Abstract
A proportion of patients who develop regional and distant recurrences of melanoma after a pathologically negative sentinel lymph node (SN) biopsy are reported to have enhanced signals for melanoma associated mRNA when sensitive molecular approaches such as RT-PCR are used to evaluate their SN tissue. The significance of these findings remains controversial because the cellular source of the augmented signals cannot be known because the nodal tissue is destroyed during preparation for RT-PCR. Nonetheless it is claimed that the source of the augmented signal is covert metastatic melanoma cells. To determine whether there are histologically occult metastases in SN and whether there are sources of augmentable melanoma-associated mRNA other than melanoma cells we applied the reverse transcriptase in situ polymerase chain reaction (RT in situ PCR) to formalin-fixed paraffin-embedded nodal tissue. This approach amplifies small amounts of melanoma-associated mRNA and permits identification of the cells that express that mRNA. Cells containing MART-1 mRNA were detected in 6/21 SN (29%) and 2/16 NSN (13%) that were tumor-negative on hematoxylin and eosin and immunohistochemical assessment for S-100, MART-1 and HMB-45. In patients with microscopic evidence of melanoma in their SN, MART-1 mRNA positive cells were identified in 2/7 NSN (29%) that were histologically tumor-free. MART-1 mRNA positive cells were also detected in tumor negative SN sections from 6/7 (86%) nodes that had tumor present in areas of the node not represented in the studied sections. Some cells that expressed MART-1 mRNA that was diffusely distributed in the cytoplasm appeared to be melanoma cells while others resembled macrophages. The latter cells expressed augmented mRNA on granules that were intermixed with melanin granules. In other cases MART-1 mRNA positive macrophage-like cells contained nuclei and nucleoli more typical of melanoma cells and may represent the macrophage-melanoma hybrids that have been previously reported. Combination of RT in situ PCR for MART-1 mRNA and immunohistochemistry for CD68 revealed that CD68 was co-localized in some cells that expressed MART-1 mRNA.
Some lymph nodes that are tumor negative by histology and immunohistochemistry contain cells that express mRNA for MART-1. Some of these cells may be interpreted as “stealth” melanoma cells in which, despite the presence of MART-1 mRNA there is an absence of immunohistochemically detectable MART-1 protein. Other cells that contain MART-1 mRNA are clearly not melanoma cells or may represent melanoma hybrids. These findings should be taken into account when interpreting and applying the results of RT-PCR analysis of nodal (and other) tissues.
Keywords: MART-1, macrophage, melanoma, metastasis, micrometastasis, sentinel lymph node
Introduction
Malignant melanomas have a high potential for metastasis and usually spread first to the regional lymph nodes. Lymphatic mapping and sentinel node (SN) biopsy (46) are recently developed procedures that are widely used in the management of melanoma patients. SN evaluation permits accurate staging of patients and determines the need for additional nodal surgery. Patients who have a melanoma-positive SN currently undergo complete dissection of the affected lymph node basin (completion lymphadenectomy), while patients with a histologically negative SN receive no additional nodal surgery. Approximately 20% of patients with a primary melanoma ≥1 mm in thickness have metastatic tumor in microscopically assessed (hematoxylin and eosin (H&E) and immunohistochemistry) sections from the SN (44, 45). Accurate determination of SN tumor status requires evaluation of multiple sections and immunohistochemistry for multiple melanocyte lineage-associated antigens (14). However, a portion of patients assessed as having a metastasis-negative SN on the basis of H&E histology and immunohistology subsequently develop regional (and distant) recurrences. Such patients have been claimed to reflect false-negative sentinel node biopsies. The false negative rate of SN (the number of false negative SN divided by the sum of the number of positive SN and that of false negative SN) has been reported to be 11-18% (7, 13, 39, 45, 58). False-negativity has been attributed to technical problems associated with identification of the true SN by nuclear medicine and surgery and errors in tissue sampling and interpretation during pathological assessment (34). Anomalies of tumor biology and variation in the timing and routing of spread of tumor cells from the primary site to the SN (34, 47) may also explain some “false negative” SN. Molecular analyses including the reverse transcriptase polymerase chain reaction (RT-PCR) has been reported to detect enhanced signal for melanoma-associated mRNA that may indicate micrometastases (3, 5, 22, 27, 44). This has been considered to indicate the presence of truly occult melanoma cells that are difficult to detect microscopically or missed by the relatively limited sampling that is used to evaluate SN in most pathology protocols. It is, however, also possible that the technique may be too sensitive, augmenting signals for mRNA in cells other than melanoma cells and providing results that are thus “false positive”. Evaluation of the results of RT-PCR studies is particularly challenging as the cellular source of the augmented mRNA signals cannot be directly determined in the RT-PCR approach since the tissue is disrupted during the preparative stages of the technique.
The reverse transcriptase in situ polymerase chain reaction (RT in situ PCR) amplifies and visualizes small amounts of specific mRNA and allows identification of the nature of the cells that contain the amplified mRNA. We have successfully modified the RT in situ PCR approach to detect melanoma-associated genes in melanoma cell lines (26, 40) and formalin-fixed paraffin-embedded tissues (25, 31). mRNA expression reflecting the presence of melanoma-associated genes can be detected microscopically in lymph nodes containing metastases by this technique (31). To detect cells expressing MART-1 (melanoma associated antigen recognized by T cells-1: MLANA) mRNA, including histologically occult metastatic melanoma cells, we applied the RT in situ PCR to SN and non-sentinel nodes (NSN) that were tumor-negative by standard histopathological and immunohistochemical evaluations.
