Abstract
The global distribution of soil animals and the relationship of below-ground biodiversity to above-ground biodiversity are not well understood. We examined 17,516 environmental 18S rRNA gene sequences representing 20 phyla of soil animals sampled from 11 locations covering a range of biomes and latitudes around the world. No globally cosmopolitan taxa were found and only 14 of 2,259 operational taxonomic units (OTUs) found were common to four or more locations. Half of those were circumpolar and may reflect higher connectivity among circumpolar locations compared with other locations in the study. Even when OTU assembly criteria were relaxed to approximate the family taxonomic level, only 34 OTUs were common to four or more locations. A comparison of our diversity and community structure data to environmental factors suggests that below-ground animal diversity may be inversely related to above-ground biodiversity. Our data suggest that greater soil inorganic N and lower pH could explain the low below-ground biodiversity found at locations of high above-ground biodiversity. Our locations could also be characterized as being dominated by microarthropods or dominated by nematodes. Locations dominated by arthropods were primarily forests with lower soil pH, root biomass, mean annual temperature, low soil inorganic N and higher C:N, litter and moisture compared with nematode-dominated locations, which were mostly grasslands. Overall, our data indicate that small soil animals have distinct biogeographical distributions and provide unique evidence of the link between above-ground and below-ground biodiversity at a global scale.
Keywords: cosmopolitan species, endemism
The influence that geography, biome type, above-ground vegetation, and soil quality have on the distribution and biodiversity of below-ground small soil organisms is poorly understood at the global scale. Some have argued that distribution patterns of eukaryote species may be dependent on size (1), with smaller eukaryote species more easily dispersed and thus cosmopolitan species common. However, it has been demonstrated by others (2, 3) that in general, unicellular eukaryote organisms such as protists and diatoms disperse over shorter geographical distances than larger multicellular organisms such as annelids, nematodes, and bivalves in both marine and terrestrial environments. A global molecular study of freshwater cladocerans found that continental and regional endemism is common and cosmopolitan species are rare (4). It was found that taxa described in the morphological literature as cosmopolitan are not homogeneous at the molecular level, suggesting that taxa previously identified as cosmopolitan can be an artifact resulting from inaccurate taxonomy, the presence of morphological stasis, or phenotype plasticity. Among small soil animals, molecular studies of nematodes in regions of high above-ground biodiversity (5) and low above-ground biodiversity (6) found high degrees of endemism at the regional level with a general lack of cosmopolitan taxa. All of these studies support the hypothesis that cosmopolitan species are rare among small soil animals, but these molecular studies were conducted using narrowly defined taxa.
Dispersal ability is a major factor determining geographical distributions of organisms. Whereas dispersal mechanisms for aquatic and above-ground organisms have been described, it is unclear what mechanisms drive dispersal of small exclusively below-ground organisms. Low dispersal ability would explain high endemism in small soil animals, but at least some dispersal mechanisms exist. For example, it has been suggested that soil nematodes that parasitize plant roots could disperse within plant rhizome fragments (7) following soil erosion.
The diversity of plants and vertebrate animals generally increases from the poles to the equator (8). A field study of oribatid mites (9) in soil found that species richness increased from high latitudes to more temperate regions, but did not increase further in the tropics, suggesting a negative correlation. A metaanalysis of several studies of soil taxa showed soil fauna exhibit different compositions at low taxonomic resolution on the basis of biomass of mites, collembolans, enchytraeids, nematodes, and earthworms under seven biomes, including desert, tundra, temperate grassland, temperate deciduous and coniferous forest, and tropical forest (10). However, no studies have examined the distribution of small soil animals at the species level at a global geographic scale. Such a study would allow the determination of the relative contribution of cosmopolitan and endemic species to community structure at high taxonomic resolution. It would also detect any relationship between above-ground and below-ground biodiversity at a global scale. Therefore, we used a molecular approach to analyze samples from 11 locations worldwide at or near the species level to determine whether biogeography influences the distribution of small soil animals at a global scale and whether global soil animal distribution is influenced by environmental factors, biome type, above-ground diversity, and latitude.
Results
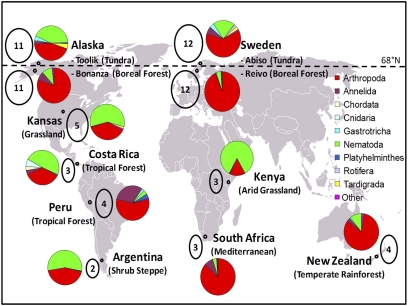
A total of 17,516 rRNA gene sequences (GenBank accession nos. JN135477–JN152992) were obtained from 42 of the 44 plots across the 11 geographic locations (Table 1). At least 297 sequences were obtained from 40 of the 44 plots. One plot from the Argentina shrub steppe (AR) location yielded only 236 sequences, and no sequences were obtained from two other Argentinean plots. One of the North American Kansas grassland (KZ) plots yielded only 44 sequences. An average of 1,595 sequences, with a range of 533–2,621 was collected from each of the 11 geographic locations. The 17,516 sequences were assembled into 2,259 operational taxonomic units (OTUs) of 99% similarity (Table S1) using Sequencher software (Gene Codes). The overall community structure at each location at the phylum level is shown in Fig. 1. Nematodes and arthropods made up 88% of the sequences, whereas the other 12% represented 18 other phyla. Nematodes dominated the soil communities at four locations and arthropods (mostly mites) dominated at six locations, whereas nematodes and arthropods codominated at one location. The percentage of nematodes from each of 42 plots was positively correlated with soil pH (r2 = 0.40, P < 0.0001) and negatively correlated with soil C:N ratio (r2 = 0.26, P = 0.0005). The percentage of arthropods was positively correlated with soil C:N ratio (r2 = 0.41, P < 0.0001) and negatively correlated with soil pH (r2 = 0.30, P = 0.0002) (Fig. S1). T-test comparisons of environmental parameters comparing the arthropod-dominated locations to the nematode-dominated locations indicated that arthropod-dominated locations were of higher latitude and had greater litter standing biomass, root biomass, soil moisture, soil C, and soil C:N ratio (Table S2). Arthropod-dominated locations had lower mean annual temperature, soil temperature, soil bulk density, soil pH, soil NO3, and soil inorganic N than did the nematode-dominated locations (Table S2).
