Abstract
Parasites that exploit multiple hosts often experience diversifying selection for host-specific adaptations. This can result in multiple strains of host specialists coexisting within a single parasitic species. A long-standing conundrum is how such sympatric host races can be maintained within a single parasitic species in the face of interbreeding among conspecifics specializing on different hosts. Striking examples are seen in certain avian brood parasites such as cuckoos, many of which show host-specific differentiation in traits such as host egg mimicry. Exploiting a Zambian egg collection amassed over several decades and supplemented by recent fieldwork, we show that the brood parasitic Greater Honeyguide Indicator indicator exhibits host-specific differentiation in both egg size and egg shape. Genetic analysis of honeyguide eggs and chicks show that two highly divergent mitochondrial DNA lineages are associated with ground- and tree-nesting hosts, respectively, indicating perfect fidelity to two mutually exclusive sets of host species for millions of years. Despite their age and apparent adaptive diversification, however, these ancient lineages are not cryptic species; a complete lack of differentiation in nuclear genes shows that mating between individuals reared by different hosts is sufficiently frequent to prevent speciation. These results indicate that host specificity is maternally inherited, that host-specific adaptation among conspecifics can be maintained without reproductive isolation, and that host specificity can be remarkably ancient in evolutionary terms.
Keywords: coevolution, gentes, phylogeography, W chromosome hypothesis
Species interactions can be powerful generators of evolutionary diversity (1, 2). In host–parasite relationships, different hosts can exert divergent selection for parasitic specialization. Although this sometimes results in speciation in the parasitic lineage (3–5), in other cases multiple strains of host specialists coexist within a single parasitic species. A textbook example is seen in avian brood parasites such as cuckoos, in which the individuals of a single parasitic species specialize on different hosts and show phenotypic differentiation in traits such as host egg mimicry (6, 7). How phenotypically distinct host races [also known as “gentes” in the context of avian brood parasitism (8)] can evolve and be maintained over evolutionary time in the face of interbreeding among conspecifics has long remained paradoxical (9–12).
Genetic data have provided important insights by allowing a test of whether lineages of parasitic females consistently specialize on particular host species. Studies of the common cuckoo Cuculus canorus (13) and the shiny cowbird Molothrus bonariensis (14), both of which show host-specific phenotypic differentiation in egg traits (7, 15), have revealed subtle genetic differentiation in mitochondrial DNA (mtDNA) haplotype frequencies in relation to host use. These results are potentially consistent with the long-standing hypothesis that egg mimicry might be controlled by genes on the maternally inherited W chromosome [Punnett's sex chromosome hypothesis (10, 16)], thereby allowing host-specific adaptation of female lineages despite interbreeding among males and females reared by different hosts. In both cuckoos and cowbirds, however, females with identical or closely related mtDNA haplotypes were associated with different hosts, indicating frequent host shifts over evolutionary time (13, 14). Parentage analyses based on both microsatellite loci (12, 17) and radio telemetry (18) indicate that female common cuckoos occasionally lay eggs in the nest of more than one host species. A recent study showing slight but significant genetic structure among male common cuckoos associated with different hosts (17) suggests an alternative model in which egg mimicry evolves through strong selection on autosomal genes in partially isolated local populations. Thus, the stability and evolutionary longevity of cuckoo gentes and their adaptation to specific hosts remains uncertain.
Clearer patterns of genetic diversification in relation to host use are perhaps most likely in tropical species, which may experience relatively greater climatic stability (19–21) and hence longer periods of host–parasite coevolution (22); the vast majority of avian brood parasites are tropical in distribution (23). In this study, we investigate both phenotypic and genotypic diversification in relation to host use in the Greater Honeyguide Indicator indicator. This Afrotropical species is perhaps best known for its remarkable mutualistic interactions with human honey gatherers (24), but is also an obligate brood parasite that exploits a range of host species at any given geographical location (25). Parasitized hosts suffer high fitness costs because the young honeyguide stabs host young to death as soon as they hatch, using needle-sharp hooks at the tips of its bill (25, 26).