Materials and methods
Fifty one tissue blocks from lymph nodes (28 SN, 23 NSN) from patients with malignant melanoma were retrieved from the surgical pathology archives of UCLA and evaluated with Institutional Review Board permission. Patients had been treated by sentinel lymph node biopsy and completion lymphadenectomy if the SN contained metastatic tumor. Pathological examination of sentinel lymph node biopsy was performed by using published criteria (14). H&E and immunohistochemically stained (S-100 protein, HMB-45, MART-1) slides from all lymph nodes were re-reviewed to confirm the presence or absence of metastases (AJC and DRW). Microscopically melanoma-free SN (n=21) and NSN (n=16) were further evaluated. Histologically tumor-negative NSN from patients with tumor-positive SN were also evaluated (n=7). Tumor-free microscopic sections from tumor-positive SN were identified and evaluated (n=7). As a negative control, we examined 5 tumor-free axillary lymph nodes from patients with breast cancer (3 SN and 2 NSN).
RT in situ PCR
Pretreatment
Formalin-fixed, paraffin-embedded tissue sections (4μm thickness) were de-paraffinized and hydrated through xylene and a series of graded ethanols. The slide-mounted tissue sections were digested with trypsin (0.2-0.4 mg/ml) in phosphate-buffered saline (PBS) for periods that ranged from 30 to 60 min at 37°C (Roche Diagnostics, Mannheim, Germany) for optimization, followed by digestion with RNase-free DNase I (EPICENTRE Biotechnologies, Madison, WI) in PBS at 37°C overnight.
One step RT-PCR
Single step RT in situ PCR approach was performed using a technique based on descriptions in recent articles (2, 31, 32, 48). The Gene Amp EZ rTth (recombinant Thermus thermophilus) RNA PCR Kit (Applied Biosystems, Foster City, CA) was employed. The oligonucleotide primer sequences (Invitrogen, Carlsbad, CA) for MART-1 mRNA were derived from a previous paper (55). The forward primer, 5’-CACGGCCACTCTTACACCAC-3’ and the reverse primer, 5’-GGAGCATTGGGAACCACAGG-3’ yielded a product of 254 bp (Table 1). Each slide was loaded with the following mixture: 15μl of 5× EZ rTth buffer, 9μl of Mn(OAc)2, 2.4μl each of dATP/dCTT/dGTT/dTTP, 3μl of rTth DNA polymerase (Applied Biosystems), 0.9μl digoxigenin-11-dUTP, 2.4μl of 2% bovine serum albumin (BSA), 1.3μl of RNase inhibitor (Roche), 2.3μl of each primer, and 30.7μl diethylpyrocarbonate (DEPC)-treated water. The slides were covered with HybriWell sealing covers (Research Products International, Mount Prospect, IL) and placed on the Gene Amp In Situ PCR System 1000 thermal cycler (PerkinElmer, Foster City, CA). cDNA syntheses were performed at 60°C for 30 min. After initial denaturation at 94°C for 3 min, the cDNA was amplified by 30 cycles of annealing at 55°C for 1 min and denaturation at 94°C for 30 sec. After RT-PCR, the slides were washed in 0.1× saline-sodium citrate with 2% BSA at 60°C for 15 min and rinsed in Tris-buffered saline (TBS) three times for 5 min.
Table 1.
Primers used for MART-1 mRNA detection.
| Target | Gene | Sequences | Amplicon size | |
|---|---|---|---|---|
| MART-1 | MLANA | Forward | 5’-CACGGCCACTCTTACACCAC-3’ | 254 bp |
| Reverse | 5’-GGAGCATTGGGAACCACAGG-3’ |
Immunodetection of RT-PCR products
Following incubation with an alkaline phosphatase–conjugated anti-digoxigenin antibody (Roche; 1:200) for 30 min, the RT-PCR product was developed with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Zymed Laboratories, South San Francisco, CA) for 15 min. After chromogen development, selected slides were stained by eosin.
Controls
Negative control preparations were prepared by: 1) omission of the primer pair for MART-1; 2) omission of the Tth DNA polymerase. Sections of metastatic melanomas in lymph nodes served as a positive control. Formalin-fixed paraffin-embedded sections of cultured human melanoma cells (M7, M14 and M24; courtesy of Dr Donald L. Morton, John Wayne Cancer Institute, Santa Monica, CA), expressing MART-1 mRNA (31), also served as a positive control.
Double labeling using RT in situ PCR for MART-1 mRNA and immunohistochemistry for CD68
Double staining using RT in situ PCR for MART-1 mRNA and immunohistochemistry for CD68 antigen was employed on 6 representative MART-1 mRNA-positive cases. After the RT in situ PCR products were developed with NBT/BCIP, immunohistochemistry for CD68 was performed. Endogenous peroxidase activity was blocked by exposure to 3% hydrogen peroxide for 30 min. Heat-induced epitope retrieval was performed by boiling in citrate buffer, pH 6.0 for 30 min. Monoclonal antibody for CD68 (clone: KP-1; Dako, Glostrup, Denmark) was diluted in PBS at 1:20. Sections were incubated with the primary antibody overnight at 4°C. The subsequent development of antibody-bridge labeling was made by the streptavidin-biotin-peroxidase method using biotinylated anti-mouse antibody (Vector Laboratories, Burlingame, CA) and streptavidin horseradish peroxidase (Zymed Laboratories). The sections were then reacted in with aminoethyl carbazole chromogen (AEC; Zymed Laboratories)
Results
Cells containing MART-1 mRNA were detected in 6/21 SN (29%) and 2/16 NSN (13%) that were tumor-negative on examination of H&E and immunohistochemistry stained slides. In cases with a positive SN, MART-1 mRNA positive cells were found in 2/7 NSN (29%) that were histologically negative. MART-1 mRNA positive cells were also detected in microscopically tumor-free areas of 6/7 (86%) tumor-positive SN (Table 2).
Table 2.