Table 1.
Brief description of 11 sampling locations
| Latitude and longitude | Site | Symbols | Soil types | Biome classification | Dominant plants |
| 68°N, 19°E | Abisko, Sweden (NERC site) | AB | Spodosol | Tundra | Empetrum hermaphroditum, Vaccinium, lichen |
| 68°N, 150°W | Toolik Lake LTER, Alaska | TK | Gelisol | Tundra | Eriophorum vaginatum |
| 66°N, 19°E | Reivo, Sweden (NERC site) | RE | Spodosol | Boreal forest | Pinus sylvestris, Vaccinium myrtillus, Empetrum hermaphroditum, Pleurozium schreberi |
| 64°N, 148°W | Bonanza, LTER, Alaska | BZ | Inceptisol | Boreal forest | Picea mariana |
| 39°N, 97°W | Konza Prairie LTER, Kansas | KZ | Molisol | Grassland | Ambrosia, Symphotrichum |
| 10°N, 84°W | La Selva, Costa Rica | CR | Inceptisol and ultisol | Tropical forest | Pentaclethra macroloba |
| 1°S, 37°E | Kenya, Kapiti Ranch | KY | Vertisol | Arid grassland | Themeda triandra, Digitaria macroblephara, Penisetum mezianum |
| 13°S, 70°W | Peru, Los Amigos Biological Station | PU | Ultisol and inceptisol | Tropical forest | Bertholletia excelsa and other Lecythidaceae |
| 34°S, 18°E | Cape Peninsula, South Africa | SA | Spodosol | Mediterranean | Erica (heather), Elegia caspidata, Elegia filacea |
| 42°S, 171°E | New Zealand, forests | NZ | Inceptisol | Warm temperate rainforest | “Mixed podicarp-hardwood,” Dacrydium cupressinum, Quintinia acutifolia, Weinmannia racemosa |
| 45°S, 70°W | Argentina, INTA Río Mayo Station | AR | Aridisol | Shrub steppe | Mulinum spinosum, Adesmia campestris, Senecio filaginoides, Stipa speciosa, Stipa humilis, Poa ligularis |
LTER, Long Term Ecological Research; INTA, Instituto Nacional de Tecnología Agropecuaria; NERC, National Environment Research Council.
Fig. 1.
Distribution and composition of OTUs at the 11 locations of this study. The number of widely distributed OTUs (those found at four or more locations) present at each location is shown in each oval. The pie charts indicate the phylum level composition of soil animals at each location. Note that only 14 of 2,259 OTUs were found at four or more locations. The horizontal dashed line indicates the 68°N latitude.
A cluster analysis of the community structure of the 11 locations by abundance of OTUs is shown in Fig. S2A and by presence/absence of OTUs in Fig. S2B. The cluster analysis by abundance showed that the animal communities of the two boreal forest locations were more similar to each other than to the nearby tundra locations. The animal communities of the two tundra locations were more similar to each other than to the boreal forest locations. The cluster analysis by presence/absence indicated that the animal communities of the Alaska tundra and boreal forest locations were more similar to each other than to the Sweden tundra and boreal forest locations. In this analysis the animals of the Sweden tundra and boreal forest locations were more similar to each other than to the animals of the Alaska tundra or boreal forest locations. The other locations were similar in both types of cluster analysis.