Greater honeyguides parasitize hole-nesting host species that produce eggs of varying color, size, and shape. Host species are drawn primarily from the Coraciiformes and Upupiformes, some of which have eggs that differ in coloration from the invariably white eggs of honeyguides (Fig. 1). Color differences, however, are likely to be imperceptible in the darkness of deep tree holes and terrestrial burrows. A more important aspect of host–parasite matching may be tactile cues provided by egg size and shape (27–29), which we might thus predict to show host-specific specialization. In this study, we first test for evidence of phenotypic differentiation in egg size and shape among honeyguides parasitizing different hosts. We then test for evidence of genetic differentiation in relation to host use, with particular interest in comparing patterns of differentiation in maternally inherited mtDNA vs. biparentally inherited nuclear DNA. Fidelity of female lineages to particular hosts combined with assortative mating of males and females raised by the same host species would be reflected in both genomes and could lead to speciation. In contrast, if host-specific female lineages mate randomly with respect to the host species of males, we would expect to find host-related genetic structure in mtDNA only.
Fig. 1.
Host and Greater Honeyguide eggs from representative parasitized clutches for each host species in Zambia. Host eggs are in the left column and parasite eggs are in the right column. Subtle host-specific differentiation in honeyguide egg size and shape (Fig. 2) is visible even to the human eye: note the small honeyguide egg from a little bee-eater nest and the rounder egg from a striped kingfisher nest. Clutches were selected from near the middle of the honeyguide egg size and shape distributions. (Photos courtesy of M. D. Anderson, B. Danckwerts, and W. R. Tarboton.)
Results
Are Greater Honeyguide Eggs Specialized for Different Hosts?
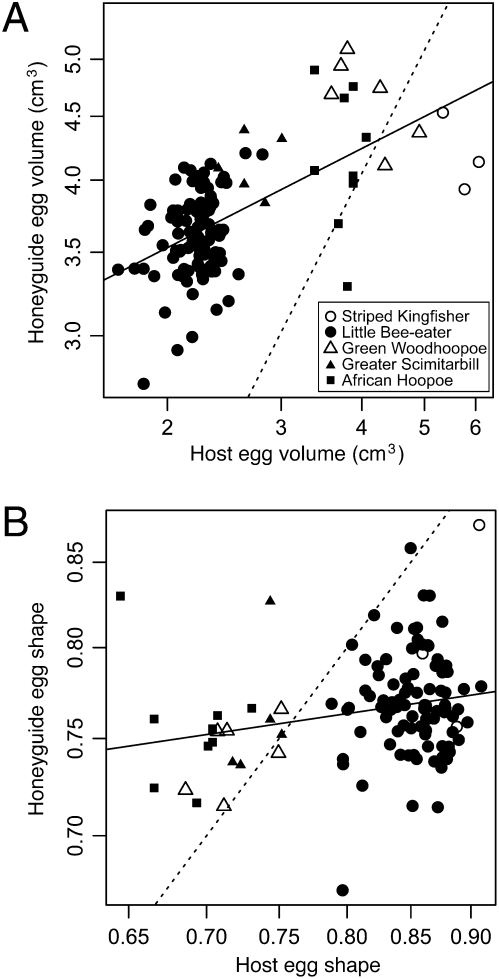
The five common host species at our study site in Zambia (Methods) occurred syntopically and varied markedly in both egg size (ANOVA: R2 = 0.906, F = 447.294,185, P < 0.001) and egg shape (R2 = 0.842, F = 245.584,185, P < 0.001; Table S1). Correspondingly, Greater Honeyguide eggs laid in the nests of different hosts also varied significantly in size and shape (volume: R2 = 0.507, F4,112 = 28.83, P < 0.001; shape: R2 = 0.142, F4,112 = 4.63, P = 0.002; Table S1), and this variation was positively correlated with variation in host egg traits (generalized estimating equations: volume: slope ± SE = 0.264 ± 0.049, Z = 5.37, P < 0.001; shape: slope ± SE = 0.111 ± 0.035, Z = 3.21, P = 0.001; Fig. 2). Although subtle, these correlations are perceptible to the human eye (Fig. 1). Thus, a single parasitic species has apparently evolved host-specific variation in traits highly relevant to host–parasite interactions (27–29), suggesting the existence of phenotypically differentiated host races or gentes in the Greater Honeyguide.
Fig. 2.
Size (A) and shape (B) of Greater Honeyguide eggs in relation to those of host eggs in the same nest. Data points are individual parasitized clutches from the Choma District of Zambia, with different host species indicated (A, Inset). The regression lines (solid lines) result from generalized estimating equations that take into account nonindependence of parasitized clutches within host species. The dotted lines show the expectation if there were perfect correspondence between parasite and host egg traits, emphasizing that honeyguides do not perfectly match their hosts’ eggs. Note that the axes are displayed on a log scale so the untransformed units are displayed.