MART-1 mRNA positivity in histology-negative SN and NSN
| MART-1 mRNA positive/total | % | ||
|---|---|---|---|
| SN | (SN: histology-negative) | 6/21 | 29% |
| NSN | (SN, NSN: histology-negative) | 2/16 | 13% |
| NSN | (SN: histology-positive, NSN: histology-negative) | 2/7 | 29% |
| SN | (SN: histology-positive; Microscopically tumor-free areas were examined.) | 6/7 | 86% |
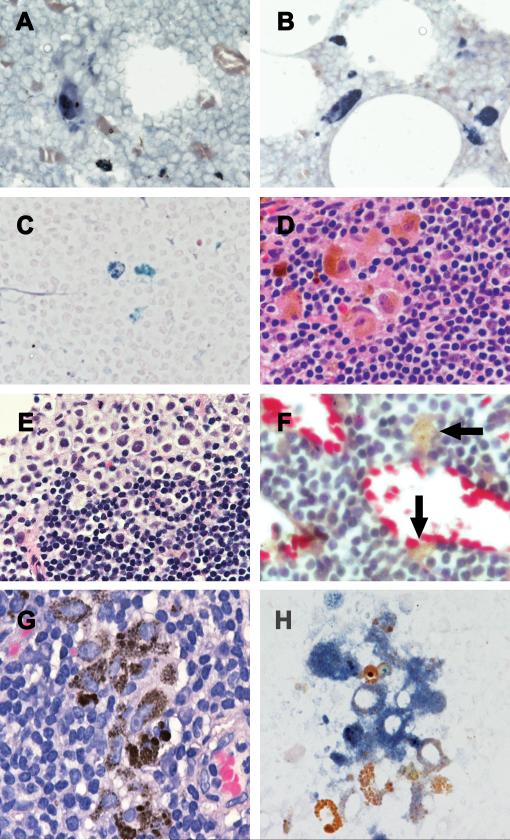
Scattered, single isolated cells in the nodal parenchyma that contained diffuse cytoplasmic MART-1 mRNA appeared to be (stealth) melanoma cells-melanoma cells that despite containing MART-1 mRNA did not show immunohistochemically detectable MART-1 protein (Figure 1A-C). With knowledge of the location of the RT in situ PCR positive cells, review of the original H&E slides allowed identification of melanoma cells in the areas of the node remote from the RT in situ PCR positive cells (Figure 1D&E). On the basis of size, nuclear characteristics and scattered melanin these cells were consistent with melanoma cells. Some MART-1 mRNA positive cells contained microdispersed melanin granules, a pattern that is typical of melanogenic cells, but lacked a nucleus which would have permitted determination of their malignant or benign status by H&E (Figure 1F). In other cases, MART-1 mRNA-positive cells were associated with small clusters of macrophage-like cells that contained nuclei and nucleoli regarded as more typical of melanoma cells as well as the fine micro-dispersed melanin granules typical of melanogenic cells (Figure 1G). These cells were adjacent to typical melanophages (Figure 1H) that had smaller, sometimes reniform nuclei, inconspicuous nucleoli and abundant coarse brown (aggregated) pigmented granules. Typical melanophages did not express MART-1 mRNA. Yet other MART-1 mRNA-expressing cells, singly dispersed and scattered in the nodal parenchyma, had typical macrophage morphology with coarse brown pigmented granules (compound melanosomes) characteristic of melanophages (Figure 2A&B). Immunohistochemistry on the corresponding SN/NSN tissue sections revealed that these MART-1 mRNA-expressing cells were negative for melanocytic markers except in 2 cases where positive signals for both HMB-45 and MART-1 were detected. In these two cases, singly dispersed and isolated immunoreactive cells that were HMB-45 and MART-1 and positive, with non-atypical nuclei and coarse melanin granules, were readily identified as melanophages (Figure 2C&D). Single isolated cells that were positive for MART-1 mRNA were detected in the corresponding RT in situ PCR slides.
Figure 1.
“Stealth” melanoma cells identified by enhanced signal for MART-1 mRNA detected by RT in situ PCR (blue/purple color) (Panels A-C). In some original H&E slides, melanoma cells were present in areas separate from the RT in situ PCR-detected “stealth” melanoma cells (Panels D&E). Some MART-1 mRNA positive cells contained microdispersed melanin granules, a pattern that is typical of melanogenic cells, but lacked a nucleus which would have permitted determination of their malignant or benign status by H&E (Panel F). Small clusters of macrophage-like cells containing nuclei and nucleoli more typical of melanoma cells and microdispersed melanin granules (Panel G). These cells show positivity for MART-1 mRNA by the RT in situ PCR (blue/purple color) (Panel H). MART-1 mRNA is not seen in adjacent melanophages with coarse brown pigmented granules.
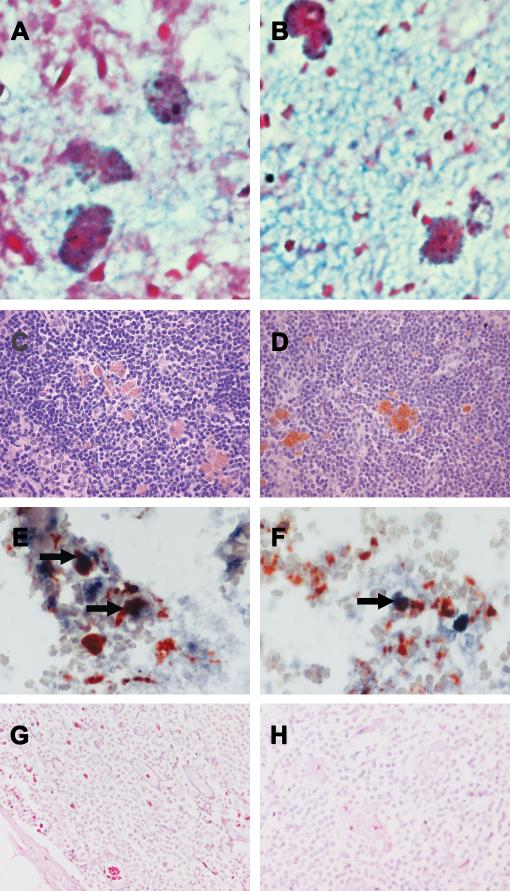
Figure 2.
