Abstract
Rod and cone opsin genes are expressed in a mutually exclusive manner in their respective photoreceptor subtypes in the mammalian retina. Previous transgenic mouse studies showed that functional interactions between the distal enhancer and proximal promoter of rhodopsin and long/medium-wavelength (L/M) opsin genes are essential for regulating their cell-type–specific transcription. We have used chromosomal conformation capture assays in mouse retinas to investigate the molecular mechanism responsible for this interaction. Here we show that each opsin gene forms intrachromosomal loops in the appropriate photoreceptor subtype, while maintaining a linear configuration in other cell types where it is silent. The enhancer forms physical contacts not only with the promoter but also with the coding regions of each opsin locus. ChIP assays showed that cell-type–specific target binding by three key photoreceptor transcription factors—cone–rod homeobox (CRX), neural retina leucine zipper (NRL), and nuclear receptor subfamily 2, group E, member 3 (NR2E3)—is required for the appropriate local chromosomal organization and transcription of rod and cone opsins. Similar correlations between chromosomal loops and active transcription of opsin genes were also observed in human photoreceptors. Furthermore, quantitative chromosomal conformation capture on human retinas from two male donors showed that the L/M enhancer locus control region (LCR) loops with either the L or M promoter in a near 3:1 ratio, supporting distance-dependent competition between L and M for LCR. Altogether, our results suggest that the photoreceptor transcription factor network cooperatively regulates the chromosomal organization of target genes to precisely control photoreceptor subtype-specific gene expression.
Keywords: neuronal gene expression, enhancer-promoter interaction, chromatin modulation, epigenetic regulation
Rods and cones are two types of image-forming photoreceptors that constitute 70% of retinal cells in mice and humans. Rods, representing 95–97% of the photoreceptors, are sensitive to dim light and responsible for night vision. Cones, although constituting only 3–5% of the photoreceptors, sense bright light and mediate color discrimination and visual acuity. These functional differences are attributed, at least in part, to the unique visual pigments made by rods [rhodopsin (Rho)] or cones (cone opsins). Cones can be divided into subtypes based on light-wavelength sensitivity that are characteristically distributed in different species. In humans, cones sensitive to long (L), medium (M), and short (S) wavelengths, each expressing a distinct cone opsin, are enriched within the macular region (1, 2). The mouse retina expresses only two cone opsins, M and S, which are distributed in opposing gradients along the dorsal–ventral axis, forming spatially distinct populations of M, S, and hybrid M/S cones (3, 4).
Photoreceptor subtype-specific opsin expression is regulated by interactions between DNA cis-regulatory elements and a network of photoreceptor transcription factors (5, 6). The homeodomain protein cone–rod homeobox (CRX) is required for differentiation and maintenance of both rods and cones (7, 8). Crx-deficient mice (Crx−/−) fail to develop visual function because of defective photoreceptor transcription (9). The rod transcription factor neural retina leucine zipper (NRL) (10) is essential for specifying rod cell fate. Nrl-deficient mice (Nrl−/−) fail to express rod genes and silence cone genes in rods, turning all rods to cones (11). Acting downstream of NRL, the nuclear receptor subfamily 2, group E, member 3 (NR2E3) promotes rod differentiation by activating rod genes but repressing cone genes (12–14). The Nr2e3-deficient mouse Nr2e3rd7/rd7 (15) develops hybrid rod/S-cone cells that coexpress Rho and S opsin (12, 16). Cone transcription factors, including thyroid hormone receptor β2 (TRβ2) (17), retinoid X receptor γ (RXRγ) (18), and retinoic acid receptor-related orphan receptor α (RORα) (19), regulate cone opsin expression and establish an appropriate S/M cone ratio and mosaic. How each factor accomplishes its specific role is unclear. We previously reported that CRX promotes histone acetylation in opsin regulatory regions (20). Here we extend these studies to reveal the role of CRX, NRL, and NR2E3 in opsin chromosomal organizations.