Do Greater Honeyguides Show mtDNA Genetic Divergence in Relation to Host Use?
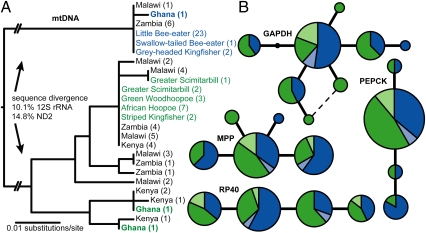
To test for genetic divergence among honeyguide females parasitizing different host species, we sequenced mtDNA (Methods) from samples of known host origin: shell membranes from honeyguide eggs in parasitized clutches and blood or tissue from parasitic nestlings (Table S2). We found that two highly divergent mtDNA lineages (genetic distance = 14.8% for the ND2 gene) were perfectly associated with two groups of host species (Fig. 3A): those breeding in terrestrial burrows (principally the little bee-eater Merops pusillus, but also the swallow-tailed bee-eater M. hirundineus and the gray-headed kingfisher Halcyon leucocephala) and those breeding in tree cavities (the green woodhoopoe Phoeniculus purpureus, greater scimitarbill Rhinopomastus cyanomelas, African hoopoe Upupa africana, and striped kingfisher Halcyon chelicuti). This implies that female honeyguide lineages have been perfectly faithful to ground- or tree-nesting hosts, respectively, for millions of years (Discussion).
Fig. 3.
Mitochondrial and nuclear DNA relationships among honeyguides using different host species. (A) Mitochondrial phylogeny based on partial 12S rRNA gene sequences. Genetic divergence for the ND2 gene was measured for a representative sample of individuals with divergent 12S sequences. (B) Haplotype networks for four nuclear loci. Area is proportional to allele frequency; each line represents a single difference in DNA sequence; dark blue and dark green are birds from Zambia with divergent mtDNA lineages (n = 28 individuals, including 17 individuals of confirmed host origin); light blue and light green are birds from Ghana (n = 3). ΦST values reported in the text are based on samples from Zambia only. A fifth nuclear locus (TGFB2) had three rare SNPs, each present in only one or two of the sampled individuals.
Do Greater Honeyguides Show Nuclear DNA Genetic Divergence in Relation to Host Use?
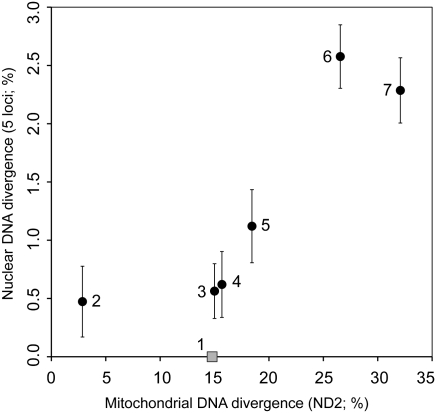
We speculated that the highly divergent mtDNA lineages in Greater Honeyguides might represent previously overlooked, cryptic species and thus expected to find a commensurate level of host-specific divergence at nuclear loci (Methods). Surprisingly, however, Greater Honeyguides at our Choma, Zambia, study site belonging to divergent mtDNA lineages showed no evidence of host-specific sequence divergence or differentiation in allele frequencies at five nuclear loci (ΦST = ∼0.0, P > 0.19; Fig. 3B). The data instead suggest that host-specialist Greater Honeyguide females mate randomly with respect to the host origin of males. Recently diverged populations typically show lower levels of genetic differentiation at nuclear loci compared with mtDNA due to different rates of lineage sorting between the two genomes. To test whether the lack of nuclear differentiation that we observed is simply an artifact of lineage sorting, we compared the relative levels of mtDNA and nuclear divergence in Greater Honeyguides with that observed in six additional pairs of honeyguide lineages that are comparably divergent in mtDNA sequence. Fig. 4 shows that the observed depth of mtDNA divergence between Greater Honeyguide lineages is similar to or larger than that observed between other honeyguide species pairs, which also show relatively higher levels of nuclear divergence. This result confirms that the lack of nuclear divergence in Greater Honeyguides is not simply the result of different rates of lineage sorting for nuclear and mtDNA.
Fig. 4.