MART-1 mRNA (blue/purple color) is seen by RT in situ PCR in melanophages with coarse pigment granules (Panels A&B). Immunohistochemistry for MART-1 shows a weakly positive signal in scattered large cells with “normal” nuclei and coarse melanin granules, consistent with melanophages (Panel C). Immunohistochemistry for HMB-45 shows a positive signal in scattered large cells, consistent with melanophages (Panel D). Double staining of MART-1 mRNA (blue/purple color) and CD68 antigen (red color) shows that some cells co-express both MART-1 mRNA and CD68 (arrow), while MART-1 mRNA and CD68 are separately expressed by other cells. (Panels E&F). Sentinel node (Panel G) and NSN (Panel H) from a breast cancer patient are negative for MART-1 mRNA.
There was one example of nodal nevus in the examined lymph nodes, but the nevus cells were not detected by RT in situ PCR for MART-1 mRNA.
Double staining for MART-1 mRNA by RT in situ PCR and CD68 antigen by immunohistochemistry showed that 40% of the cells that contained MART-1 mRNA co-expressed MART-1 mRNA and CD68. Cells coexpressing MART-1 mRNA and CD68 were identified in areas where there were abundant CD68-positive cells. Cells expressing MART-1 mRNA but not CD68 were co-localized in the same area as CD68-positive cells and cells that coexpressed MART-1 mRNA and CD68 (Figure 2E&F). As NBT/BCIP (blue/purple) and ACE (red) were used as chromogens, positive staining was clearly distinguishable from brown cytoplasmic melanin.
None of the five lymph nodes (SN and NSN) from patients with breast cancer showed positivity for MART-1 mRNA or MART-1 antigen (Figure 2G&H). None of these lymph nodes showed inclusions of melanocytic nevus cells.
Discussion
Identification of melanoma cells in H&E stained sections of sentinel nodes is usually straightforward when they form masses or clusters of tumor cells. However, early metastases may be seen as single melanoma cells, making their detection in H&E sections more challenging as such cells can show a considerable range of morphological variation, and are often amelanotic. Lightly pigmented or unpigmented melanoma cells may closely simulate sinus and other histiocytes that are often abundant in lymph nodes. Extended histopathological search for small metastatic deposits by serial sectioning and immunohistochemistry reportedly detects occult melanoma cells in up to 32% of SN (39). Micrometastases may be missed if the wrong part of a node is examined or if tumor cells do not express melanoma-associated markers detectable by immunohistochemistry.
There are reports of patients with an intermediate prognosis who have an enhanced signal for mRNA for melanoma-associated genes detected by RT-PCR, in nodes that showed no evidence of melanoma cells on microscopic evaluation (44). The RT in situ PCR technique is valuable in basic research and clinical science and permits detection of mRNA in specifically identifiable cells and tissues. In this study, we identified MART-1 mRNA-positive cells in SN and NSN in which prior search for cells expressing MART-1 protein by immunohistochemistry was negative. Some of these cells are truly occult melanoma cells that despite containing MART-1 mRNA do not have detectable cytoplasmic MART-1 protein. Such “stealth” melanoma cells were also detected in microscopically tumor-free NSN from patients with a tumor-positive SN When a completion lymph node dissection (CLND) is performed for patients with a positive SN, tumor is detected in the NSN of 10 to 20% of patients (21, 24, 43, 57). We showed that MART-1 mRNA-positive cells were also present in microscopically negative NSN. The frequency of tumor-positive NSN may increase when nodal tissues are evaluated by sensitive molecular analyses (4). The biological significance of nodes found to contain tumor by special and extended evaluation remains unknown. Equally the biological potential and significance of “stealth” melanoma cells remains to be determined.
Human tumors express antigens recognized by autologous T lymphocytes. Tumour progression is often accompanied by alterations in the expression of such “differentiation” antigens, in some instances leading to tumor escape from immunosurveillance. Although MART-1 is detectable in most melanomas and is considered a suitable target for immunomodulation therapy, metastatic melanomas often shows heterogeneous expression of MART-1 (15, 28, 33). Melanocyte-specific microphthalmia-associated transcription factor (Mitf-M) regulates the expression of melanocyte-associated genes, and downregulation of Mitf-M results in the loss or reduced expression of melanoma-associated antigens in melanoma cells (16, 36). Loss of melanoma-associated antigens may also be induced by soluble factors produced by melanoma cells (37), including oncostatin M (19). Thus, when attempting to detect micrometastases in SNs by molecular methods, attention should be paid to the possibility of downregulation of genes for tumor-associated antigen. The use of multiple melanoma markers can significantly improve the specificity of molecular testing (65), but conversely may affect sensitivity, because alteration of melanoma-associated antigens has implications for tumor growth and metastasis and these melanoma-associated antigens are downregulated in metastatic melanoma cells.