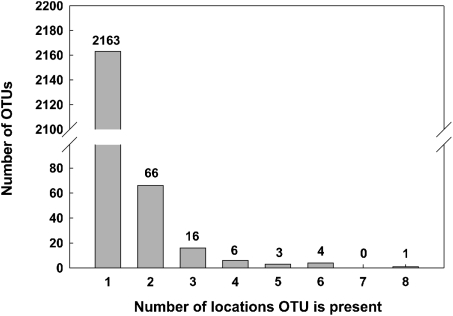
Most of the OTUs (78.5%) were represented by only a single sequence from a single location (Fig. 2 and Table S1). The geographic distribution of sequences within OTUs represented by more than one sequence was examined to determine which OTUs were present at multiple locations. We found that the vast majority of OTUs were present at just a single location (2,163/2,259 or 95.8%). Only one OTU of 2,259 was present at eight locations, none at seven locations, five were common to six or more locations, 14 were common to four or more locations, and 96 OTUs were common to two or more locations (Fig. 2, Table S1). The 96 OTUs common to two or more locations represented only 4.2% of the OTUs, but represented 52.4% of the 17,516 sequences. We found that four circumpolar locations (64°–68° latitude: Sweden tundra (AB), Sweden boreal forest (RE), Alaska boreal forest (BZ), and Alaska tundra (TK) together accounted for seven of the 14 OTUs found at more than four locations (Fig. 1). We used ANOVA to compare the average number of widespread OTUs (present at four or more locations) within the 42 plots representing all 11 locations (Table S3). This showed that three of four circumpolar locations (BZ, RE, and TK) had significantly more (P < 0.05) widespread OTUs than did the other locations with the exception of circumpolar location (AB), which had significantly more widespread OTUs than several locations (PU, SA, CR, and AR). An analysis of 40 plots representing 11 locations shows an increase in OTUs common to four or more locations at high latitudes compared with lower latitudes (Fig. S3). There was little or no correlation between the number of OTUs common to four or more locations and Shannon diversity (r2 = 0.07), dominance (r2 = 0.01), richness (r2 = 0.10), or Hurlbert's probability of an interspecific encounter (PIE) (r2 = 0.03) or between latitude and Shannon diversity (r2 = 0.12).
Fig. 2.
Distribution of OTUs found at multiple locations.
The 14 OTUs found at four or more locations by the Sequencher analyses were identified by comparison with known sequences in GenBank (Fig. S4). Most of the sequences were mites, three in the taxon Oribatida, one in Endeostigmata, and one in Astigmata. Another mite sequence was not closely related to other GenBank mite sequences. Nematodes were represented by five sequences: Two were in the taxon Plectida and one each in Rhabditida, Enoplida, and Dorylaimida. One sequence appeared to be a member of Collembola (Arthropoda), one sequence represented Tardigrada, and another was a member of Annelida. In several cases, the sequences in this study had exact matches in GenBank. These included one OTU that matched the oribatid mite Northrus truncates and another OTU that matched the nematode Plectus tenuis. One OTU found at four locations had no close matches, but phylogenetic analysis (Fig. S4) suggests it may belong to a family of mites not currently represented in GenBank.
The analysis of the 17,516 sequences revealed a high degree of variability in OTU numbers and Shannon diversity indices. We found very low correlations between diversity indices and environmental parameters (r2 < 0.20), and no correlation between diversity indices and latitude (r2 = 0.01) was observed. The analysis of community structure indicated significant differences among the 11 locations [analysis of similarity (ANOSIM), P = 0.001] with a three-dimensional stress value of 0.10 based on nonmetric multidimensional scaling (NMDS) analysis of the animal OTU compositions of the 42 plots (Fig. S5). A biotic-environmental (BIO-ENV) analysis revealed a moderate correlation between community structure and latitude (r2 = 0.43).
Four of our locations correspond to regions identified as areas of especially high above-ground biodiversity (11). We compared the Shannon diversity indices of the small soil animal OTUs in the plots from those four locations (CR, PU, SA, and NZ) with those of the plots from the other seven locations. A t-test revealed a significantly (P < 0.01) lower mean of Shannon diversity (2.04) and richness (39.5) for the four locations representing higher above-ground biodiversity than the mean of Shannon diversity (2.66) and richness (60.9) for the other seven locations. Hurlbert's PIE and dominance were not significantly different among the two groups.
Discussion
Identification of Widely Distributed OTUs.
The sequencing method used here allowed the presence of organisms to be compared between locations at a high resolution and minimized problems associated with identifying cryptic morphospecies among geographically distant locations. This “relative” identification of sequences among locations provides a powerful tool to study geographic species distribution, as done in this study. Because the 18S rRNA gene has been sequenced from an ever-increasing number of organisms, we could provisionally identify sequences to the genus and possibly to the species level. The 99% match criteria used to group sequences into OTUs with 18S rRNA sequences generally underestimates the number of species present (6). We reexamined an earlier dataset of 890 known nematode 18S rRNA sequences from GenBank reported by Wu et al. (6) and found that 79% of the OTUs (99% match criterion) contained sequences that matched a single species in GenBank, whereas 21% of the OTUs matched more than a single species and 9% of the OTUs matched more than one genus. Similar results were found with 229 known mite sequences. This means that although the 99% match criterion groups sequences from the same species 79% of the time, the other 21% of the time it groups sequences from more than one species. Therefore, our identifications, even with exact matches to known GenBank sequences, will sometimes not provide an absolute identification, especially if the sequence belongs to a closely related group of species or genera. We chose the 18S rRNA gene because of the large number of sequences in GenBank, the reliability in its amplification, and its usefulness in taxonomy. More variable genes such as the mitochondrial genes used in barcoding would likely identify even fewer potentially cosmopolitan OTUs in our samples. Our use of a relatively large portion (519 bp) of the 18S rRNA gene allowed a broader phylogenetic analysis than short highly variable barcode sequences.
