Abstract
Biallelic inactivation of cancer susceptibility gene BRCA1 leads to breast and ovarian carcinogenesis. Paradoxically, BRCA1 deficiency in mice results in early embryonic lethality, and similarly, lack of BRCA1 in human cells is thought to result in cellular lethality in view of BRCA1's essential function. To survive homozygous BRCA1 inactivation during tumorigenesis, precancerous cells must accumulate additional genetic alterations, such as p53 mutations, but this requirement for an extra genetic “hit” contradicts the two-hit theory for the accelerated carcinogenesis associated with familial cancer syndromes. Here, we show that heterozygous BRCA1 inactivation results in genomic instability in nontumorigenic human breast epithelial cells in vitro and in vivo. Using somatic cell gene targeting, we demonstrated that a heterozygous BRCA1 185delAG mutation confers impaired homology-mediated DNA repair and hypersensitivity to genotoxic stress. Heterozygous mutant BRCA1 cell clones also showed a higher degree of gene copy number loss and loss of heterozygosity in SNP array analyses. In BRCA1 heterozygous clones and nontumorigenic breast epithelial tissues from BRCA mutation carriers, FISH revealed elevated genomic instability when compared with their respective controls. Thus, BRCA1 haploinsufficiency may accelerate hereditary breast carcinogenesis by facilitating additional genetic alterations.
Keywords: breast cancer 1, early onset; human cell gene targeting; somatic cell knock-in
Women carrying heterozygous mutations in the cancer susceptibility gene BRCA1 (1) are estimated to have a lifetime breast cancer risk of up to 85% (2). Wild-type BRCA1 alleles are lost in the majority of these tumors, further underscoring BRCA1's crucial tumor suppressor function (3). Paradoxically, biallelic disruption of BRCA1 confers early embryonic lethality in mouse models (4–6), and similarly, lack of functional BRCA1 in mature cells is thought to result in proliferation defects or cell death in view of BRCA1's essential function in maintaining genomic integrity (7–9).
Although it remains unclear how BRCA1 tumor cells survive biallelic BRCA1 inactivation during tumorigenesis, evidence suggests a role for accumulated genetic alterations, such as p53 mutations, in overcoming cell lethality (6, 10, 11). Because BRCA1 tumors presumably follow the two-hit theory that explains the accelerated carcinogenesis in familial cancer syndromes (12), this requirement for an extra genetic “hit” in BRCA1-mediated tumorigenesis appears contradictory. A proposed mechanism that could facilitate such genetic alterations is genetic instability resulting from the inactivation of a single BRCA1 allele (i.e., BRCA1 haploinsufficiency) (7, 13, 14). BRCA1 haploinsufficiency has also been suggested by several assays using breast epithelial cells from BRCA1 carriers (15–21). However, to date there has been no rigorous, defined experimental system to address BRCA1 haploinsufficiency in human breast epithelial cells. Genetically engineered mouse models have been extensively studied to elucidate BRCA1's function. However, in contrast to human BRCA1 carriers, most animals bearing heterozygous BRCA1 mutations do not demonstrate increased spontaneous breast or ovarian tumor formation (22–24), with the exception of a heterozygous mouse strain that is susceptible to other types of neoplasms (25). Thus, in the current study, we created a genetically defined experimental system derived from human breast epithelial cells and used it to address BRCA1 haploinsufficiency in humans.
Results
We initially carried out gene targeting in human cell lines and generated isogenic cell models harboring heterozygous mutant BRCA1 (Fig. 1). Two noncancerous breast epithelial cell lines were used for this study: the spontaneously immortalized MCF-10A cell line and a cell line immortalized by hTERT introduction (hTERT-IMEC) (26). These cell lines exhibit a basal-like phenotype (26, 27) (i.e., a gene expression signature similar to mammary basal epithelial cells from which BRCA1 breast tumors are thought to originate). In addition, relative to cancer cells, these cell lines are genetically stable with wild-type p53. One of the most common pathogenic BRCA1 mutations, 185delAG, which results in a 2-bp deletion at the coding region close to the N terminus, was heterozygously introduced in these cell lines via gene targeting (Fig. 1A).
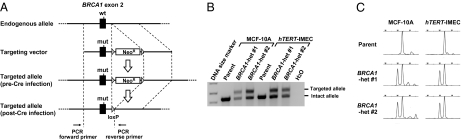
Fig. 1.