Alternative cellular sources for mRNA for melanoma-associated genes in lymph nodes include macrophages that have phagocytosed materials from melanoma cells. MART-1 mRNA was detected in macrophages (melanophages) containing intracellular granules of melanin. Co-localization of CD68 and the RT in situ PCR enhanced signal for MART-1 mRNA was found, further supporting the view that these cells are macrophages. Melanophage accumulation is typically observed at the primary site in the early stage of melanoma evolution. Single isolated melanophages and groups of melanophages forming small granulomas often mimic micrometastasis in regional lymph nodes. Large irregularly shaped intracellular granules of pigment, comprised of aggregated melanosomes are morphological characteristics that help distinguish melanophages from melanoma cells where, in most instances, the melanosomes are small, regular in size and shape and diffusely dispersed. Electron microscopic examination has revealed that melanophages contain partially or fully melanized melanosomes that may be located within phagosomes. Melanosomes can also be found in dermal melanophages in postinflammatory hyperpigmentation (42, 63) or in tumor cells in cutaneous neoplasms other than melanoma such as pigmented squamous cell carcinoma and pigmented basal cell carcinoma (63). Melanosomes often exhibit immunoreactivity for HMB-45 (56), and HMB-45-positive melanophages have been demonstrated by immunoelectron microscopy. On the other hand, MART-1 is not localized to melanosomes but it is located in electron-lucent cytoplasmic vacuoles and vesicles in melanocytes (64). Our study revealed that the MART-1 epitope is also be detectable in melanophages, possibly indicating phagocytosed materials retained in their cytoplasm. However, we can not entirely exclude the possibility that the highly sensitive molecular biology technique that we employed may augment acquired nucleotides in the cytoplasm of macrophages that have phagocytosed melanoma cell components. MART-1- or HMB-45-positive melanophages may be difficult to differentiate from melanoma cells on light microscopy.
Melanoma-macrophage hybridization is another possible explanation for cells that express characteristics of both melanoma cells and macrophages. Until recently, the hypothesis that fusion of cancer cells and normal cells may facilitate metastasis has received limited attention. It has been suggested that cancer cells may spontaneously fuse with host cells to produce hybrids (17, 18, 35, 49, 50). Evidence in support of cancer cell fusion with macrophages or other bone marrow-derived cells has been reported from studies of animal models of human cancer (6, 9-11, 49, 52, 53, 59). Hybrid cells formed from macrophages and melanoma cells may possess the malignant potential of the melanoma cells and the migratory phenotype of the macrophages. Such hybrid cells contain coarse intracellular melanin granules of the type typically seen in melanophages (53). In the tumor microenvironment, macrophages often constitute a major component of the tumor stroma. Macrophages are conventionally regarded as promoters of tumor progression (68) and are recruited to melanoma by expression of chemokines including CCL2 (MCP-1). However, it is difficult to further track the underlying macrophages and demonstrate hybridization with tumor cells in human tissues. A recent study reported that epithelial cancers arise by transdifferentiation of bone marrow stem cells, a process that did not involve hybridization since these cells were diploid without evidence of fusion of donor and recipient cells (23). Transdifferentiation of bone marrow stem cells to cancer cells may be another possible explanation of tumor dormancy and cancer cell metastasis.
In contrast, melanoma cells, particularly metastatic melanoma cells may exhibit an aberrant immunophenotype and may express many of the known lysosomal and phagocytic markers, such as CD68, α1 antitrypsin and α1 chymotrypsin that are typical of macrophages. They also frequently show immunoreactivity with macrophage markers including, MAC387 and HAM56 (1, 20, 38, 51). Some melanoma cells express MHC class II molecules that are normally present on antigen presenting cells and activated T cells. Metastatic melanoma cells may show macrophage-like phagocytic activities that involve cytoplasmic vesicles (41). Macrophage-like properties have also been observed in gliomas (29), which can preferentially express melanoma-associated antigens such as gp100, MART-1, tyrosinase, TRP-2, MAGE-A1 and MAGE-A3 (12, 54, 67). It remains unclear how melanoma cells develop aberrant macrophage-like immunophenotypes. The possibility that they are the result of melanoma-macrophage hybridization is clearly worthy of further consideration.
Another pitfall of the use of molecular biology techniques is that the “tumor-associated” genes used for detection of melanoma cells by RT-PCR are not absolutely specific to melanoma cells. These genes are mostly genes for melanocyte-lineage antigens that are also expressed by normal melanocytes and benign nevus cells. Nevus cells in lymph nodes represent a well known potential diagnostic pitfall in the RT-PCR analysis (61, 66). Histologically, nodal nevi appear as aggregates of nevus cells in the lymph node capsule and trabeculae, and are a potential source of confusion with metastatic melanoma. Discrimination of benign nevocytes from metastatic melanoma requires close consideration of cytologic features of the cells which are negative or at most faintly positive for HMB-45 (8).
Though infrequent, benign inclusions in lymph nodes represent the presence of heterologous elements not normally found in lymph nodes. These include salivary inclusions in peri-parotid lymph nodes, thyroid inclusions in cervical lymph nodes, mammary gland inclusions in axillary lymph nodes, or endometrial glandular inclusions in abdominal and pelvic lymph nodes (30, 60, 62). The occurrence of such inclusions provides an additional potential pitfall in the use of molecular biology to detect cancer metastases.
The prognostic significance of histology-negative RT-PCR-positive sentinel nodes remains debatable. There are no objective data to support or contradict complete lymph node dissection for histology-negative, molecular-positive sentinel nodes. A definitive answer must await completion of MSLT-2, a multicenter randomized trial in the United States, Europe and Australia (44). Although the clinical importance of the lymphatic system is well recognized, the actual molecular mechanisms involved remain poorly understood. As part of the elucidation of these mechanisms improvements in the techniques and criteria involved in the detection of (early) lymph node metastasis of melanoma will be of great importance. These investigations will determine the clinical significance of “stealth” melanoma cells and the other MART-1 mRNA positive cells.
Acknowledgements
This work was supported by NIH/NCI grant P01 CA 29605 (administered by John Wayne Cancer Institute, Saint John's Health Center, Santa Monica, CA).