In the current study, we found sequences in our samples that were considerably different from known sequences in GenBank. For example, the sequence labeled ABC006 in the phylogenetic analysis of Fig. S4 did not group closely with a known mite but is associated with a clade that includes oribatid and endeostigmatid mites and likely represents a family of mites not yet represented in GenBank. Most soil studies focus on the morphological identification of nematodes and mites and may not identify lesser known groups. Our molecular approach clearly identified an OTU that likely represents a tardigrade of the genus Diphascon as a potentially widely distributed organism (Fig. S4). All of the OTUs we identified as common to four or more locations were provisionally identified as taxa known to be bacteriovores, herbivores, fungivores, or predators, and none appeared likely to be animal parasites, which limits the possibility that the more widely distributed OTUs were dispersed by animal hosts. Some mites, nematodes, and tardigrades are known to be dispersed by wind and may have a dormant stage that aids dispersal. However, there does not appear to be a common factor affecting dispersal among the widely distributed OTUs identified in this study.
Geographic Distribution of OTUs.
We did not find any OTUs common to all 11 locations and only 14 of 2,259 OTUs were present in four or more locations in this study. As a result, we conclude that cosmopolitan taxa are extremely rare at the global level among small soil animals. Most OTUs in our study have a narrow geographical range. We found that 95.8% of the 2,259 OTUs were present at only a single location, suggesting that endemism is prevalent in soil animal communities. We used a 99% similarity criterion for clustering sequences into OTUs that served as a proxy for the species level (6), but even when the similarity criterion was relaxed to 95% (a proxy for the family level) the number of OTUs common to four or more locations only increased from 14 to 34 (Table S1). This is an unexpected outcome that suggests that cosmopolitan families are nearly as rare as cosmopolitan species.
It could be argued that with more intense sampling, cosmopolitan taxa could be more common. If we had collected more sequences per sample and more soil from additional plots from a greater number of locations across the world, the number of OTUs present would also be expected to rise. This is a fundamental problem of sampling and community structure analysis, but the data presented here are by far the largest and most comprehensive dataset for small soil animals at the global level. Moreover, large-scale pyrosequencing of bacterial 16S rRNA genes from soil locations in North and South America (12) suggest that adding more sequences will not necessarily identify more cosmopolitan OTUs. Rather, it is likely that adding more sequences would add mostly rare OTUs and singletons that would not contribute to the number of cosmopolitan taxa.
Another way of analyzing the sequences is to use phylogenetic distance, the inverse of similarity, where, for example, 99% sequence similarity would be roughly comparable to a phylogenetic distance of 0.01. Therefore, ESPRIT software (13) was used to assemble OTUs on the basis of phylogenetic distance criterion. At a phylogenetic distance of 0.01 we found only five OTUs present at four or more locations using 0.01 phylogenetic distance and 16 OTUs using a phylogenetic distance of 0.03, considerably lower than with the comparable Sequencher analysis. The ESPRIT method appears to be more conservative in identifying OTUs common to several locations than Sequencher for very similar sequences, but the two methods provide similar results at a distance of 0.05 (ESPRIT) and a similarity of 95% (Sequencher), where both methods found 34 OTUs present at four or more locations (Table S1), again suggesting that widely distributed taxa are rare, even at the family level.
It has been suggested that widely distributed taxa are simply the most common taxa present at multiple locations (14). In our dataset, OTUs common to four or more sites were represented by as few as 43 and as many as 1,005 sequences, and we found no correlation between the number of sequences found within a widely distributed OTU and the number of locations at which members of each OTU were found (r2 = 0.14). As a consequence, we conclude that widely distributed OTUs do not simply represent the most common OTUs in the dataset. There was no measurable correlation between the number of OTUs present at four or more locations and Shannon diversity, Hurlbert's PIE, dominance, or richness among plots. This is likely due to the extreme rarity of widely distributed OTUs where they may not contribute significantly to the overall community structure at the locations. If cosmopolitan species are rare, and endemism is prevalent, it should be difficult to correlate instances of cosmopolitan species to measures of diversity. One would expect to find a correlation between cosmopolitanism and diversity indices only if endemism were low and cosmopolitanism were high.
In our study, the number of sampling sites was limited to 11 locations for practical reasons. Many of the locations had no clear connectivity with each other, and cosmopolitanism would not be expected to be significant. However, the map shown in Fig. 1 reveals that most of the OTUs common to four or more sites were found at high latitude locations where some connectivity is expected (tundra and boreal forest in Alaska and Sweden). These four circumpolar locations (AB, TK, RE, and BZ in Table S3) may lack significant barriers to connectivity, and the higher number of OTUs common to four or more sites might be due to broad similarities in soils, climate, latitude, or landform. However, AB can be considered alpine rather than arctic, differentiating it from TK, RE, and BZ in Table S3. The presence of paired datasets between Alaska and Sweden, each with a site from tundra and boreal forest, allowed a close examination of the relative importance of biome type where there may be a lack of significant barriers to connectivity. The cluster analyses in Fig. S2 show that the soil animal communities of the four circumpolar locations group together. Clustering by abundance generally grouped locations together by biome type (Fig. S2A), whereas clustering by presence/absence of OTUs partitioned the four circumpolar locations by geographic location instead of biome type (Fig. S2B). This suggests that both the lack of significant barriers to connectivity and similarity of biome type are important factors in cosmopolitanism.
Below-Ground Biodiversity and Environmental Factors.