Knock-in of BRCA1 185delAG mutation in MCF-10A and hTERT-IMEC. (A) A scheme for the knock-in process. Primers shown at the bottom were used for the PCR amplification in B. (B) PCR encompassing the targeted genomic locus. Targeted and intact alleles yield PCR products of 517 bp and 429 bp in size, respectively. Cell clones post Cre-loxP recombination were hereafter used in this study. Primers used for PCR are denoted in A. (C) Capillary electrophoresis of RT-PCR products encompassing the 2-bp deletion site in exon 2. Two neighboring peaks in BRCA1 heterozygous clones represent PCR products of 186 bp and 188 bp in size, respectively.
We established two independently derived knock-in clones for both MCF-10A and hTERT-IMEC cell lines (hereafter termed BRCA1-het #1 and #2). Correct targeting was demonstrated by PCR amplification of the targeted genomic locus (Fig. 1B). Proper transcription from targeted alleles was confirmed by capillary electrophoresis and direct sequencing of RT-PCR products (Fig. 1C and Fig. S1). In addition, we also isolated cell clones that underwent random integration of the targeting vectors within their genomes (hereafter termed “control”). These clones were used as control clones in all subsequent experiments. We also attempted to create MCF-10A and hTERT-IMEC cell clones carrying homozygous BRCA1 185delAG mutations by targeting the remaining wild-type alleles in BRCA1-het clones. However, despite several repeated attempts, we only obtained cell clones undergoing random integration of the targeting vector to the genome. This finding is consistent with the proposed model of cellular lethality because of biallelic BRCA1 inactivation (7–9), although the possibility remains that unknown technical reasons may have accounted for the inability to obtain null clones.
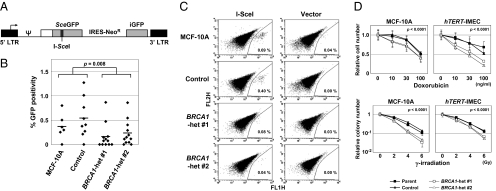
To address BRCA1 haploinsufficiency in humans, we employed several assays using our gene-targeted cell lines. Because it has been well established that BRCA1 plays a pivotal role in homologous recombination (HR) repair of DNA double-strand breaks (also termed “gene conversion”) (28), we initially evaluated HR efficiency in our MCF-10A-derived clones by stably transducing an EGFP reporter construct detecting HR (BABE-HR) (Fig. 2A). Through subsequent introduction of the restriction enzyme I-SceI, the SceGFP fragment present in BABE-HR is cleaved and then repaired by HR using an iGFP fragment as a template. This DNA recombination results in the reconstitution of a functional EGFP, which can be detected by fluorescence flow cytometry. Because of the technical difficulties of targeting the EGFP reporter construct to the same genomic position in all MCF-10A–derived isogenic clones, we isolated multiple single-cell subclones from each MCF-10A clone after BABE-HR transduction, transfected these subclones with the I-SceI–expressing plasmid, and then statistically compared their percentage of GFP positivities. This analysis revealed a significant decrease in HR repair in BRCA1 heterozygous clones compared with their wild-type counterparts (Fig. 2 B and C).
Fig. 2.
DNA homologous recombination repair is suppressed in BRCA1 heterozygous clones. (A) Retroviral vector-based GFP reporter construct BABE-HR measuring homologous recombination efficiency. SceGFP, inactive EGFP in which an 18-bp sequence is substituted with an I-SceI recognition site; iGFP, an internal portion of EGFP; arrows, transcription start sites; ψ, a packaging sequence. (B and C) HR repair assay. Multiple single-cell clones established after BABE-HR infection into MCF-10A-derived isogenic clones were transfected with an I-SceI–expressing plasmid, and GFP-positive ratios were determined by flow cytometry. Shown are a graphic representation of the entire results (B) and dot plots and percentage of GFP positivities in representative single-cell clones (C). Horizontal bars in B are averages. (D) Sensitivity of isogenic cell clones to Doxorubicin (Upper, n = 5) and γ-irradiation (Lower, n = 3). Data are shown relative to those of nontreated cells (mean ± SD).