Footnotes
Disclosure:
The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdelatif OM, Khankhanian NK, Crosby JH, et al. Malignant fibrous histiocytoma and malignant melanoma: the role of immunohistochemistry and electron microscopy in the differential diagnosis. Mod Pathol. 1989;2:477–485. [PubMed] [Google Scholar]
- 2.Adam PJ, Boyd R, Tyson KL, et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J Biol Chem. 2003;278:6482–6489. doi: 10.1074/jbc.M210184200. [DOI] [PubMed] [Google Scholar]
- 3.Blaheta HJ, Ellwanger U, Schittek B, et al. Examination of regional lymph nodes by sentinel node biopsy and molecular analysis provides new staging facilities in primary cutaneous melanoma. J Invest Dermatol. 2000;114:637–642. doi: 10.1046/j.1523-1747.2000.00925.x. [DOI] [PubMed] [Google Scholar]
- 4.Bonin S, Niccolini B, Calacione R, et al. Molecular analyses of sentinel lymph nodes: an open question. J Eur Acad Dermatol Venereol. 2002;16:34–39. doi: 10.1046/j.1468-3083.2002.00360.x. [DOI] [PubMed] [Google Scholar]
- 5.Bostick PJ, Morton DL, Turner RR, et al. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol. 1999;17:3238–3244. doi: 10.1200/JCO.1999.17.10.3238. [DOI] [PubMed] [Google Scholar]
- 6.Busund LTR, Killie MK, Bartnes K, et al. Spontaneously formed tumorigenic hybrids of Meth A sarcoma cells and macrophages in vivo. Int J Cancer. 2003;106:153–159. doi: 10.1002/ijc.11210. [DOI] [PubMed] [Google Scholar]
- 7.Carlson GW, Page AJ, Cohen C, et al. Regional recurrence after negative sentinel lymph node biopsy for melanoma. Ann Surg. 2008;248:378–386. doi: 10.1097/SLA.0b013e3181855718. [DOI] [PubMed] [Google Scholar]
- 8.Carson KF, Wen DR, Li PX, et al. Nodal nevi and cutaneous melanomas. Am J Surg Pathol. 1996;20:834–840. doi: 10.1097/00000478-199607000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty AK, Funasaka Y, Ichihashi M, et al. Upregulation of alpha and beta integrin subunits in metastatic macrophage-melanoma fusion hybrids. Melanoma Res. 2009;19:343–349. doi: 10.1097/CMR.0b013e32832fe121. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty AK, Sodi S, Rachkovsky M, et al. A spontaneous murine melanoma lung metastasis comprised of host x tumor hybrids. Cancer Res. 2000;60:2512–2519. [PubMed] [Google Scholar]
- 11.Chakraborty AK, Yamaga S. Differential gene expression in genetically matched mouse melanoma cells with different metastatic potential. Gene. 2003;315:165–175. doi: 10.1016/s0378-1119(03)00736-4. [DOI] [PubMed] [Google Scholar]
- 12.Chi DD, Merchant RE, Rand R, et al. Molecular detection of tumor-associated antigens shared by human cutaneous melanomas and gliomas. Am J Pathol. 1997;150:2143–2152. [PMC free article] [PubMed] [Google Scholar]
- 13.Clary BM, Brady MS, Lewis JJ, et al. Sentinel lymph node biopsy in the management of patients with primary cutaneous melanoma: review of a large single-institutional experience with an emphasis on recurrence. Ann Surg. 2001;233:250–258. doi: 10.1097/00000658-200102000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochran AJ, Roberts A, Wen DR, et al. Update on lymphatic mapping and sentinel node biopsy in the management of patients with melanocytic tumours. Pathology. 2004;36:478–484. doi: 10.1080/00313020412331282726. [DOI] [PubMed] [Google Scholar]
- 15.Cormier JN, Hijazi YM, Abati A, et al. Heterogeneous expression of melanoma-associated antigens and HLA-A2 in metastatic melanoma in vivo. Int J Cancer. 1998;75:517–524. doi: 10.1002/(sici)1097-0215(19980209)75:4<517::aid-ijc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 16.Du J, Miller AJ, Widlund HR, et al. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol. 2003;163:333–343. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 18.Duelli D, Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer. 2007;7:968–976. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- 19.Durda PJ, Dunn IS, Rose LB, et al. Induction of “antigen silencing” in melanomas by oncostatin M: down-modulation of melanocyte antigen expression. Mol Cancer Res. 2003;1:411–419. [PubMed] [Google Scholar]
- 20.Facchetti F, Bertalot G, Grigolato PG. KP1 (CD 68) staining of malignant melanomas. Histopathology. 1991;19:141–145. doi: 10.1111/j.1365-2559.1991.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 21.Govindarajan A, Ghazarian DM, McCready DR, et al. Histological features of melanoma sentinel lymph node metastases associated with status of the completion lymphadenectomy and rate of subsequent relapse. Ann Surg Oncol. 2007;14:906–912. doi: 10.1245/s10434-006-9241-3. [DOI] [PubMed] [Google Scholar]
- 22.Goydos JS, Patel KN, Shih WJ, et al. Patterns of recurrence in patients with melanoma and histologically negative but RT-PCR-positive sentinel lymph nodes. J Am Coll Surg. 2003;196:196–204. doi: 10.1016/S1072-7515(02)01758-1. [DOI] [PubMed] [Google Scholar]