Comparison of mitochondrial and nuclear divergence between mitochondrial sister clades in the honeyguide phylogeny. Nuclear divergence (±SEM) is corrected for within group nucleotide diversity. (1) Divergent Greater Honeyguide Indicator indicator lineages; (2) scaly-throated honeyguide I. variegatus vs. spotted honeyguide I. maculatus; (3) Willcocks's honeyguide I. willcocksi vs. lesser I. minor and least honeyguides I. exilis; (4) yellow-footed honeyguide Melignomon eisentrauti vs. Zenker's honeyguide M. zenkeri; (5) Wahlberg's honeyguide Prodotiscus regulus vs. green-backed honeyguide P. zambesiae; (6) scaly-throated and spotted honeyguides vs. Willcocks's and lesser and least honeyguides; and (7) Malaysian honeyguide I. archipelagicus vs. Greater Honeyguide.
Did Greater Honeyguide Lineages Evolve in Allopatry?
One possible explanation for the origin and maintenance of the two divergent Greater Honeyguide mtDNA lineages is that they evolved in allopatry and only recently came into contact in the region of our study site in Zambia. To test this, we sampled Greater Honeyguide museum skins from Zambia as well as from other parts of Africa (Malawi, Kenya, and Ghana). In three of these four areas (Zambia, Malawi, and Ghana), we recovered both mtDNA lineages (Fig. 3A). The presence of both lineages in both southern Africa (Zambia, Malawi) and ∼4,000 km away in West Africa (Ghana) represents a highly unusual phylogeographic pattern that renders a simple allopatric explanation unlikely (30). In addition, nuclear polymorphisms are shared both between individuals with divergent mtDNA lineages and between geographic regions (Fig. 3B). This suggests substantial autosomal gene flow even on a continental scale. Although we cannot fully exclude the possibility that the two lineages initially arose in allopatry (cf. 31), secondary contact would have to be relatively ancient, as sufficient time has elapsed for at least one lineage to have crossed the African continent. Regardless of the geographic origins of the two divergent honeyguide lineages, their coexistence in sympatry in Zambia has clearly been maintained by the host specificity of female lineages, likely for considerable periods of evolutionary time.
Discussion
In this study, we have shown that the Greater Honeyguide comprises two deeply divergent female lineages that differ in host use. The data indicate that these female lineages have remained perfectly faithful to one of two host groups for significant stretches of evolutionary time, without any successful switches between the two host groups. In contrast, the complete lack of genetic structure at nuclear loci suggests that these ancient lineages do not represent cryptic species. Male and female honeyguides reared by different hosts must interbreed sufficiently often to prevent divergence at neutral nuclear loci, and ancient host-specific female lineages apparently have been maintained in the absence of any detectable reproductive isolation.
The precise age of the two ancient female-specific lineages is open to debate owing to uncertainty about the mtDNA molecular clock, which has recently been suggested to show accelerated short-term rates of molecular evolution (32). In the absence of independently derived calibration points, and given that Piciformes have relatively high rates of mtDNA sequence evolution (33), we used maximum rate estimates to conservatively estimate a minimum divergence time between the two lineages. Even using the fastest estimated rates of mtDNA sequence evolution in birds (33–35), the 14.8% mtDNA divergence between Greater Honeyguide lineages corresponds to an evolutionary age of ∼3 My. For comparison, the common cuckoo C. canorus host races (gentes) appear to have diverged on the order of ∼80,000 y ago (13) and have shown imperfect host fidelity since that time (13, 17). Hence, even a highly conservative date estimate places the Greater Honeyguide host races an order of magnitude older, providing additional evidence for the age and diversifying potential of species interactions in the tropics (21, 36).
Female Greater Honeyguides specialized either on hosts nesting in tree cavities or on those nesting in terrestrial burrows, the latter requiring the traversal of a long, narrow, subterranean tunnel by the parasitic female. Imprinting on host nest type is a potential behavioral mechanism explaining the fidelity of honeyguide females to one of two host groups. Indeed, structurally and/or ecologically distinct host nest types might be an important factor generating diversifying selection in generalist brood parasites (14, 37).
The perfect host fidelity of Greater Honeyguides over the course of millions of years should provide precisely the circumstances under which host-specific adaptations could accumulate and be stably passed on via the female-specific W chromosome. A direct test of Punnett's sex chromosome hypothesis (10, 16) would require controlled breeding experiments and/or extensive pedigree data for individuals of known egg traits, neither of which has thus far been possible in any brood parasitic species. Nonetheless, there is clearly much greater potential for sex-linked divergence in honeyguides than in parasitic birds that show only subtle host-specific genetic differentiation (13, 14, 17).