opsin cis-regulatory regions include an enhancer and promoter. The human L and M opsin genes form a head-to-tail array on the X chromosome. They share an enhancer 3-kb upstream of L, a so-called locus control region (LCR) similar to that found in the β-globin locus (21). L/M LCR is required for expressing either gene; its deletion results in blue cone monochromacy, a disorder in which both L and M cone functions are defective (22, 23). Transgenic mice carrying human L/M arrays showed that expression of either L or M is a stochastic event resulting from competition between the L and M promoters for the LCR, which favors the promoter proximal to the LCR (24, 25). Similarly, Rho expression requires functional pairing between its Rho enhancer region (RER) and promoter (26). The molecular basis for opsin enhancer–promoter pairing is thought to be mediated by physical contact between the two regions (24, 25). Chromosomal conformation capture (3C) assays (27) revealed that the β-globin locus forms intrachromosomal loops, where the LCR contacts different β-like genes for transcription during erythroid cell development (28, 29). Using 3C to reveal opsin intrachromosomal loops in both mouse and human retinas, we found that rod and cone opsin loci adopt different chromosomal organizations in rods vs. cones. Each opsin locus contains an upstream looping organizer that contacts the promoter and intragenic regions in the respective photoreceptor subtype actively transcribing it. This looping organization correlates with transcription levels and requires the combinatorial action of at least three photoreceptor transcription factors: CRX, NRL, and NR2E3.
Results
Adapting 3C Protocols for Whole Mouse/Human Retinas.
3C detects in vivo physical contacts between distant chromatin segments (27). The chromatin of living cells is fixed with formaldehyde to crosslink interacting sites, digested with a restriction enzyme (e.g., Bpm1), and ligated in situ to covalently join linked DNA fragments, which are then analyzed and quantified by PCR. 3C requires ≥1 × 107 cells, of which the target cell type of interest represents ≥70–80% (30). Because rods constitute 70% of retinal cells, we performed 3C with whole retinas of WT mice for chromosomal loops in rods. Loops in cones were analyzed with Nrl−/− mice. Crx−/− and Nr2e3rd7/rd7 mice provided defective rods and hybrid rods/S cones, respectively. 3C on human retinas used manually dissected subregions to achieve rod- and cone-dominant cell populations. Each sample consisting of 1 × 107 cells was analyzed for rod and cone opsins by using PCR primers flanking Bpm1 fragments along the full length of each locus (Figs. 1A, 2Aand , 3A, Fig. S3 A–C, and Tables S1 and S2). The pan photoreceptor marker Rbp3 provided a positive control for all retinal samples, and BAC clone DNA cut with Bpm1 and randomly ligated provided positive PCR controls (Table S3). The results were interpreted and compared based on the specific cell type enriched in each sample.
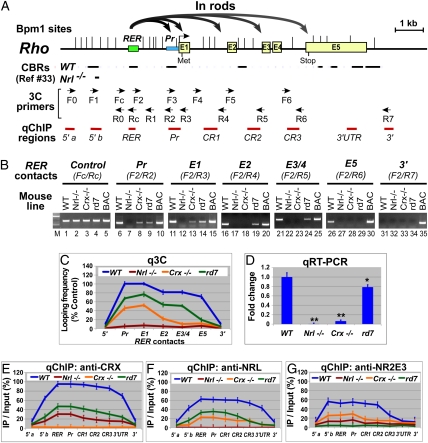
Fig. 1.
Active murine Rho locus displays chromosomal loops between its enhancer (RER) and promoter/coding regions and is widely bound by CRX, NRL, and NR2E3. (A) Diagram of the Rho locus, showing gene structure, Bpm1 restriction sites (|), positions of CRX binding regions (CBRs) detected by ChIP-seq (33), 3C primers, and ChIP-PCR (qChIP) regions. Curved gray arrows represent chromosomal loops in rods detected by 3C shown in B and C. (B) Gel images of 3C performed on P14 retinas of the indicated mouse strains with Rho-containing BAC samples as positive controls for PCR. Chromatin segments and corresponding 3C primer pairs are indicated above the gel images (Pr, Rho promoter; E1–E5, exons). (C) qPCR quantification of samples (q3C) relative to control (Fc/Rc) from B. (D) qRT-PCR of Rho mRNA. *P < 0.05, **P < 0.001. (E–G) qChIP assays using antibodies to CRX, NRL, and NR2E3, respectively.