- 23.Guest I, Ilic Z, Ma J, et al. Direct and indirect contribution of bone marrow-derived cells to cancer. Int J Cancer. 2010;126:2308–2318. doi: 10.1002/ijc.24946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guggenheim M, Dummer R, Jung FJ, et al. The influence of sentinel lymph node tumour burden on additional lymph node involvement and disease-free survival in cutaneous melanoma--a retrospective analysis of 392 cases. Br J Cancer. 2008;98:1922–1928. doi: 10.1038/sj.bjc.6604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Cheng L, Wen DR, et al. Detection of tyrosinase mRNA in formalin-fixed, paraffin-embedded archival sections of melanoma, using the reverse transcriptase in situ polymerase chain reaction. Diagn Mol Pathol. 1998;7:10–15. doi: 10.1097/00019606-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Guo J, Wen DR, Huang RR, et al. Detection of multiple melanoma-associated markers in melanoma cell lines by RT in situ PCR. Exp Mol Pathol. 2003;74:140–147. doi: 10.1016/s0014-4800(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 27.Hilari JM, Mangas C, Xi L, et al. Molecular staging of pathologically negative sentinel lymph nodes from melanoma patients using multimarker, quantitative real-time RT-PCR. Ann Surg Oncol. 2009;16:177–185. doi: 10.1245/s10434-008-0183-9. [DOI] [PubMed] [Google Scholar]
- 28.Hofbauer GF, Kamarashev J, Geertsen R, et al. Melan A/MART-1 immunoreactivity in formalin-fixed paraffin-embedded primary and metastatic melanoma: frequency and distribution. Melanoma Res. 1998;8:337–343. doi: 10.1097/00008390-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Huysentruyt LC, Mukherjee P, Banerjee D, et al. Metastatic cancer cells with macrophage properties: evidence from a new murine tumor model. Int J Cancer. 2008;123:73–84. doi: 10.1002/ijc.23492. [DOI] [PubMed] [Google Scholar]
- 30.Ioachim HL, Medeiros LJ. Ioachim's lymph node pathology. Lippincott, Williams & Wilkins; Philadelphia: 2009. [Google Scholar]
- 31.Itakura E, Huang RR, Wen DR, et al. RT in situ PCR detection of MART-1 and TRP-2 mRNA in formalin-fixed, paraffin-embedded tissues of melanoma and nevi. Mod Pathol. 2008;21:326–333. doi: 10.1038/modpathol.3801008. [DOI] [PubMed] [Google Scholar]
- 32.Itakura E, Huang RR, Wen DR, et al. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod Pathol. 2011;24:801–809. doi: 10.1038/modpathol.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kageshita T, Kawakami Y, Hirai S, et al. Differential expression of MART-1 in primary and metastatic melanoma lesions. J Immunother. 1997;20:460–465. doi: 10.1097/00002371-199711000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Karim RZ, Scolyer RA, Li W, et al. False negative sentinel lymph node biopsies in melanoma may result from deficiencies in nuclear medicine, surgery, or pathology. Ann Surg. 2008;247:1003–1010. doi: 10.1097/SLA.0b013e3181724f5e. [DOI] [PubMed] [Google Scholar]
- 35.Kluk MJ, Grant-Kels JM, Kerr P, et al. Melanoma on the move: the progression of melanoma: novel concepts with histologic correlates. Am J Dermatopathol. 2004;26:504–510. doi: 10.1097/00000372-200412000-00011. [DOI] [PubMed] [Google Scholar]
- 36.Kono M, Dunn IS, Durda PJ, et al. Role of the mitogen-activated protein kinase signaling pathway in the regulation of human melanocytic antigen expression. Molecular cancer research. 2006;4:779–792. doi: 10.1158/1541-7786.MCR-06-0077. [DOI] [PubMed] [Google Scholar]
- 37.Kurnick JT, Ramirez-Montagut T, Boyle LA, et al. A novel autocrine pathway of tumor escape from immune recognition: melanoma cell lines produce a soluble protein that diminishes expression of the gene encoding the melanocyte lineage melan-A/MART-1 antigen through down-modulation of its promoter. J Immunol. 2001;167:1204–1211. doi: 10.4049/jimmunol.167.3.1204. [DOI] [PubMed] [Google Scholar]
- 38.Leader M, Patel J, Collins M, et al. Anti-alpha 1-antichymotrypsin staining of 194 sarcomas, 38 carcinomas, and 17 malignant melanomas. Its lack of specificity as a tumour marker. Am J Surg Pathol. 1987;11:133–139. doi: 10.1097/00000478-198702000-00007. [DOI] [PubMed] [Google Scholar]
- 39.Li LXL, Scolyer RA, Ka VSK, et al. Pathologic review of negative sentinel lymph nodes in melanoma patients with regional recurrence: a clinicopathologic study of 1152 patients undergoing sentinel lymph node biopsy. Am J Surg Pathol. 2003;27:1197–1202. doi: 10.1097/00000478-200309000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Li PX, Cheng L, Wen DR, et al. Demonstration of cytoplasmic tyrosinase mRNA in tissue-cultured cells by reverse transcription (RT) in situ polymerase chain reaction (PCR) and RT PCR in situ hybridization. Diagn Mol Pathol. 1997;6:26–33. doi: 10.1097/00019606-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Lugini L, Lozupone F, Matarrese P, et al. Potent phagocytic activity discriminates metastatic and primary human malignant melanomas: a key role of ezrin. Lab Invest. 2003;83:1555–1567. doi: 10.1097/01.lab.0000098425.03006.42. [DOI] [PubMed] [Google Scholar]
- 42.Mahmood MN, Lee MW, Linden MD, et al. Diagnostic value of HMB-45 and anti-Melan A staining of sentinel lymph nodes with isolated positive cells. Mod Pathol. 2002;15:1288–1293. doi: 10.1097/01.MP.0000037313.33138.DF. [DOI] [PubMed] [Google Scholar]
- 43.McMasters KM, Wong SL, Edwards MJ, et al. Frequency of nonsentinel lymph node metastasis in melanoma. Ann Surg Oncol. 2002;9:137–141. doi: 10.1007/BF02557364. [DOI] [PubMed] [Google Scholar]