Although we did not observe a correlation between latitude and Shannon diversity, richness, dominance, or Hurlburt's PIE, there was a moderate Spearman correlation (0.43) between latitude and community structure as measured by the PrimerE BIO-ENV analysis. Other environmental factors, such as mean annual precipitation and soil inorganic N concentration, had minimal Spearman correlations to community structure (<0.38). These low correlation values likely reflect the extreme endemism and lack of common OTUs among locations. Another study (15) reported a decrease in the diversity of soil nematodes at higher latitudes, which we did not observe. However, we sampled soils to a latitude of 68°N, whereas the study that showed a drop in animal diversity included data from more extreme latitudes such as Antarctica (77°S) and Spitsbergen (79°N).
We compared the broad community structure at the different locations and discovered that six of 11 locations were dominated by arthropods, four locations were dominated by nematodes, and the remaining site was codominated by arthropods and nematodes (Fig. 1). This pattern could partially be a reflection of locations that are primarily grassland versus forest. For example, four of six mite-dominated locations were coniferous (Alaska and Sweden) or tropical (Peru and Costa Rica) forests, whereas three of four locations dominated by nematodes were grassland [AR, Kenya grassland (KY), and KZ]. Forests usually have lower soil pH than grassland (16), and microarthropods are typically the dominant soil animals in forest ecosystems (17, 18), which was reflected in our results. We analyzed our data by comparing grassland [AB, AR, KY, KZ, South Africa Mediterranean grassland (SA), and TK] to primarily forest locations [BZ, Costa Rica tropical forest (CR), New Zealand temperate rainforest (NZ), Peru tropical forest (PU), and RE] and found the average dominance of microarthropods was higher in forest (70%) than grassland (48%, P = 0.02), and that average dominance of nematodes was higher in grassland (42%) than in forest (15%, P = 0.005). To further investigate, we compared environmental factors between the arthropod-dominated locations and the nematode-dominated locations (Table S2). Although it is difficult to identify causal links on the basis of these correlations, they do enable us to identify some general patterns. For example, microarthropod-dominated sites had lower soil bulk density than nematode-dominated sites, and microarthropod dominance was associated with several factors related to resource quantity and quality, such as high root biomass, more litter, greater soil C content, and C:N ratios, suggesting that mites may be more bottom-up controlled than are nematodes. This is consistent with previous studies that show that microarthropods can be found at higher density in untilled agricultural fields and temperate forests, which generally have high levels of soil organic matter, whereas nematodes are found at high density in tilled fields (19) and microarthropod density can be lower in tropical forests where organic carbon is lower (19, 20). However, experimental studies report mixed effects of increased resources on mites and nematodes, with both groups often increasing in abundance (21–24). We also noticed that microarthropod-dominated locations were significantly higher in latitude, with lower temperatures and higher moisture than nematode-dominated sites.
We also found that locations with higher soil pH favored nematodes and locations with lower soil pH favored arthropods (Table S2 and Fig. S1). This is consistent with a study of 284 agroecosystems along a pH gradient in the Netherlands (25), where nematode abundance was positively—and microarthropod abundance negatively— correlated with soil pH.
Relationships Between Above-Ground and Below-Ground Diversity.
Previous studies have found a positive correlation between above-ground plant and below-ground biodiversity (e.g., ref. 26). However, our results indicate that Shannon diversity and richness of soil animals are lower at four locations identified by Myers et al. (11) as biodiversity hotspots, on the basis of above-ground diversity, compared with seven locations that were not identified as having high above-ground biodiversity (Table S4). This indicates that on a global scale, there may be an inverse relationship between above-ground plant biodiversity and soil animal biodiversity and that soil animal community structure appears to be linked to above-ground plant biodiversity. It has been proposed that relationships between above-ground and below-ground diversity can be positive, negative, or neutral (27, 28). Positive correlations can be caused by increased heterogeneity of litter inputs to soil or the formation of more diverse soil microhabitats associated with higher plant diversity, whereas negative or neutral relationships can be caused by the presence of different abiotic constraints above ground compared with below ground or by negative interactions between organisms found above ground and below ground (27–29).
Four of our locations coincide with above-ground biodiversity hot spots. Three of these (CR, NZ, and PU) are dense tropical or temperate rainforests, whereas one (SA) is a Mediterranean grassland in South Africa. The seven sites of lower above-ground biodiversity were arctic sites (AB, BZ, RE, and TK), arid grassland (KY), shrub steppe (AR), or prairie (KZ). We compared environmental parameters at locations that coincide with above-ground biodiversity hotspots to those of lower above-ground biodiversity (Table S4). We found that soil inorganic N concentration, temperature and precipitation, and litter mass were all significantly greater in above-ground biodiversity hotspots than at the other sites. These factors are likely responsible for the result seen in the MDS analysis (Fig. S5) of below-ground community structure, where locations that coincide with above-ground biodiversity hotspots are partitioned from locations of lower above-ground biodiversity. The higher mean temperature and rainfall at locations of high above-ground biodiversity could contribute to enhanced decomposition of the larger amounts of litter present and thus lead to higher availability of soil N. High soil inorganic N availability is associated with lower microbial abundance and has negative effects on fungi, often leading to bacterial-dominated soils, which are typically of lower diversity (30). Studies of soil mites (31) suggest that high soil inorganic N increases the abundance, but not diversity, under disturbance conditions. In another study (32), it was found that in subtropical monsoon evergreen broadleaf forests, increased soil inorganic N caused a significant decrease in biodiversity, group abundance, and density of small soil animals. Because of the mixed results of studies relating soils with high inorganic N to changes in below-ground biodiversity, there appears to be varied effects in different soil ecosystems where other factors such as the quality and quantity of litter, soil heterogeneity, the nature of the soil animal communities, and their interactions with plants may play important roles. For example, it has been shown that dune grasses (Ammophila arenaria) of different genotype recruit distinct above-ground invertebrate communities and those with large above-ground invertebrate communities are less likely to have a diverse below-ground root-feeding nematode community (33). Our molecular method measures relative abundance to calculate biodiversity of soil animals but does not measure density, absolute abundance, or biomass of soil animals, making our data difficult to compare with morphological-based studies, yet provides a useful view of below-ground biodiversity.