We also evaluated homology-mediated repair capacities in both MCF-10A and hTERT-IMEC isogenic clones using another EGFP reporter construct, which detects both HR and single-strand annealing repair (pEGFPx2-C1) (Fig. S2A). Of note, BRCA1 is shown to be involved in both of these homology-mediated repair mechanisms (29). Assays were conducted based on transient transfection of I-SceI–cleaved pEGFPx2-C1, precluding the concern that the genomic positions of the integrated reporter constructs affect the results. Fluorescence flow cytometric analyses revealed a significant decrease in homology-mediated repair in both MCF-10A and hTERT-IMEC BRCA1 heterozygous cell lines compared with wild-type counterparts (Fig. S2B).
In accord with reduced capacity for homology-mediated repair, cell survival assays demonstrated that BRCA1 heterozygous clones have increased sensitivity to genotoxic stress conferred by either the chemotherapeutic drug Doxorubicin or γ-irradiation, both of which are known to cause DNA damage that is primarily repaired by HR (Fig. 2D). In contrast, we failed to detect a difference in the cytotoxicity of the poly(ADP-ribose) polymerase (PARP) inhibitor NU1025 between BRCA1 heterozygous and wild-type clones (Fig. S3). Similarly, the PARP inhibitor ABT-888 currently being evaluated in clinical trials also had no difference in sensitivity in our isogenic clones. Notably, PARP inhibitors are thought to exert cell killing selectively in S-phase, unlike γ-irradiation and Doxorubicin, which exert cytotoxic effects in all phases of the cell cycle, including resting cells (30, 31). Thus, it may be that any elevated sensitivity to PARP inhibitors in BRCA1 heterozygous clones is counterbalanced by G0/G1-dominant cell-cycle profiles and reduced proliferation capacities in these clones (see below). Overall, the data from DNA repair and cell survival assays demonstrated modest but significant impairment of DNA repair in BRCA1 heterozygous cells.
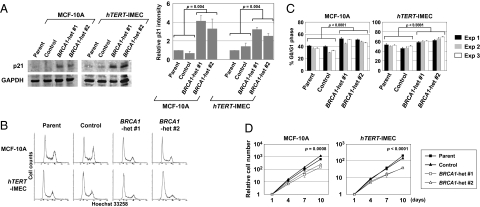
We next explored other cellular properties previously described in BRCA1-deficient mouse embryos, such as deregulated expression of p21 protein and impaired proliferation (4). When BRCA1 heterozygous human breast epithelial cells were propagated in exponential growth conditions and compared with their wild-type counterparts, p21 protein was up-regulated in immunoblot analyses similar to BRCA1-deficient mouse embryos (Fig. 3A), and cells were slightly but significantly more accumulated in the G0/G1 phase of the cell cycle (Fig. 3 B and C). Slower cell growth in BRCA1 heterozygous clones in comparison with their wild-type counterparts was also shown by proliferation assays (Fig. 3D). In contrast, although G2 checkpoint deficiency upon γ-irradiation is a notable property of BRCA1-deficient cells (32), there was no attenuation in either induction or maintenance of G2 checkpoint responses after γ-irradiation in BRCA1 heterozygous cell clones (Fig. S4). We also found no transformed phenotypes in BRCA1 heterozygous clones by an EGF-free cell culture, as well as an anchorage independent growth assay. Collectively, these results demonstrated that cells with a single mutant BRCA1 allele exhibit similar yet muted properties, as has been described for BRCA1-null cells.
Fig. 3.
BRCA1 heterozygous clones have similar yet muted properties with BRCA1-null cells. (A) Western blot analysis of p21 in exponentially growing cells. Shown are a representative blot (Left) and densitometric analyses of three independent experiments (Right). Data were normalized to GAPDH and shown relative to each parental cell line (mean ± SD). (B and C) Cell-cycle profiling by flow cytometry. Representative results (B) and G0/G1 populations (mean ± SD; n = 3) in three independent experiments (C). (D) Cell proliferation assay. Cell numbers are indicated relative to those at day 1 (mean ± SD; n = 3).