- 44.Morton DL, Hoon DSB, Cochran AJ, et al. Lymphatic mapping and sentinel lymphadenectomy for early-stage melanoma: therapeutic utility and implications of nodal microanatomy and molecular staging for improving the accuracy of detection of nodal micrometastases. Ann Surg. 2003;238:538–549. doi: 10.1097/01.sla.0000086543.45557.cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morton DL, Thompson JF, Cochran AJ, et al. Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med. 2006;355:1307–1317. doi: 10.1056/NEJMoa060992. [DOI] [PubMed] [Google Scholar]
- 46.Morton DL, Wen DR, Wong JH, et al. Technical details of intraoperative lymphatic mapping for early stage melanoma. Arch Surg. 1992;127:392–399. doi: 10.1001/archsurg.1992.01420040034005. [DOI] [PubMed] [Google Scholar]
- 47.Nieweg OE, Estourgie SH. What is a sentinel node and what is a false-negative sentinel node? Ann Surg Oncol. 2004;11:169s–173s. doi: 10.1007/BF02523623. [DOI] [PubMed] [Google Scholar]
- 48.Nuovo GJ. Co-labeling using in situ PCR: a review. J Histochem Cytochem. 2001;49:1329–1339. doi: 10.1177/002215540104901101. [DOI] [PubMed] [Google Scholar]
- 49.Pawelek JM. Tumour cell hybridization and metastasis revisited. Melanoma Res. 2000;10:507–514. doi: 10.1097/00008390-200012000-00001. [DOI] [PubMed] [Google Scholar]
- 50.Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 51.Pernick NL, DaSilva M, Gangi MD, et al. “Histiocytic markers” in melanoma. Mod Pathol. 1999;12:1072–1077. [PubMed] [Google Scholar]
- 52.Rachkovsky M, Sodi S, Chakraborty A, et al. Melanoma x macrophage hybrids with enhanced metastatic potential. Clin Exp Metastasis. 1998;16:299–312. doi: 10.1023/a:1006557228604. [DOI] [PubMed] [Google Scholar]
- 53.Rupani R, Handerson T, Pawelek J. Co-localization of beta1,6-branched oligosaccharides and coarse melanin in macrophage-melanoma fusion hybrids and human melanoma cells in vitro. Pigment Cell Res. 2004;17:281–288. doi: 10.1111/j.1600-0749.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 54.Saikali S, Avril T, Collet B, et al. Expression of nine tumour antigens in a series of human glioblastoma multiforme: interest of EGFRvIII, IL-13Ralpha2, gp100 and TRP-2 for immunotherapy. J Neurooncol. 2007;81:139–148. doi: 10.1007/s11060-006-9220-3. [DOI] [PubMed] [Google Scholar]
- 55.Sarantou T, Chi DD, Garrison DA, et al. Melanoma-associated antigens as messenger RNA detection markers for melanoma. Cancer Res. 1997;57:1371–1376. [PubMed] [Google Scholar]
- 56.Schaumburg-Lever G, Metzler G, Kaiserling E. Ultrastructural localization of HMB-45 binding sites. J Cutan Pathol. 1991;18:432–435. doi: 10.1111/j.1600-0560.1991.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 57.Scheri RP, Essner R, Turner RR, et al. Isolated tumor cells in the sentinel node affect long-term prognosis of patients with melanoma. Ann Surg Oncol. 2007;14:2861–2866. doi: 10.1245/s10434-007-9472-y. [DOI] [PubMed] [Google Scholar]
- 58.Scoggins CR, Martin RCG, Ross MI, et al. Factors associated with false-negative sentinel lymph node biopsy in melanoma patients. Ann Surg Oncol. 2010;17:709–717. doi: 10.1245/s10434-009-0858-x. [DOI] [PubMed] [Google Scholar]
- 59.Sodi SA, Chakraborty AK, Platt JT, et al. Melanoma x macrophage fusion hybrids acquire increased melanogenesis and metastatic potential: altered N-glycosylation as an underlying mechanism. Pigment Cell Res. 1998;11:299–309. doi: 10.1111/j.1600-0749.1998.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 60.Stansfeld AG, D'Ardenne AJ. Lymph node biopsy interpretation. Churchill Livingstone; Edinburgh: 1992. [Google Scholar]
- 61.Starz H, Haas CJ, Schulz GM, et al. Tyrosinase RT-PCR as a supplement to histology for detecting melanoma and nevus cells in paraffin sections of sentinel lymph nodes. Mod Pathol. 2003;16:920–929. doi: 10.1097/01.MP.0000086074.55963.24. [DOI] [PubMed] [Google Scholar]
- 62.Strauchen JA. Diagnostic histopathology of the lymph node. Oxford University Press; New York: 1998. [Google Scholar]
- 63.Szpak CA, Shelburne J, Linder J, et al. The presence of stage II melanosomes (premelanosomes) in neoplasms other than melanomas. Mod Pathol. 1988;1:35–43. [PubMed] [Google Scholar]
- 64.Taatjes DJ, Pang Y, von Turkovich M, et al. Ultrastructural localization of diagnostically relevant melanoma-specific antibodies by immunoelectron microscopy. J Histotechnol. 2003;26:263–266. [Google Scholar]
- 65.Takeuchi H, Morton DL, Kuo C, et al. Prognostic significance of molecular upstaging of paraffin-embedded sentinel lymph nodes in melanoma patients. J Clin Oncol. 2004;22:2671–2680. doi: 10.1200/JCO.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tatlidil C, Parkhill WS, Giacomantonio CA, et al. Detection of tyrosinase mRNA in the sentinel lymph nodes of melanoma patients is not a predictor of short-term disease recurrence. Mod Pathol. 2007;20:427–434. doi: 10.1038/modpathol.3800754. [DOI] [PubMed] [Google Scholar]
- 67.Zhang JG, Eguchi J, Kruse CA, et al. Antigenic profiling of glioma cells to generate allogeneic vaccines or dendritic cell-based therapeutics. Clin Cancer Res. 2007;13:566–575. doi: 10.1158/1078-0432.CCR-06-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]