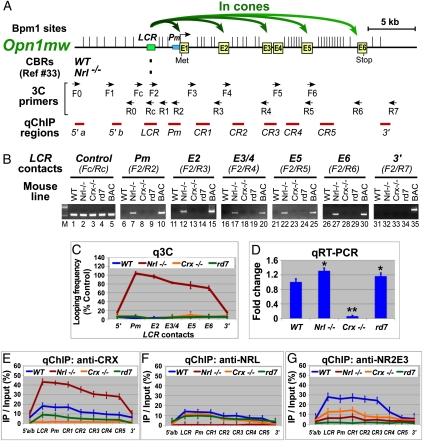
Fig. 2.
Active murine Opn1mw (M) locus displays chromosomal loops between its enhancer (LCR) and promoter/coding regions and is widely bound by CRX. (A) Diagram of the M locus, showing gene structure, Bpm1 restriction sites (|), positions of CBRs detected by ChIP-seq, 3C primers, and ChIP-PCR (qChIP) regions. Curved green arrows represent M chromosomal loops in cones detected by 3C shown in B and C. (B) Gel images of 3C performed on P14 retinas of the indicated mouse strains with M-containing BAC samples as positive controls. Chromatin segments and corresponding 3C primer pairs are indicated above the gel images (Pm, M promoter; E1–E6, exons). (C) qPCR quantification of samples (q3C) relative to control (Fc/Rc) from B. (D) qRT-PCR of M mRNA. *P < 0.05, **P < 0.001. (E–G) qChIP assays using antibodies to CRX, NRL, and NR2E3, respectively.
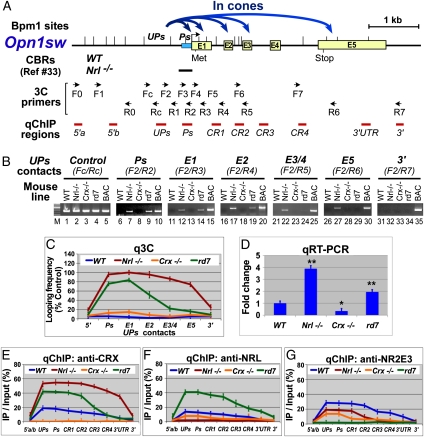
Fig. 3.
Active murine Opn1sw (S) locus displays chromosomal loops between an upstream region UPs and promoter/coding regions and is widely bound by CRX. (A) Diagram of the S locus, showing gene structure, Bpm1 restriction sites (|), positions of CBRs detected by ChIP-seq, 3C primers, and ChIP-PCR (qChIP) regions. Curved blue arrows represent S chromosomal loops in cones detected by 3C shown in B and C. (B) Gel images of 3C performed on P14 retinas of the indicated mouse strains with S-containing BAC samples as positive controls. Chromatin segments and corresponding 3C primer pairs are indicated above the gel images (Ps, S promoter; E1–E5, exons). (C) qPCR quantification of samples (q3C) relative to control (Fc/Rc) from B. (D) qRT-PCR of S mRNA. *P < 0.05, **P < 0.001. (E–G) qChIP assays using antibodies to CRX, NRL, and NR2E3, respectively.
Murine opsin Loci Display Looping Organizations in Their Corresponding Photoreceptor Subtypes.
Rho.