To study how impaired DNA repair function caused by BRCA1 haploinsufficiency impacts genomic integrity in human breast epithelial cells, we analyzed copy number (CN) changes, defined as the gain or loss of a given gene, in 389 cancer-related genes with the aid of a focused SNP array. BRCA1 heterozygous MCF-10A clones and their wild-type counterparts were exposed to 6 Gy or mock γ-irradiation. A total of 30 single-cell subclones were then isolated from each γ-irradiated cell clone, as denoted in Tables S1 and S2, maintained for 4 wk, and analyzed with SNP arrays. Signal-intensity data were obtained for 1,223 of 1,421 markers in the Cancer SNP Panel, and the CNs of 389 of 408 cancer-related genes in the panel were determined. Of the SNP markers, 399 were informative for genotyping and enabled loss of heterozygosity (LOH) assessment in 186 genes.
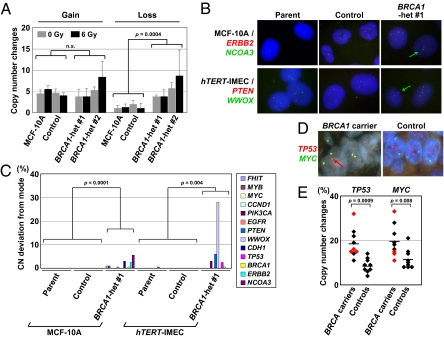
The vast majority of the 389 genes have CN scores within 1.5 to 2.5, and no amplification or high CN gain was found in this dataset. Nonetheless, BRCA1 heterozygous MCF-10A clones with or without 6 Gy γ-irradiation showed a significantly higher degree of CN loss compared with their wild-type counterparts (Fig. 4A and Table S1). Because this was significant in both nonirradiated (P = 0.001) and 6-Gy irradiated (P = 0.006) conditions upon individual analyses, the compromised genomic integrity is at least partially inherent to BRCA1 heterozygous cells rather than a function of increased sensitivity to γ-irradiation, presumably reflecting a critical role for BRCA1 against genomic DNA replication stress (33). Intriguingly, no difference in CN gain was detectable between BRCA1 heterozygous and wild-type clones. In line with observed preferential CN loss in BRCA1 heterozygous clones, the increase of LOH regions was also found in both 6-Gy and mock-irradiated BRCA1 heterozygous clones compared with their respective wild-type counterparts, although the difference was significant only in 6-Gy irradiated samples (Table S2).
Fig. 4.
Increased genomic instability in BRCA1 heterozygous cells in vitro and in vivo. (A) MCF-10A–derived isogenic clones were exposed to 6 Gy or mock γ-irradiation and subjected to single-cell cloning. Single-cell subclones were analyzed with SNP arrays after 4 wk of culture, and genes exhibiting CN changes (< 1.5 or > 2.5) were counted (mean ± SD). (B and C) FISH analyses of 13 genomic loci in isogenic cell clones. (B) Representative FISH images. Red and green spots represent ERBB2 and NCOA3 gene loci in MCF-10A clones and PTEN and WWOX loci in hTERT-IMEC clones, respectively. Green arrows indicate cells carrying single-copy genes labeled with green fluorescence. (C) Percentages of cells with allelic counts deviated from the modes. (D and E) FISH analyses of TP53 and MYC gene loci in noncancerous breast epithelial tissues from BRCA mutation carriers and controls undergoing reduction mammoplasty. (D) Representative FISH images for a BRCA1 mutation carrier and a control. A red arrow indicates a single TP53 gene copy. (E) Percentage of cells undergoing CN changes. Red dots denote BRCA2 mutation carriers. Horizontal bars represent averages.
To examine genomic alterations in a more stringent fashion—that is, at a single-cell resolution—we subsequently analyzed the isogenic cell clones with FISH. FISH visualizes individual alleles in each cell, and thereby enables the detection of subtle genomic CN changes in a small subset of cell populations that may be otherwise overlooked. Because of its high resolution, FISH can delineate genomic instability (measurable rate of genomic changes) in addition to aneuploidy or allelic imbalance (state of genomic changes) (34). For this analysis, we used probes to assess 13 genomic loci that are known to undergo frequent CN changes in sporadic or BRCA1 mutant breast cancer (Table S3). As expected, deviation of an allele count from the mode was found in only 1 of 10,388 (< 0.01%) cells in wild-type controls. In stark contrast, a total of 108 of 5,200 (2.08%) cells in BRCA1 heterozygous clones derived from MCF-10A and hTERT-IMEC exhibited allele counts distinct from the modal populations, affecting 11 of 13 genomic loci (Fig. 4 B and C and Table S4). In BRCA1 heterozygous clones, CN losses (88 cells) were dominant to CN gains (20 cells), concordant with our SNP array data (Fig. 4A and Table S1). In this FISH analysis, the BRCA1 gene locus at chromosome 17q21.31 was retained in a total of 1,200 cells, with the exception of one cell each in BRCA1 heterozygous MCF-10A and hTERT-IMEC clones (Table S4). Similarly, in our SNP array analysis, the BRCA1 gene locus was retained and exhibited no CN changes in all of the examined single-cell subclones (Table S2). The retention of the BRCA1 gene in BRCA1 heterozygous single-cell subclones was further confirmed by a PCR amplification of the targeted gene locus followed by analytical electrophoreses. These results strongly suggest that the observed genomic instability phenotype in BRCA1 heterozygous clones was indeed attributable to BRCA1 heterozygosity and not from a fraction of BRCA1-deficient cells emerging within BRCA1 heterozygous populations.
