Complexes of ruthenium have recently entered clinical trials owing to their potent anticancer activity (1), and those that are photoactive have shown great promise over the past 4 decades in a variety of applications that involve their interaction with DNA, including sensing, signaling, diagnostics, and therapeutics (2, 3). Numerous publications have appeared that focus on the determination of the manner in which these types of complexes interact with dsDNA using various techniques, but definitive crystal structure data with atomic resolution have only now been collected for the noncovalent interaction of duplex DNA with a polypyridyl ruthenium complex, Λ-[Ru(TAP)2(dppz)]2+ (TAP, 1,4,5,8-tetraazaphenathrene; dppz, dipyrido[3,2-a:2′,3′-c]phenazine), appearing in PNAS (4).
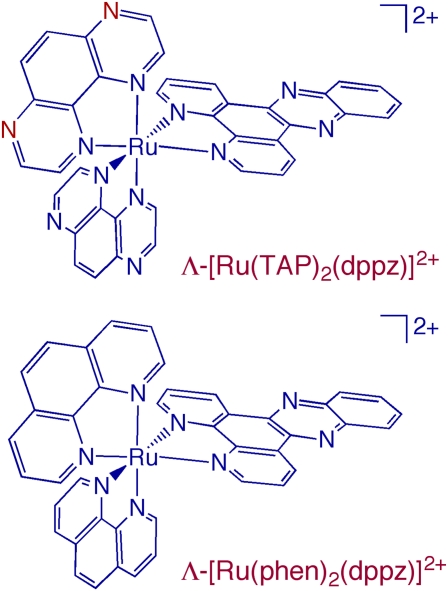
It is evident from Fig. 1 that Λ-[Ru(TAP)2(dppz)]2+ is structurally very similar to Λ-[Ru(phen)2(dppz)]2+ (phen, 1,10-phenanthroline), because the TAP and phen ligands differ only by the replacement of two nitrogen atoms in the former (shown in red) with –CH groups in the latter. [Ru(phen)2(dppz)]2+ was dubbed the “DNA light-switch” complex because it is nonemissive in aqueous media but is strongly luminescent in the presence of dsDNA (5). This complex has also been crucial for gaining understanding of charge transport over long distances mediated by dsDNA (2). The photophysical properties and DNA interactions of [Ru(phen)2(dppz)]2+ have been investigated extensively for the past 2 decades (5). Although the fact that the dppz ligand of this complex intercalates between the DNA bases was not disputed, much work was conducted to address questions concerning the possibility of the existence of additional binding modes, to elucidate structural details of such interactions, and to determine whether the binding took place from the major or minor groove. The unexpected DNA binding motif of Λ-[Ru(TAP)2(dppz)]2+ unveiled in the crystal structure may lead to improved understanding of earlier data collected for [Ru(phen)2(dppz)]2+ and will undoubtedly build the framework for further studies. It will also likely provide insight into how related complexes bind and react with DNA, as well as the observed cellular responses to such compounds.
Fig. 1.
Schematic representation of the molecular structures of Λ-[Ru(TAP)2(dppz)]2+ and Λ-[Ru(phen)2(dppz)]2+.
The crystal structure of Λ-[Ru(TAP)2(dppz)]2+ with the duplex of the self-complementary sequence d(TCGGCGCCGA) reveals two different noncovalent binding modes in a 1:1 (complex:single strand) stoichiometry. Each Ru(II) complex is bound to the duplex through intercalation of the dppz ligand but unexpectedly also interacts with another double-stranded oligonucleotide by partial intercalation of one of its TAP ligands. In this manner, each Λ-[Ru(TAP)2(dppz)]2+ complex is simultaneously bound to two duplexes. The intercalation of the dppz ligand occurs at the terminal G9–A10 steps, although the adenine is flipped out and forms a reverse Watson–Crick base pair with T1 of another duplex. This intercalative mode is expected from numerous studies performed with this complex and the structurally related [Ru(phen)2(dppz)]2+ (Fig. 1). Whereas the dppz is intercalated at G9–A10 of one duplex, one of the TAP ligands of the complex is partially intercalated between G3 and G4 bases of another duplex. An important point of the findings is that this partial intercalation results in significant kinking of the duplex, 51°, and this distortion is structurally very similar to that of the major covalent intrastrand adduct of cisplatin, 1,2-GpG cross-link, with bound HGM1 (6). Another point of interest is that each ruthenium complex is bound simultaneously to two duplexes. Although this may be due to packing forces in the crystal, it may be relevant to conditions in the cell nucleus, where high concentrations of genomic material are present, such that these interactions may also be possible and may play a role in cellular response.
It is well established that the changes to the dsDNA structure effected by platination are crucial for inhibition of transcription and other cellular responses (7, 8). Therefore, it may be expected that if the DNA bending that results from the binding of Λ-[Ru(TAP)2(dppz)]2+ is thermodynamically and kinetically stable (slow on/off rates), the complex may also elicit biological consequences similar to those of platinum drugs. Although many ruthenium complexes being investigated for therapeutic applications often include an intercalating ligand, such as dppz, the crystal structure by Hall et al. (4) indicates that perhaps the focus should be shifted away from intercalators if inhibition of transcription is a desired goal. With the knowledge that semiintercalation results in such double-helix deformation, new complexes may now be designed to maximize this particular interaction.