3C assays on WT rods revealed physical contacts of Rho enhancer RER with the Rho promoter (Pr) and exon 1 (E1) (Fig. 1B, lanes 6 and 11). Interestingly, RER also contacts other Rho coding regions (E2, lane 16; E3/E4, lane 21; and E5, lane 26), but the looping frequency decreases as the distance from RER increases (Fig. 1C, blue line). No RER loops were detected with regions 3′ (Fig. 1B, lane 31; Fig. 1C, 3′) or 5′ (Fig. 1C, 5′; Fig. S1A, F0/R0 and F1/R0) to the Rho locus, suggesting that looping is a local organization within the gene boundaries. RER is the Rho looping organizer because no PCR product was seen with forward primers other than F2 (Fig. S1A). No loops were detected in Nrl−/− cones (Fig. 1B, Nrl−/− lanes; Fig. 1C, red line), where Rho is silenced (Fig. 1D). In Crx−/−-defective rods, where Rho is transcribed at 10% of WT levels (Fig. 1D), only RER–Pr and RER–E1 loops were seen (Fig. 1B, lanes 8 and 13) with a frequency of about half that of WT rods (Fig. 1C, orange vs. blue lines). In the rd7 retina, where the rod/S-cone hybrid cells express Rho at slightly reduced levels (Fig. 1D), the frequency of all loops is reduced (Fig. 1C, green vs. blue lines) and the last RER–E5 loop is undetectable (Fig. 1B, lane 29; Fig. 1C, green line). Thus, RER looping frequency appears to correlate with Rho transcriptional levels. We conclude that RER-organized looping is an active chromosomal organization for Rho transcription.
Opn1mw (M).
M chromosomal loops were detected only in Nrl−/− retina (Fig. 2B, Nrl−/− lanes), where LCR makes contacts with the M promoter (Pm) and other coding regions (E2–E6) but not the 3′ or 5′ regions (Fig. S1B). The looping frequency decreases as the distance from LCR increases (Fig. 2C, red line). No M loops were detected in WT (Fig. 2B, WT lanes; Fig. 2C, blue line) and Crx−/− rods (Fig. 2B, Crx−/− lanes; Fig. 2C, orange line) or rd7 hybrid cells (Fig. 2B, rd7 lanes; Fig. 2C, green line), suggesting that M has a linear configuration in rods, where it is silenced. Thus, LCR-organized chromosomal looping is an active conformation for M.
Opn1sw (S).
Unlike Rho and M, the S locus does not have a defined enhancer. However, 3C detected multiple S loops in Nrl−/− cones (Fig. 3B, Nrl−/− lanes), where S is highly expressed (Fig. 3D, Nrl−/− bar). All S loops are anchored on UPs, a region upstream of the S promoter (Ps). In cones, UPs makes contacts with Ps and S coding regions E1–E5 (Fig. 3B, Nrl−/− lanes) but not with the 3′ region (Fig. 3B, lane 32). No loops were detected when using other forward primers upstream or downstream of UPs (Fig. S1C), suggesting that UPs is the S loop organizer. The UPs looping frequency decreases as the distance from UPs increases (Fig. 3C, red line). S loops were undetectable in WT or Crx−/− rods (Fig. 3B, WT and Crx−/− lanes) so that S has a linear organization in rods where S is silenced. Interestingly, some S loops were detected in rd7 hybrid cells (Fig. 3B, rd7 lanes), although at lower frequency and shorter range than those seen in Nrl−/− cones (Fig. 3C, green vs. red line), consistent with S mRNA levels in rd7 and Nrl−/− retinas (Fig. 3D, rd7 vs. Nrl−/−). Thus, UPs-directed looping represents an active chromosomal organization for S.
Other rod and cone genes.
Using the same four retinal 3C preparations, we also analyzed Rpb3, a “CRX-independent” gene expressed by both rods and cones (5, 9), as well as Gnat1 and Gnat2, coding for rod- and cone-specific transducin α, respectively. All three genes display looping organization in the respective photoreceptor subtype(s) in which they are expressed (Fig. S2), suggesting that looping, in general, represents a transcriptionally active organization for photoreceptor-specific genes.
Appropriate opsin Chromosomal Organization Requires Network Transcription Factors.
Quantitative ChIP (qChIP) assays were performed on retinas of WT and mutant mice to visualize the binding of three photoreceptor transcription factors to each opsin locus.
CRX is more abundant on any expressed opsin locus than on the same locus when it is silenced (Figs. 1E, 2E, and 3E), correlating with 3C-detected looping organizations. Thus, more CRX is found on Rho in the rods of WT retinas than in cones of Nrl−/− retinas (Fig. 1E, blue vs. red line). This binding pattern is reversed for M and S loci (Figs. 2E and 3E, red vs. blue line). Within active opsin loci, CRX levels correlated with looping frequencies for each gene subregion (Figs. 1E, 2E, and 3E). CRX binding is also altered by Nr2e3 deficiency, correlating with changes in opsin looping patterns and transcription in rd7 retinas.