Finally, we carried out a FISH analysis against nontumorigenic breast tissue specimens to determine if genomic instability in BRCA1 heterozygous mutant cells is also detectable in vivo. Because of a limited number and amount of available BRCA1 specimens, this analysis used five and four noncancerous breast tissues from known BRCA1 and BRCA2 carriers, respectively, and two genomic loci among the most frequently altered in BRCA1 tumors—the TP53 and MYC gene loci (11, 35)—were then analyzed. For controls, we used nine breast specimens from women with no personal or family history of breast cancers who had undergone reduction mammoplasties. As seen in Fig. 4 D and E, the TP53 and MYC gene loci showed significantly more CN changes in breast epithelia from BRCA carriers compared with control samples. Virtually all of the CN changes detected in this analysis were losses of a single allele, consistent with our SNP array and FISH data for isogenic cell clones. It should be noted that control specimens also displayed a low level of CN changes and, specifically, CN loss. This result is because of the limitations of FISH using tissue samples. In this assay, some nuclei are cut into multiple sections, and thus a given section does not always contain all of the targets for a given FISH probe in a cell. The resultant false CN loss likely constitutes the background of the system. Chromosomal aberrations in breast tissues from BRCA1 carriers have been previously suggested by LOH and array-based comparative genomic hybridization analyses that used pools of cells as samples (17, 21). Our current FISH data provide unambiguous evidence for this by demonstrating a low degree, but significant elevation of genomic instability in nontumorigenic breast epithelia from BRCA carriers.
Discussion
Our study demonstrates that an inactivating mutation of a single BRCA1 allele leads to haploinsufficiency, which results in genomic instability in human breast epithelial cells. Genomic instability may then promote additional genetic changes in BRCA1 heterozygous cells. Because BRCA1-null cells seem to require additional genetic changes for survival (4–9), it is tempting to speculate that genetic changes accumulated in BRCA1 heterozygous cells enable cancer progenitor cells to evade cell death that would otherwise occur upon loss of the wild-type BRCA1 allele. Given this extra genetic requirement, BRCA1-mediated breast carcinogenesis may be considered in a distinct manner from other familial cancer syndromes that follow the original two-hit theory (12), in which consecutive deletion of two alleles suffices to accelerate tumorigenesis (Fig. S5). Although BRCA1 haploinsufficiency plays a pivotal role in the current proposed model, it should be noted that heterozygous BRCA1 inactivation itself is likely to be insufficient for complete breast carcinogenesis, because the vast majority of BRCA1 tumors undergo loss of the second wild-type BRCA1 allele (3). We suggest that genomic alterations resulting from BRCA1 haploinsufficiency is an early but not sufficient step of BRCA1-mediated breast carcinogenesis.
It has previously been proposed that the second wild-type BRCA1 allele is lost before other somatic alterations, and the resultant BRCA1-null cells, specifically in breast and ovarian tissues, have prolonged survival, allowing them to acquire additional mutations that support cell proliferation (8, 9). Although this model is not formally excluded, our data and the work of others are better explained by the model proposed in our study. As an example, our FISH analysis in BRCA1 heterozygous cell clones (Table S4) and a previous genome-wide array comparative genomic hybridization analysis using breast biopsies from BRCA1 carriers (21), suggest that the loss of the second wild-type BRCA1 allele is an infrequent genomic alteration in BRCA1 heterozygous cells, and thus likely preceded by other genomic CN changes and genetic alterations. However, because our study analyzed only a limited number of clinical specimens, further studies involving larger series of normal breast tissues, as well as preneoplastic lesions from BRCA1 carriers, will be needed to better elucidate the mechanisms of BRCA1-mediated carcinogenesis.
