding this particular ZFN set into fertilized porcine embryos. However, no positive gene targeting was identified from a total of 18 piglets or aborted fetuses (data not shown), indicating that ZFNs validated in yeast or tumor-derived cell lines may not be active in the porcine genome. It is apparently risky to use this direct embryo injection method in large animals because of long gestation periods and the high cost of large animal recipients. Thus, we have re-validated all three ZFN sets in a parthenogenetically activated (PA) embryo microinjection (MI) assay. ZFN-encoding mRNAs were MI into the cytoplasm of one-cell stage PA porcine embryos, followed by individual embryo PCR sequencing in the morulae or blastocyst stage using primers amplify across the target sites (Supplementary information, Table S3). Interestingly, ZFN set #2 displayed high activity, while ZFN set #1 and set #3 showed no activity in this assay (Figure 1G and Supplementary information, Table S1). Thus, the plasmids of ZFN set #2 were co-transfected with pcDNA3.1 (providing a neomycin-resistance gene) into male porcine fibroblasts by electroporation. After selecting with G418 (1 mg/ml for 1 week), 119 cell clones were screened by PCR sequencing and 5 cell clones were identified as carrying different mutations in the targeted Ppar-γ gene (Figure 1C and 1G and Supplementary information, Table S2).
Figure 1.
ZFN-mediated Ppar-γ gene disruption in pigs. (A) Binding sites (underlined) of the zinc-finger nucleases produced. (B) A schematic diagram of the Ppar-γ gene structure. (C) Sequencing diagram of a mutant with a mono-allelic insertion, as indicated by an arrow, at the ZFN-targeting site. The upper line shows the wild-type sequence while the lower line shows the mutant sequence. The diagram shows a double curve after insertion. (D) DNA and amino acid sequences of Ppar-γ proximal to the ZFN target site in cell clone #15 (“*” in red indicates stop codon sites after a frame shift resulted from an insertion at the 56 codon of Ppar-γ1). (E) Piglets confirmed to be Ppar-γ heterozygous knockout. (F) PPAR-γ western blotting. In the left panel, piglets #2 and #3 are mutant piglets while piglets #1, #4 and #5 are wild-type controls. Piglets #1 and #2 are littermates while piglets #3, #4, and #5 are littermates. In the right panel, HeLa cells transfected with wild-type Ppar-γ1 cDNA (HeLa+wt) or with mutated Ppar-γ1 cDNA (HeLa+mut) were used for PPAR-γ western blotting. The untransfected cells were used as the control (HeLa). γ 1 and γ 2 represent the two isoforms of PPAR-γ and GAPDH was used as the internal control. (G) The efficiency of ZFN-mediated targeting in pigs. The yeast assay data were provided by the ZFN manufacturer. ND: not determined; NA: not applicable; MC assay: mammalian cultured cell line-based assay; Muts: Mutants.
Cells in clone #15 with a mono-allelic single-nucleotide (A) insertion at exon 1 were used to clone Ppar-γ heterozygous KO pigs by nuclear transfer using the method described previously 2, 3 and detailed in Supplementary information Data S1. This single-nucleotide insertion was predicted to result in premature termination of PPAR-γ translation (Figure 1D). A total of 1 340 cloned embryos were introduced into 8 surrogate mothers, and 4 of them developed to term with a total of 10 live piglets delivered. Sequence analysis revealed that 2 piglets carried the predicted ZFN-induced mutation in their genome (Figure 1E and 1G). Using western blotting for determination of PPAR-γ protein expression in the peritesticular adipose tissue of these cloned piglets, we found a relatively lower expression of both PPAR-γ1 and PPAR-γ2, two isoforms of this receptor (Figure 1B), in mutant piglet #2 when compared with the wild-type littermate piglet #1 (Figure 1F). Similarly, PPAR-γ1 and PPAR-γ2 levels were lower in mutant piglet #3 when compared with its littermate control piglets #4 and #5 (Figure 1F). Since this particular mutation is heterozygous and the expression of PPAR-γ in newborn piglets is variable, direct western blotting is not considered to be an ideal tool for quantification. For example, PPAR-γ expression is greater in mutant piglet #3 than in the non-littermate wild-type piglet #1. To further confirm that the Ppar-γ mutation produced in our study has truly resulted in genetic disruption, both mutant and wild-type Ppar-γ cDNA, which were reverse-transcribed from Ppar-γ-mutant piglets, were cloned into a eukaryotic expression plasmid (pCAG) and then transfected into HeLa cells. Western blotting analysis of these transfected cells clearly showed disruption of Ppar-γ gene expression by this mutation and further confirmed the successful generation of Ppar-γ heterozygous KO pigs by a combined approach of ZFN technology and SCNT (Figure 1F). Next, bioinformatics tools were used to address potential ZFN off-targets, and among 37 predicted off-target sites sequenced in the mutant piglets, we detected two events. However, these two off-target events are in an intron and an intergenic sequence (Supplementary information, Tables S4 and S5), and therefore should not influence gene expression. Although we cannot exclude other potential off-target events, such events could be separated from the desired mutation by backcrossing to the parental strain, as mentioned previously in the Geurts et al. 6 paper.
Pigs, known to have physiological and anatomical similarities with humans, are considered to be more suitable experimental models than rodents for studying human diseases and are also considered to be potential donors for xenotransplantation. However, the difficulty of precise genetic modification in pigs as well as in other large animals has prevented sophisticated gene function studies and, thus, hampered their application in biomedical research and drug discovery. In the current study, we have successfully achieved, for the first time, the generation of an endogenous gene KO in pigs using the ZFN technology combined with SCNT. Our approaches established in this study increased the efficiency of gene targeting in primary pig cells from less than 10−6 to greater than 4%. Taken together, we have provided a high-efficiency platform for generating gene-targeted transgenic pigs for translational medicine and drug development. Furthermore, the platform we developed here can support the generation of KO models in other large animals lacking established ES cell lines.
Acknowledgments
We thank Prof Weiwang Gu (Southern Medical University) for generously providing the Tibetan mini pig tissues for fibroblast isolation. The work is partially supported by grants from the National High Technology Research and Development Program of China (2006AA02A103), the National Basic Research Program of China (973 Program) (2009CB941001, 2011CB944204), the Hundred Talents Project of Chinese Academy of Sciences (A0858), the Natural Science Foundation of Guangdong Province, China (9251066302000001), and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R-227) to LL, grant from the National Natural Science Foundation of China (31071172) to DY and grants from the National Institutes of Health (HL68878 and HL89544) to YEC. YEC is an established investigator of the American Heart Association (0840025N).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Parthenogenetically activated embryo microinjection (PA-MI) assay
ZFN set #2 induced-mono-allelic mutations
Primer pairs used for sequence analysis and cDNA amplification
Off-target analysis
Primer sequences used for off-target analysis
Materials and Methods
References
- Schnieke AE, Kind AJ, Ritchie WA, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- Lai L, Kolber-Simonds D, Park KW, et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- Rogers CS, Stoltz DA, Meyerholz DK, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumera KJ, Trautman JK, Bozas A, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parthenogenetically activated embryo microinjection (PA-MI) assay
ZFN set #2 induced-mono-allelic mutations
Primer pairs used for sequence analysis and cDNA amplification
Off-target analysis
Primer sequences used for off-target analysis
Materials and Methods