NRL and NR2E3, as rod trans-activators, both preferentially bind to Rho over cone opsins. The highest amounts are seen on Rho enhancer and promoter regions where chromosomal loops occur most frequently (Fig. 1 F and G, blue lines). Their binding to Rho is reduced by Crx deficiency (Fig. 1 F and G, orange vs. blue line), correlating with reduced Rho loops (Fig. 1C, orange vs. blue line) and transcription (Fig. 1D, Crx−/− vs. WT). Interestingly, Rho-bound CRX and NRL are reduced by 50% in the Nr2e3-deficient rd7 retina (Fig. 1 E and F, green vs. blue line), coinciding with reduced Rho loops and transcription (Fig. 1 C and D, rd7 vs. WT). Altogether, these data support cooperativity among all three factors on Rho.
NRL and NR2E3 also both function as repressors for cone genes in rods, but they show different binding patterns for cone opsins in WT rods: NRL has limited binding activity for M and S (10–15% occupancy) (Figs. 2F and 3F, blue line), whereas NR2E3 binds well to both M and S (∼30% occupancy) (Figs. 2G and 3G, blue line), suggesting that NR2E3 directly mediates silencing of cone opsins in rods. Consistent with this idea, in NR2E3 deficiency (rd7), the amount of NRL and CRX on S is more than doubled (Fig. 3 E and F, green vs. blue line), which is associated with ectopic S chromosomal loops and transcription in the rd7 retina (Fig. 3 C and D, rd7 vs. WT). Thus, NR2E3 appears to achieve its repression function by modulating chromatin configuration. Interestingly, the amount of CRX or NRL bound to M does not increase in the rd7 retina (Fig. 2 E and F, green vs. blue line), where no ectopic M loops (Fig. 2C, green vs. blue) or large increase in M transcripts (Fig. 2D, rd7 vs. WT) are observed. Thus, M silencing in rods appears not to depend on NR2E3.
Human OPSIN Loci Also Adopt Differential Chromosomal Organizations in Rods vs. Cones.
To test whether active human OPSIN loci also undergo looping organizations, we performed 3C assays on human retinas of two male donors, dissected into three separate regions with different rod and cone distributions. Rod/cone-expressed RBP3 was used as a positive 3C control. RHO RER loops were detected in rod-rich far periphery (FP) and, to a lesser degree, in peripheral macula lutea (PML) with mixed rods/cones but rarely in cone-rich macula lutea (ML) samples (Fig. S3 A, D, and E). Cone genes L and M show the opposite pattern: L/M LCR loops were detected mostly in cone-rich ML and, to a lesser degree, in PML but rarely in rod-rich FP samples (Fig. S3 B, D, and E). No looping organizers other than the RHO RER and L/M LCR were detected (Fig. S4 A and B). In contrast, RBP3 displays similar levels of enhancer-organized loops in all subregions (Figs. S3 C, D, and E and S4C). Thus, as with their mouse counterparts, human OPSINS adopt looping organization in the respective photoreceptor subtype expressing them.
We used quantitative PCR (qPCR) to compare frequencies for LCR contacting L versus M promoter in ML and PML samples. Fig. S3 D and E show that LCR loops with L/M promoter in a near 3:1 ratio in both donors (2.8:1 in each). This ratio is maintained at coding region E5 (Fig. S3F), thus reflecting expression.
CRX Induces Human OPSIN Chromosomal Loops in Y79 Retinoblastoma Cells.
Cultured Y79 retinoblastoma cells, although expressing RBP3, do not express OPSINS because of insufficient CRX expression. Transiently expressing a recombinant CRX in these cells induces OPSIN expression (20). To test whether recombinant CRX changes OPSIN organization from the silent linear status to active loops, we performed 3C on Y79 cells transiently transfected with expression vectors with or without CRX. In Y79 cells receiving an empty vector, RHO, L, and M loci all had a linear configuration (Fig. S5 A and B, −CRX lanes), whereas active RBP3 adopted a looping organization (Fig. S5C, −CRX lanes). When Y79 cells received a CRX expression vector (+CRX lanes), all three OPSIN loci showed active loops, corresponding to CRX-induced OPSIN transcription in these cells (20).