In fact, the DNA bending that results from semiintercalation discovered by Hall et al. (4) may be able to explain some prior results on the inhibition of transcription by related octahedral rhodium(III) complexes. In one example, the nonintercalating [Rh(bqdi)2(phen)]3+ (bqdi, 1,2-benzoquinone diimine) inhibited transcription in vitro nearly as well as the intercalating complex [Rh(phi)2(phen)]3+ (phi, 9,10-phenanthrenequinone diimine) (9). The transcription inhibition by the two complexes was 12- and 30-fold better than for the corresponding [Rh(phen)2(L)]3+ (L, bdqi, phi), respectively. These results could not be easily explained at the time; however, it is possible that the phen ligand in the [Rh(L)2(phen)]3+ (L, bqdi, phi) complexes semiintercalates in a manner similar to TAP and bends the double helix, leading to suppression of transcription. This interaction may be stabilized by hydrogen bonding from the diimine ligands that is not present in the bis–phen compounds. The inhibition of transcription in vitro observed for dirhodium complexes possessing polypyridyl ligands may also require reinvestigation, because semiintercalation is also possible in these systems (10, 11).
In contrast to the bending of the double helix associated with inhibition of transcription, undisrupted nucleobase π-stacking interactions are required for long-range charge transport (CT) mediated by DNA (2). There is now evidence that long-range DNA CT may play a role in important cellular functions, including signaling that involves DNA-binding proteins and protection of the genome from oxidative damage (2, 12, 13). When the redox-active transition metal complexes [Ru(phen)2(dppz)]2+ and [Rh(phi)2(phen)]3+ are covalently tethered to the ends of a duplex, only the intercalation of the dppz and phi ligands, respectively, is expected (2). In systems where the transition metal complexes are free in solution, however, semiintercalation that disrupts the DNA π-stack may be possible. It could then be envisioned that semiintercalating complexes may be designed specifically to disrupt the biological cellular protection mechanisms and may thus act as therapeutics when targeted to disease, such as tumors.
TAP complexes of ruthenium, including [Ru(TAP)2(dppz)]2+, are able to photochemically oxidize and cross-link DNA
The crystal structure by Hall et al. indicates that perhaps the focus should be shifted away from intercalators.
(14). The latter is a consequence of initial guanine oxidation by the excited state of the complex, whereby the covalent adduct of one of the TAP ligands and guanine is generated upon irradiation in the related complex [Ru(TAP)2(phen)]2+ (14). The semiintercalation of one of the TAP ligands between adjacent guanine bases discovered in the crystal structure by Hall et al. (4) may be important in the interpretation of the earlier cross-linking results and in the design of new, more efficient complexes. Such agents for photoinduced covalent DNA modification and cross-linking may prove useful as photodynamic therapy agents (15).
If [Ru(phen)2(dppz)]2+ is able to engage in DNA interactions similar to those of the corresponding TAP complex, some reinterpretation of prior work on the subject will be required. In addition, new related complexes may be designed such that they target DNA but do not require intercalation, instead seeking structural motifs that enhance the stabilization of the semiintercalative binding mode. Overall, the work by Hall et al. (4) represents a critical advance that is of importance in fields that include interactions of transition metal complexes with DNA, sensing and signaling, chemotherapy, and photodynamic therapy.
Acknowledgments
Work on the photochemistry and interactions of ruthenium complexes with nucleic acids in my laboratory is supported by National Science Foundation Grant CHE-0911354.
Footnotes
The author declares no conflict of interest.
See companion article on page 17610.
References
- 1.Antonarakis ES, Emadi A. Ruthenium-based chemotherapeutics: Are they ready for prime time? Cancer Chemother Pharmacol. 2010;66:1–9. doi: 10.1007/s00280-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genereux JC, Boal AK, Barton JK. DNA-mediated charge transport in redox sensing and signaling. J Am Chem Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao R, et al. Nuclear targets of photodynamic tridentate ruthenium complexes. Dalton Trans. 2009;48:10926–10931. doi: 10.1039/b913959a. [DOI] [PubMed] [Google Scholar]
- 4.Hall PH, et al. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc Natl Acad Sci USA. 2011;108:17610–17614. doi: 10.1073/pnas.1108685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeglis BM, Pierre VC, Barton JK. Metallo-intercalators and metallo-insertors. Chem Commun (Camb) 2007;(44):4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 7.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 9.Fu PKL, Bradley PM, Turro C. Stabilization of duplex DNA structure and suppression of transcription in vitro by bis(quinone diimine) complexes of rhodium(III) and ruthenium(II) Inorg Chem. 2003;42:878–884. doi: 10.1021/ic020338p. [DOI] [PubMed] [Google Scholar]
- 10.Aguirre JD, Lutterman DA, Angeles-Boza AM, Dunbar KR, Turro C. Effect of axial coordination on the electronic structure and biological activity of dirhodium(II,II) complexes. Inorg Chem. 2007;46:7494–7502. doi: 10.1021/ic700708g. [DOI] [PubMed] [Google Scholar]
- 11.Aguirre JD, et al. Anticancer activity of heteroleptic diimine complexes of dirhodium: A study of intercalating properties, hydrophobicity and in cellulo activity. Dalton Trans. 2009;48:10806–10812. doi: 10.1039/b915357h. [DOI] [PubMed] [Google Scholar]
- 12.Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton JK, Olmon ED, Sontz PA. Metal complexes for DNA-mediated charge transport. Coord Chem Rev. 2011;255:619–634. doi: 10.1016/j.ccr.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias B, Kirsch-De Mesmaeker A. Photo-reduction of polyazaaromatic Ru(II) complexes by biomolecules and possible applications. Coord Chem Rev. 2006;250:1627–1641. [Google Scholar]
- 15.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]