As proposed previously (13), if BRCA1 haploinsufficiency is exclusive to specific tissue types, such as breast and ovarian epithelia, it may provide an explanation for the restricted tissue distribution of BRCA1 tumorigenesis. In this regard, a number of studies have addressed BRCA1 haploinsufficiency in lymphoid cells, Epstein-Barr virus-immortalized lymphoid cell lines, and fibroblasts derived from BRCA1 carriers. Many of these studies described phenotypes specific to the cells carrying heterozygous BRCA1 mutations (36–40). However, other studies failed to detect differences between cells from BRCA1 carriers and controls (41–43), and thus BRCA1 haploinsufficiency in the above-mentioned cell types has been controversial (13, 44, 45). This controversy may be reflective of the fact that neither lymphocytes nor connective tissues are susceptible to cancer development in BRCA1 carriers.
In this study we showed that BRCA1 heterozygous cells exhibited higher sensitivity to the genotoxic agent Doxorubicin compared with wild-type counterparts. However, in clinical use, anticancer drugs, including Doxorubicin, seem not to demonstrate higher tissue toxicity in BRCA1 carriers in comparison with patients with sporadic breast cancers. One possible explanation for this can be the potential tissue specificity of BRCA1 haploinsufficiency. If BRCA1 is not haploinsufficient in normal tissues susceptible to chemotherapies, such as hematopoietic cells and intestinal epithelium, BRCA1 carriers would not suffer from excessive adverse effects relative to control populations. Another possibility may be that the observed hypersensitivity to Doxorubicin in BRCA1 heterozygous cells may not be severe enough to lead to a recognizable increase in tissue toxicity in BRCA1 carriers.
Finally, genomic instability because of a heterozygous BRCA1 mutation may have clinical implications. It has been reported that p53 alterations partially rescue cellular lethality caused by BRCA1 deficiency (6, 10). The identification of similar genetic alterations, particularly activating genetic changes, could be potentially exploited to develop prophylactic therapies for BRCA1-related cancers. Thus, further studies are warranted to better elucidate the properties and consequences of BRCA1 haploinsufficiency.
Materials and Methods
Knock-In of 185delAG BRCA1 Mutation.
To construct the targeting vector, homology arms were amplified by PCR and ligated to an adeno-associated viral plasmid. The resulting targeting vector was transduced into MCF-10A and hTERT-IMEC, as previously described (46, 47). BRCA1 gene-targeted clones were isolated by PCR-based screening and processed for Cre-loxP recombination to remove neomycin-resistance gene cassettes, as previously described (46–48). Primer sequences for PCR are provided in Table S5.
Retroviral Vector-Based HR Repair Assay.
Retroviral plasmid pBABE-HR was constructed as described in SI Materials and Methods. To evaluate HR repair efficiency, isogenic cell clones were transduced with retroviral reporter BABE-HR and subjected to single-cell subcloning when selected with G418 (Invitrogen). Resulting multiple single-cell clones were transfected with an I-SceI expression plasmid pCBASce (49) or an empty vector pCAG in the absence of G418, and analyzed by flow cytometry after 3-d incubation. HR efficiency was determined by GFP-positive ratio after pCBASce transfection and normalized to transfection efficiency.
SNP Array.
Multiple single-cell subclones were isolated from MCF-10A isogenic clones immediately after 6-Gy or mock irradiation, as shown in Tables S1 and S2. After 4 wk of propagation, single-cell subclones were analyzed with a Cancer SNP Panel (Illumina) according to the manufacturer's instructions. Fluorescence signals from hybridized slides were scanned with Illumina BeadArray Reader and quantified with Beadstudio software. CN scores and LOH values were determined by Hidden Markov Model using the dChip program. The CN scores of individual SNP markers within a gene were averaged and used as the CN score of the corresponding gene. The gains and losses of gene copy were defined as averaged CN scores > 2.5 and < 1.5, respectively. LOH genes were identified when LOH was detected at one or more of the markers within the genes.
FISH.