Discussion
Intrachromosomal Looping Represents an Active Conformation for opsin Genes.
We present an important demonstration of physical interactions between opsin enhancer and promoter/coding chromosomal regions in mouse and human photoreceptors. Several pieces of evidence indicate that this chromosomal looping is associated with active transcription. (i) Each opsin preferentially adopts a looping conformation in the photoreceptor subtype actively transcribing that opsin. (ii) Each opsin contains a looping organizer, in most cases, a cis-regulatory region already proven important for transcription: Rho RER and L/M LCR. The exception is S UPs, which is adjacent to the promoter that drives S expression in S cones (31) but whose role in S transcription has not been described. (iii) Looping and transcription are coordinately regulated by specific transcription factors CRX, NRL, and NR2E3, whose mutations alter opsin loop patterns and transcription. Thus, we propose that precisely regulated opsin transcription depends on chromosomal organization, which in turn is regulated by the photoreceptor transcription factor network, as summarized in the models presented in Fig. 4. Three other photoreceptor genes, Rbp3, Gnat1, and Gnat2, also show transcription-related chromosomal looping, suggesting that this mechanism of regulation may be widespread in photoreceptors.
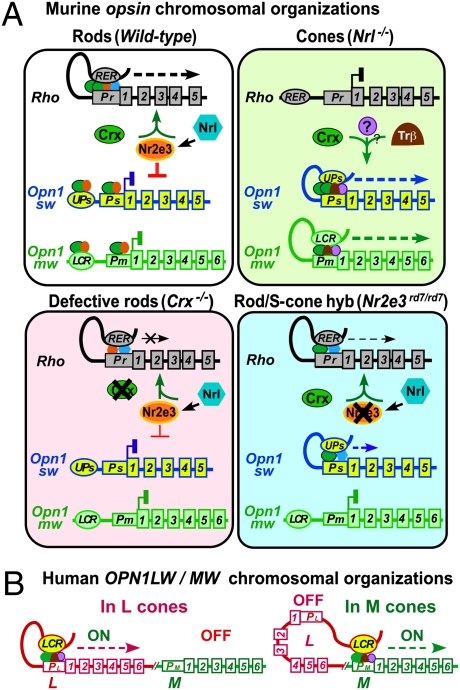
Fig. 4.
Model for opsin chromosomal organizations and cell-type–specific transcription. opsin transcription is regulated by photoreceptor transcription factor-dependent chromosomal organizations. (A) Murine opsins. In rods, Rho RER contacts Rho promoter Pr in the presence of CRX, NRL, and NR2E3 to form an initiation loop, which moves 3′ during transcriptional elongation. M and S opsin loci, silenced by NR2E3, exist in a linear organization. In cones, although silent Rho exists in a linear organization, active M and S form initiation loops where LCR or UPs contacts the respective promoter in the presence of CRX and cone factors, moving 3′ during elongation. In Crx−/− rods, defective transcription factor binding reduces Rho RER–Pr loop formation/extension and transcription. In rd7 rods, loss of NR2E3's dual function leads to defective Rho and S looping organization and hybrid cells expressing both genes. (B) Human L/M loci. In L cones, LCR loops with L promoter to allow transcription of L but not M. In M cones, LCR loops with M promoter to activate M transcription. CRX-associated photoreceptor transcription factors bound to LCR and the respective promoter are critical for establishing LCR loops during transcription initiation and elongation.
Stochastic Events and Rate-Limiting Steps in Loop Formation/Expansion and Transcription.
Each chromosomal loop detected by 3C represents the net result from a population of cells. We propose that initial contact between a looping organizer and promoter is a stochastic event, but stabilizing this loop requires association of transcription factors and the RNA polymerase II (Pol II) preinitiation complex, which is a rate-limiting step. The intragenic loops, on the other hand, reflect dynamic expansion of the initiation loop toward the 3′ end during transcription elongation, as shown in the Fig. 4 models. The 5′-to-3′ gradient of the intragenic loop frequencies suggests additional rate-limiting steps that cause loop pauses at each exon during elongation. Coupling transcription with pre-mRNA processing (32) may account for one such rate limit.