There are several possible mechanisms to explain lower below-ground animal diversity at locations with high above-ground plant diversity. It has been shown in an experimental forest system that increasing soil N results in more above-ground plant growth, which decreases the amount of C available to soil biota including bacteria, fungi, and some microarthropods (34). Soil with high levels of N may exhibit a decrease in the amount of mycorrhizal fungi, leading to a decrease in soil animals that feed on the fungi.
Conclusions
Our analyses of soils taken from a broad range of biomes and latitudes suggest cosmopolitan soil animals are extremely rare, even at the “family” level. Only 14 OTUs of 2,259 were found at four or more locations, whereas most OTUs were found at a single location. This is likely due to the lack of connectivity between locations at the global level. The sites with the most taxa in common were found in four circumpolar locations relatively close to each other and with similar climates.
Locations dominated by arthropods were primarily forests with low soil pH, high C:N ratios, high litter, higher root biomass, lower soil bulk density, and more moisture compared with nematode-dominated locations, which were mostly grasslands that were higher in nitrate, inorganic N, and had higher soil and mean annual temperatures. Numerous environmental factors related to resource quantity and quality were found to differ between microarthropod-dominated locations and nematode-dominated locations.
Our findings suggest that at the global level, there may be an inverse relationship between above-ground plant biodiversity and soil animal biodiversity. Four of our locations have been identified as above-ground biodiversity hotspots, but our results showed those locations had significantly lower soil animal biodiversity than the other locations that were not identified as above-ground biodiversity hotspots. We suggest that increased N and lower soil pH found in soil at the locations with high above-ground biodiversity could in part explain the lower soil animal biodiversity at those locations.
Materials and Methods
Locations and Sample Collection.
Eleven locations were selected at different regions around the world to represent a broad range of continents, biomes, and latitudes (Table 1). At each location, a 900-m transect consisting of four evenly spaced 10 × 10 m plots (i.e., 300 m apart) was established. The location of each transect was selected to be representative of a dominant ecosystem type in the region and was relatively undisturbed. Each plot within a transect had similar elevation, aspect, vegetation, and soil type. Twenty soil cores (3.4 cm diameter, 10 cm deep) were collected from each plot and hand mixed in a single plastic bag. Soil CO2 efflux was measured (35) using a chamber placed over three 10-cm diameter plastic collars inserted into the soil in each plot (PP Systems). Following measurement of CO2 efflux, leaf litter was collected from each 10-cm diameter collar to estimate standing litter biomass. A 200-g soil subsample was mixed with 500 mL of 95% ethanol to suspend the soil and to preserve the biological material for subsequent DNA analysis. The suspended soil/ethanol mixture was sieved (6) to exclude large animals. A total of 44 samples representing four plots along the transect at each of the 11 geographic locations were collected.
Soil Methods.
Collected soil was passed through a sieve (2 mm) to remove roots; soil was stored at 4 °C before analysis, and roots and litter were dried at 60 °C and weighed. Soil pH and electrical conductivity were measured using pH and conductivity meters (36, 37). Soil moisture was measured using a gravimetric method (38), total carbon (C) and nitrogen (N) using a C/N analyzer (39) (Carlo-Erba Instruments), and microbial biomass using a fumigation-extraction method (40). Bulk density was estimated by a core method (41).
Molecular Methods.
DNA was extracted from each sample using a modified CTAB procedure (42). Metazoan-specific primers for the 18S rRNA gene 18S11b (5′-GTCAGAGGTTCGAAGGCG-3′), which correspond to positions 1,037–1,054 of the human sequence (NR_003286 in GenBank) and 18S2a (5′-GATCCTTCCGCAGGTTCACC-3′), which corresponds to positions 1,848–1,867 of the human sequence were used to amplify approximately an 830-bp segment (6). An initial 2-min denaturing step at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min were used for PCR reactions. PCR products were cloned into libraries using Topo TA cloning kits (Invitrogen) and ∼300 single pass sequences (519 bp after trimming) were obtained from each library using a commercial sequencing facility. The animal-specific primers minimized amplification of plant, protozoa, fungi, or bacteria DNA.
Custom software that finds a highly conserved region of the 18S rRNA sequence as an anchor point and then trims upstream and downstream from that point was used to process the raw sequences. The trim points were optimized so that only the most accurate parts of the sequences were used, and a filter was used to remove poor quality sequences. This produced highly accurate sequences by reducing the length of the read to 519 bp and by discarding ∼30% of the sequences that were of poor quality.
Assigning OTUs.