Propagated isogenic cell clones were incubated in 40 mM KCl, fixed with 3:1 mixture of methanol and glacial acetic acid, and then dropped onto glass slides. Tissue microarray (TMA) was constructed using 18 formalin-fixed paraffin-embedded specimens of noncancerous breast tissues: 9 derived from BRCA1 or BRCA2 mutation carriers and 9 from controls. The absence of cancerous cells in the specimens were histopathologically confirmed. Slides were hybridized with fluorescently labeled BAC probes (cell line FISH) or commercially available probes (TMA FISH) listed in Table S3, counterstained with DAPI, and subjected to allele counting under fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank M. Jasin for providing plasmids DR-GFP and pCBASce, F. Bunz for pSEPT; L. A. Annab for the cell line 90PE6E7, M. Brown for hTERT-IMEC, and M. Seto for BAC probes. This work was supported by The V Foundation, National Institutes of Health Grant CA109274, Susan G. Komen for the Cure, the Avon Foundation, the Mary Kay Ash Charitable Foundation, the Stewart Trust Fund, the Flight Attendant Medical Research Institute (FAMRI), and the Breast Cancer Research Foundation (to B.H.P.); and FAMRI, Grants-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, Strategic Research Foundation Grant-aided Project for Private Universities from MEXT, Japan, Aichi Cancer Research Foundation, and Osaka Cancer Research Foundation (to H.K.).
Footnotes
Conflict of interest statement: B.H.P. is a paid consultant for GlaxoSmithKline and is a paid member for the scientific advisory board of Horizon Discovery, Ltd.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110969108/-/DCSupplemental.
References
- 1.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Easton DF, Ford D, Bishop DT. Breast Cancer Linkage Consortium Breast and ovarian cancer incidence in BRCA1-mutation carriers. Am J Hum Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- 3.Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 4.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 5.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 6.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 7.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 9.Elledge SJ, Amon A. The BRCA1 suppressor hypothesis: An explanation for the tissue-specific tumor development in BRCA1 patients. Cancer Cell. 2002;1:129–132. doi: 10.1016/s1535-6108(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 10.Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 11.Crook T, et al. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene. 1998;17:1681–1689. doi: 10.1038/sj.onc.1202106. [DOI] [PubMed] [Google Scholar]
- 12.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: When you don't need to go all the way. Biochim Biophys Acta - Reviews on Cancer. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: Damage threshold and the concept of ‘conditional haploinsufficiency’. Cell Cycle. 2007;6:2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- 15.King TA, et al. Increased progesterone receptor expression in benign epithelium of BRCA1-related breast cancers. Cancer Res. 2004;64:5051–5053. doi: 10.1158/0008-5472.CAN-04-1283. [DOI] [PubMed] [Google Scholar]
- 16.Mote PA, et al. kConFab Investigators Germ-line mutations in BRCA1 or BRCA2 in the normal breast are associated with altered expression of estrogen-responsive proteins and the predominance of progesterone receptor A. Genes Chromosomes Cancer. 2004;39:236–248. doi: 10.1002/gcc.10321. [DOI] [PubMed] [Google Scholar]
- 17.Larson PS, et al. Allele imbalance, or loss of heterozygosity, in normal breast epithelium of sporadic breast cancer cases and BRCA1 gene mutation carriers is increased compared with reduction mammoplasty tissues. J Clin Oncol. 2005;23:8613–8619. doi: 10.1200/JCO.2005.02.1451. [DOI] [PubMed] [Google Scholar]
- 18.King TA, et al. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510–2518. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- 19.Burga LN, et al. Altered proliferation and differentiation properties of primary mammary epithelial cells from BRCA1 mutation carriers. Cancer Res. 2009;69:1273–1278. doi: 10.1158/0008-5472.CAN-08-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellacosa A, et al. Altered gene expression in morphologically normal epithelial cells from heterozygous carriers of BRCA1 or BRCA2 mutations. Cancer Prev Res (Phila) 2010;3:48–61. doi: 10.1158/1940-6207.CAPR-09-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rennstam K, et al. Genomic alterations in histopathologically normal breast tissue from BRCA1 mutation carriers may be caused by BRCA1 haploinsufficiency. Genes Chromosomes Cancer. 2010;49:78–90. doi: 10.1002/gcc.20723. [DOI] [PubMed] [Google Scholar]