Role of Cis- and Trans-Acting Factors in Establishing opsin Intrachromosomal Loops.
Using ChIP-qPCR, we show that three transcription factors, CRX, NRL, and NR2E3, differentially bind to rod vs. cone opsin genes in a subtype-specific manner. CRX preferentially binds to each opsin in the respective photoreceptor subtype expressing it, in agreement with CRX ChIP-seq results (33) and consistent with CRX's role as a trans-activator. In contrast to the discrete binding sites detected by ChIP-seq, ChIP-qPCR revealed that CRX is bound along the entire length of each active opsin locus. This discrepancy likely reflects the different sensitivity, resolution, quantification methods, and background noise levels of these two techniques (34) as well as the ages of mice assayed, postnatal day 14 (P14) for ChIP-qPCR vs. 8 wk for ChIP-seq. We also show that CRX binds at higher frequencies to enhancer–promoter than intragenic regions, suggesting a role for CRX in stabilizing the enhancer–promoter initiation loop. CRX bound in the intragenic regions is likely associated with the transcription elongation complex moving toward 3′ as the loop expands during transcription. Supporting this hypothesis, loss of CRX significantly reduced Rho enhancer–promoter/E1 loops, and Rho intragenic loops are below detectable levels (Fig. 1C).
ChIP-qPCR revealed different binding patterns for NRL and NR2E3 on each opsin. NRL preferentially binds to Rho, but not to M and S opsin, suggesting that NRL specifies rod fate by directly activating rod genes but indirectly repressing cone genes. In contrast, NR2E3 binds to both rod and cone opsins in rods, supporting its major role in repressing cone opsins (12, 14). Loss of NR2E3 leads to S looping and enhanced NRL/CRX binding in rods, suggesting a unique role for NR2E3 in regulating S accessibility for CRX/NRL. Thus, the linear organization of silent S might result from an active repression mechanism involving epigenetic regulation. However, loss of NR2E3 does not affect M accessibility for CRX/NRL, chromatin organization, and transcription, indicating that silencing M in rods involves some other mechanism, possibly mediation by the lack of M-specific trans-activator(s) such as ligand-bound TRβ1/β2 (35). Surprisingly, in Nrl−/− retinas, we detected only a moderate (30%) increase in M transcription despite its full-scale looping. Nrl−/−-transfated cones thus have configured M for active transcription, but a limited number of M-specific activators may prevent M overexpression. Nonetheless, rod transcription factors are involved in regulating epigenetic status of all opsin genes, and both epigenetic and nonepigenetic transcription factor-mediated mechanisms are required for appropriate photoreceptor gene transcription.
LCR Preferentially Loops with L over M Gene in Humans.
As depicted in Fig. 4B, the mutually exclusive expression of human L and M genes is thought to be determined by distance-dependent competition between L and M promoters for LCR. Our 3C results are consistent with this model. The near 3:1 ratio of LCR-L vs. LCR-M loops is similar to the reported mean L/M cone ratio for Caucasian males with normal color vision (36, 37), raising the possibility that the L/M local chromatin organization determines L vs. M cone opsin expression and differentiation. However, because 3C is unable to distinguish between signals from different cone subtypes, and only two males of unknown color vision phenotype were tested, additional studies on more male donors with normal color vision are needed. Finally, although other transcription factors may also contribute to L/M looping, the Y79 transfection assays show that CRX is essential for this regulation.
Altogether, our findings indicate that intrachromosomal looping is dynamically coupled to photoreceptor gene transcription via mechanisms that are conserved among mammals. This model may also provide insights for understanding cell-type–specific gene expression in other neuronal subtypes in the central nervous system and elsewhere.