High-quality sequences were grouped into OTUs on the basis of 100, 99, 97, 95, 90, 85, and 80% sequence similarity using Sequencher version 4.7 (Gene Codes). We had previously determined that the 99% criterion for grouping 18S rRNA gene sequences approximates the number of species when analyzing GenBank sequences of mites and nematodes with documented taxonomies (6). We then examined the more widely distributed OTUs by statistical analysis of soil animal DNA sequences (6). An alternative analysis was carried out using ESPRIT software (13) to assign OTUs on the basis of a phylogenetic distance criterion of 0.01, 0.02, 0.03, and 0.05 for grouping sequences into OTUs. The phylogenetic distance criterion of 0.01 corresponds approximately to the 99% similarity criterion in Sequencher.
Identification of OTUs.
After each OTU was assigned, a sequence from that OTU was used as a query in a BLAST search. The closest matching sequence contained in GenBank for which there was documented taxonomic information was used as a provisional “identification” of the OTU for subsequent analysis. We recognize that GenBank taxonomy is not always accurate but we use it here because of the wide taxonomic scope of the project. The determination of which taxonomic groups (e.g., microarthropods or nematodes) were dominant at each site was based on the composition of OTUs identified as microarthropods or nematodes by NCBI BLAST provisional identifications.
Statistical Analysis.
The frequency of each OTU was tabulated and inputted for diversity and community structure analysis. Diversity indices including richness, Hurlbert's PIE, dominance index, and Shannon diversity index for at least 297 sequences (when available) from each sample were obtained in EcoSim (43) using the rarefaction randomization algorithm included in the program. It randomly resamples different numbers of sequences from the dataset without replacement and determines the number of OTUs in each sample. Statistica 9.1 (StatSoft) was used for univariate analysis of soil faunal diversity indices. The soil community was analyzed with the NMDS, on the basis of Bray–Curtis community similarity using PRIMER-E statistical software (PRIMER-E). The PRIMER-E software was also used for an ANOSIM, which calculated the significance of differences in community structure among plots and locations. Phylogenetic analyses were carried out using MrBayes 3.1 (44) and Mega 4 (45) software.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers M. Vandegehuchte and Z. Sylvain for their very useful comments and suggestions and J. Nkem for assisting in field work. This work was funded by the National Science Foundation Grants 0344834 (to D.H.W.) and 0344372 (to J.R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN135477–JN152992).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103824108/-/DCSupplemental.
References
- 1.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 2.Foissner W. Biogeography and dispersal of micro-organisms: A review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 3.Hillebrand H, Watermann F, Karez R, Berninger UG. Differences in species richness patterns between unicellular and multicellular organisms. Oecologia. 2001;126:114–124. doi: 10.1007/s004420000492. [DOI] [PubMed] [Google Scholar]
- 4.Xu S, Hebert PDN, Kotov AA, Cristescu ME. The noncosmopolitanism paradigm of freshwater zooplankton: Insights from the global phylogeography of the predatory cladoceran Polyphemus pediculus (Linnaeus, 1761) (Crustacea, Onychopoda) Mol Ecol. 2009;18:5161–5179. doi: 10.1111/j.1365-294X.2009.04422.x. [DOI] [PubMed] [Google Scholar]
- 5.Porazinska DL, et al. Ecometagenetics confirm high tropical rainforest nematode diversity. Mol Ecol. 2010;19:5521–5530. doi: 10.1111/j.1365-294X.2010.04891.x. [DOI] [PubMed] [Google Scholar]
- 6.Wu T, et al. Molecular profiling of soil animal diversity in natural ecosystems: Incongruence of molecular and morphological results. Soil Biol Biochem. 2009;41:849–857. [Google Scholar]
- 7.de la Peña E, Vandegehuchte ML, Bonte D, Moens M. Nematodes surfing the waves: Long-distance dispersal of soil-borne microfauna via sea swept rhizomes. Oikos. 2011 10.1111/j.1600-0706.2011.19540.x. [Google Scholar]
- 8.Willig MR, Kaufman DM, Stevens RD. Latitudinal gradients of biodiversity: Pattern, process, scale, and synthesis. Annu Rev Ecol Syst. 2003;34:273–309. [Google Scholar]
- 9.Maraun M, Schatz H, Scheu S. Awesome or ordinary? Global diversity patterns of oribatid mites. Ecography. 2007;30:209–216. [Google Scholar]
- 10.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecol Lett. 2009;12:1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 11.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 12.Fulthorpe RR, Roesch LF, Riva A, Triplett EW. Distantly sampled soils carry few species in common. ISME J. 2008;2:901–910. doi: 10.1038/ismej.2008.55. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, et al. ESPRIT: Estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JH. On the relationship between abundance and distribution of species. Am Nat. 1984;124:255–279. [Google Scholar]
- 15.Boag B, Yeats GW. Soil nematode biodiversity in terrestrial ecosystems. Biodivers Conserv. 1998;7:617–630. [Google Scholar]