- 22.Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- 23.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 24.Bouwman P, Jonkers J. Mouse models for BRCA1 associated tumorigenesis: From fundamental insights to preclinical utility. Cell Cycle. 2008;7:2647–2653. doi: 10.4161/cc.7.17.6266. [DOI] [PubMed] [Google Scholar]
- 25.Jeng YM, et al. Brca1 heterozygous mice have shortened life span and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene. 2007;26:6160–6166. doi: 10.1038/sj.onc.1210451. [DOI] [PubMed] [Google Scholar]
- 26.DiRenzo J, et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer Res. 2002;62:89–98. [PubMed] [Google Scholar]
- 27.Gordon LA, et al. Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer. 2003;106:8–16. doi: 10.1002/ijc.11172. [DOI] [PubMed] [Google Scholar]
- 28.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 29.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashworth A. A synthetic lethal therapeutic approach: Poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 31.Hill BT. Cancer chemotherapy. The relevance of certain concepts of cell cycle kinetics. Biochim Biophys Acta - Reviews on Cancer. 1978;516:389–417. doi: 10.1016/0304-419x(78)90018-5. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 33.Scully R, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 34.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 35.Grushko TA, et al. MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res. 2004;10:499–507. doi: 10.1158/1078-0432.ccr-0976-03. [DOI] [PubMed] [Google Scholar]
- 36.Foray N, et al. Gamma-rays-induced death of human cells carrying mutations of BRCA1 or BRCA2. Oncogene. 1999;18:7334–7342. doi: 10.1038/sj.onc.1203165. [DOI] [PubMed] [Google Scholar]
- 37.Rothfuss A, et al. Induced micronucleus frequencies in peripheral lymphocytes as a screening test for carriers of a BRCA1 mutation in breast cancer families. Cancer Res. 2000;60:390–394. [PubMed] [Google Scholar]
- 38.Buchholz TA, et al. Evidence of haplotype insufficiency in human cells containing a germline mutation in BRCA1 or BRCA2. Int J Cancer. 2002;97:557–561. doi: 10.1002/ijc.10109. [DOI] [PubMed] [Google Scholar]
- 39.Coupier I, et al. Fidelity of DNA double-strand break repair in heterozygous cell lines harbouring BRCA1 missense mutations. Oncogene. 2004;23:914–919. doi: 10.1038/sj.onc.1207191. [DOI] [PubMed] [Google Scholar]
- 40.Kote-Jarai Z, et al. Increased level of chromosomal damage after irradiation of lymphocytes from BRCA1 mutation carriers. Br J Cancer. 2006;94:308–310. doi: 10.1038/sj.bjc.6602912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baria K, et al. Correspondence re: A. Rothfuss et al., Induced micronucleus frequencies in peripheral blood lymphocytes as a screening test for carriers of a BRCA1 mutation in breast cancer families. Cancer Res., 60: 390–394, 2000. Cancer Res. 2001;61:5948–5949. [PubMed] [Google Scholar]
- 42.Nieuwenhuis B, et al. BRCA1 and BRCA2 heterozygosity and repair of X-ray-induced DNA damage. Int J Radiat Biol. 2002;78:285–295. doi: 10.1080/09553000110097974. [DOI] [PubMed] [Google Scholar]
- 43.Lovelock PK, et al. kConFab Investigators Prediction of BRCA1 and BRCA2 mutation status using post-irradiation assays of lymphoblastoid cell lines is compromised by inter-cell-line phenotypic variability. Breast Cancer Res Treat. 2007;104:257–266. doi: 10.1007/s10549-006-9415-5. [DOI] [PubMed] [Google Scholar]
- 44.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 45.Baeyens A, et al. Chromosomal radiosensitivity in BRCA1 and BRCA2 mutation carriers. Int J Radiat Biol. 2004;80:745–756. doi: 10.1080/09553000400017937. [DOI] [PubMed] [Google Scholar]
- 46.Konishi H, et al. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 2007;67:8460–8467. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- 47.Abukhdeir AM, et al. Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc Natl Acad Sci USA. 2008;105:288–293. doi: 10.1073/pnas.0710887105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Konishi H, et al. A PCR-based high-throughput screen with multiround sample pooling: application to somatic cell gene targeting. Nat Protoc. 2007;2:2865–2874. doi: 10.1038/nprot.2007.409. [DOI] [PubMed] [Google Scholar]
- 49.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.