Materials and Methods
Complete details of all procedures are described in SI Materials and Methods. Briefly, BAC clones were purchased from BACPAC Resource Center (http://bacpac.chori.org/). Mice were used in accordance with a protocol approved by the Washington University Animal Care and Use Committee. Human cadaver retinas were gifts from David Beebe (Washington University). 3C assays on mouse or human retinas were performed based on a protocol described by Hagège et al. (30) with modifications. Predicted 3C PCR product sizes in base pairs are listed in Table S4. Quantitative RT-PCR (qRT-PCR), ChIP, and Y79 transient transfection assays were performed essentially as described by Peng and Chen (20). Primer sets for genes are listed in Table S5.
Supplementary Material
Acknowledgments
We thank Dr. Anand Swaroop and Dr. Connie Cepko for providing Nrl−/− and Crx−/− mice; Hui Wang for technical assistance; and Anne Hennig, Nicholas Tran, and other members of S.C.'s laboratory for their comments on the manuscript. This work was supported by National Institutes of Health Grant EY12543 (to S.C.), National Institutes of Health Grant EY02687 (to Department of Ophthalmology and Visual Sciences, Washington University School of Medicine), a Lew Wasserman Merit Award (to S.C.), and unrestricted funds from Research to Prevent Blindness (to Department of Ophthalmology and Visual Sciences, Washington University School of Medicine).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109209108/-/DCSupplemental.
References
- 1.Bumsted K, Hendrickson A. Distribution and development of short-wavelength cones differ between Macaca monkey and human fovea. J Comp Neurol. 1999;403:502–516. [PubMed] [Google Scholar]
- 2.Xiao M, Hendrickson A. Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones. J Comp Neurol. 2000;425:545–559. [PubMed] [Google Scholar]
- 3.Applebury ML, et al. The murine cone photoreceptor: A single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 4.Röhlich P, van Veen T, Szél A. Two different visual pigments in one retinal cone cell. Neuron. 1994;13:1159–1166. doi: 10.1016/0896-6273(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 5.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 10.Swaroop A, et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mears AJ, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, et al. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 14.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 15.Akhmedov NB, et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng L, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 18.Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor γ is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- 19.Fujieda H, Bremner R, Mears AJ, Sasaki H. Retinoic acid receptor-related orphan receptor α regulates a subset of cone genes during mouse retinal development. J Neurochem. 2009;108:91–101. doi: 10.1111/j.1471-4159.2008.05739.x. [DOI] [PubMed] [Google Scholar]
- 20.Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navas PA, et al. Activation of the β-like globin genes in transgenic mice is dependent on the presence of the β-locus control region. Hum Mol Genet. 2002;11:893–903. doi: 10.1093/hmg/11.8.893. [DOI] [PubMed] [Google Scholar]
- 22.Ayyagari R, et al. Bilateral macular atrophy in blue cone monochromacy (BCM) with loss of the locus control region (LCR) and part of the red pigment gene. Mol Vis. 1999;5:13. [PubMed] [Google Scholar]
- 23.Wang Y, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 24.Smallwood PM, Wang Y, Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc Natl Acad Sci USA. 2002;99:1008–1011. doi: 10.1073/pnas.022629799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc Natl Acad Sci USA. 1999;96:5251–5256. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Z, Chen S, Kumar R, Zack DJ. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem. 1996;271:2667–2675. doi: 10.1074/jbc.271.5.2667. [DOI] [PubMed] [Google Scholar]
- 27.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 28.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 29.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 30.Hagège H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor β. Mol Endocrinol. 2006;20:1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 32.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbo JC, et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onishi A, Peng GH, Chen S, Blackshaw S. Pias3-dependent SUMOylation controls mammalian cone photoreceptor differentiation. Nat Neurosci. 2010;13:1059–1065. doi: 10.1038/nn.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll J, Neitz J, Neitz M. Estimates of L:M cone ratio from ERG flicker photometry and genetics. J Vis. 2002;2:531–542. doi: 10.1167/2.8.1. [DOI] [PubMed] [Google Scholar]
- 37.McMahon C, Carroll J, Awua S, Neitz J, Neitz M. The L:M cone ratio in males of African descent with normal color vision. J Vis. 2008;8:5.1–5.9. doi: 10.1167/8.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.