- 16.Chen CR, Condron LM, Davis NR, Sherlock RR. Effects of afforestation on phosphorus and biological properties in a New Zealand grassland soil. Plant Soil. 2000;220:151–163. [Google Scholar]
- 17.Ingham ER, Coleman DC, Moore JC. An analysis of food-web structure and function in a shortgrass prairie, a mountain meadow, and a lodgepole pine forest. Biol Fertil Soils. 1989;8:29–37. [Google Scholar]
- 18.Petersen H, Luxton M. A comparative analysis of soil fauna populations and their role in decomposition processes. Oikos. 1982;39:287–388. [Google Scholar]
- 19.Hendrix PF, et al. Detritus food webs in conventional and no-tillage agroecosystems. Bioscience. 1986;36:374–380. [Google Scholar]
- 20.Seastedt TR. The role of microarthropods in decomposition and mineralization processes. Annu Rev Entomol. 1984;29:25–46. [Google Scholar]
- 21.Seastedt TR, James SW, Todd TC. Interactions among soil invertebrates, microbes and plant growth in the tallgrass prairie. Agric Ecosyst Environ. 1998;24:219–228. [Google Scholar]
- 22.Mikola J, Setälä H. Productivity and trophic level biomasses in a microbial-based soil food web. Oecologia. 1998;117:396–403. doi: 10.1007/s004420050673. [DOI] [PubMed] [Google Scholar]
- 23.Chen B, Wise DH. Bottom-up limitation of predaceous arthropods in a detritus-based terrestrial food web. Ecology. 1999;80:761–772. [Google Scholar]
- 24.Wardle DA. Communities and Ecosystems: Linking the aboveground and Belowground Components. Princeton, NJ: Princeton Univ Press; 2002. [Google Scholar]
- 25.Mulder C, Van Wijnen HJ, Van Wezel AP. Numerical abundance and biodiversity of below-ground taxocenes along a pH gradient across the Netherlands. J Biogeogr. 2005;32:1775–1790. [Google Scholar]
- 26.Vanbergen AJ, et al. Scale-specific correlations between habitat heterogeneity and soil fauna diversity along a landscape structure gradient. Oecologia. 2007;153:713–725. doi: 10.1007/s00442-007-0766-3. [DOI] [PubMed] [Google Scholar]
- 27.Hooper DU, et al. Interactions between above and belowground biodiversity in terrestrial ecosystems: Patterns, mechanisms, and feedbacks. Bioscience. 2000;50:1049–1061. [Google Scholar]
- 28.De Deyn GB, Van der Putten WH. Linking aboveground and belowground diversity. Trends Ecol Evol. 2005;20:625–633. doi: 10.1016/j.tree.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Coleman DC, Crossley DA, Jr, Hendrix PF. Fundamentals of Soil Ecology. 2nd Ed. San Diego: Elsevier Academic Press; 2004. [Google Scholar]
- 30.Bardgett RD. The Biology of Soils: A Community and Ecosystem Approach. Oxford: Oxford Univ Press; 2005. [Google Scholar]
- 31.Cole L, Buckland SM, Bardgett RD. Influence of disturbance and nitrogen addition in plant and soil animal diversity in grassland. Soil Biol Biochem. 2008;40:505–514. [Google Scholar]
- 32.Xu G, Mou J, Zhou G, Fu S. Preliminary response of soil fauna to simulated N deposition in three typical subtropical forests. Pedosphere. 2006;16:596–601. [Google Scholar]
- 33.Vandegehuchte ML, de la Peña E, Bonte D. Contrasting covariation of above- and belowground invertebrate species across plant genotypes. J Anim Ecol. 2011;80:148–158. doi: 10.1111/j.1365-2656.2010.01766.x. [DOI] [PubMed] [Google Scholar]
- 34.Högberg MN, et al. Quantification of effects of season and nitrogen supply on tree below-ground carbon transfer to ectomycorrhizal fungi and other soil organisms in a boreal pine forest. New Phytol. 2010;187:485–493. doi: 10.1111/j.1469-8137.2010.03274.x. [DOI] [PubMed] [Google Scholar]
- 35.Holland EA, et al. Soil CO2, N2O and CH3 exchange. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 185–201. [Google Scholar]
- 36.Robertson GP, Sollins P, Ellis BG, Lajtha K. Exchangeable ions, pH, and cation exchange capacity. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 106–114. [Google Scholar]
- 37.Rhoades JD. Salinity: Electrical Conductivity and Total Dissolved Solids. In: Sparks DL, et al., editors. Methods of Soil Analysis Part 3—Chemical Methods. Madison, WI: Soil Science Society of America; 1996. pp. 417–436. [Google Scholar]
- 38.Jarrell WM, et al. Soil water and temperature status. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 55–73. [Google Scholar]
- 39.Sollins P, et al. Soil carbon and nitrogen pools and fractions. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 89–105. [Google Scholar]
- 40.Paul EA, Harris D, Klug MJ, Ruess RW. The determination of microbial biomass. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 291–317. [Google Scholar]
- 41.Elliot ET, Heil JW, Kelly EF, Monger HC. Soil structural and other physical properties. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard Soil Methods for Long-Term Ecological Research. New York: Oxford Univ Press; 1999. pp. 74–85. [Google Scholar]
- 42.Gawel NJ, Jarret RL. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol. 1991;9:262–266. [Google Scholar]
- 43.Gotelli NJ, Entsminger GL. Jericho, VT: Acquired Intelligence and Kesey-Bear; 2011. EcoSim: Null models software for ecology. version 7. Available at http://garyentsminger.com/ecosim/index.htm. Accessed September 29, 2011. [Google Scholar]
- 44.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.