Although Greater Honeyguides meet the necessary conditions of Punnett's sex chromosome hypothesis better than other brood parasites studied to date, phenotypic differences between historically divergent matrilines do not fully explain the host-specific differentiation in egg traits that we observed. Significant phenotypic diversification in egg traits is also evident within the honeyguide lineage specializing on tree-hole–nesting hosts (volume: R2 = 0.331, F3,21 = 3.46, P = 0.035; shape: R2 = 0.328, F3,21 = 3.42, P = 0.036). Additional mechanisms may be needed to explain this host-specific divergence in egg phenotype within a single female lineage. Differences might reflect phenotypic plasticity if honeyguides raised by larger hosts attain larger body size and subsequently lay larger eggs. We genotyped 12 adult or free-flying juvenile honeyguides with known body masses and, after controlling for sex, found that birds in the “ground-nesting” mtDNA lineage (n = 6), which is associated with smaller hosts, were significantly lighter than those associated with larger tree-nesting hosts (sex effect: slope ± SE = 10.49 ± 2.54, t = 4.13, P = 0.003; mtDNA lineage effect: slope ± SE = 10.21 ± 2.50, t = 4.08, P = 0.003). This finding, however, cannot explain variation in egg shape within the “tree-nesting” lineage, as there is no consistent overall correlation between egg size and shape either in this sample or among birds in general (38).
In conclusion, our results suggest that lineages of Greater Honeyguide females have for millions of years remained perfectly faithful to hosts with different nesting habits, but have been maintained as a single species through interbreeding with males irrespective of the male's host. These results are consistent with the hypothesis that host-specific adaptations are inherited via the female-specific W chromosome, although additional explanations are needed for the maintenance of phenotypic differentiation within one of the two genetic lineages. More generally, the maintenance of divergent mtDNA lineages within a single population and the co-occurrence of such lineages over a broad geographic area represent a highly unusual phylogeographic pattern (39). Taken together, our results show that female specificity in host use is remarkably strong and maternally inherited, whether genetically or culturally, and that host-specific adaptation within a single parasitic species can be achieved and maintained with little or no reproductive isolation among host races. Finally, the genetic data show that host specificity can be remarkably ancient in evolutionary terms and suggest that host–parasite interactions can generate and maintain cryptic biological diversity.
Methods
Data on Host and Parasite Egg Phenotype.
All data on egg dimensions came from the Choma District of Zambia's Southern Province, within a 28-km radius of Choma town (16°49′S, 26°59′E). We measured eggs collected during 1969–2002 by J.F.R.C.-R. (and subsequently bequeathed to the Natural History Museum, London) and observed in the field during 2008–2009 by C.N.S. The narrow geographical scope precluded confounding geographical variation in body size and host use and ensured that females parasitizing different host species were syntopic even on a relatively fine geographic scale. The habitat in the Choma District consists of broad-leaved woodland dominated by Brachystegia, interspersed with agricultural fields and seasonally flooded grassy depressions; available hosts were both woodland and generalist species. Greater Honeyguides regularly used five host species in this area (Table S1) (40). All bred in tree cavities except for little bee-eaters, which bred exclusively in terrestrial burrows, usually excavated in the roofs of aardvark Orycteropus afer holes. Two additional hosts, the swallow-tailed bee-eater and the gray-headed kingfisher, also bred in terrestrial burrows. Although these species were rarely parasitized, we obtained a few genetic samples of honeyguides that parasitized their nests.
Female Greater Honeyguides lay an estimated 20 eggs per season (41), and the maximum recorded longevity for Greater Honeyguides is 9 y (42), raising the possibility of pseudoreplication arising from multiple eggs laid by the same female. This is not likely to be a widespread problem because parasitized nests were found over a 40-y period in a geographical area spanning 46 × 26 km, which encompasses a large number of honeyguide females. In three cases, however, honeyguide eggs of almost identical dimensions were found in two nests of the same host species within 2 km of each other, 1–6 y apart. It is conservative to assume that the same female honeyguide was responsible for both eggs in each of these cases, so we decided a priori to exclude one randomly selected clutch from each pair; results were similar if they were included. In cases of multiple parasitism (n = 10), all parasitic eggs were included in the analysis, as they were laid by different females (42).
Egg dimensions were measured by J.F.R.C.-R. using Vernier calipers accurate to 0.1 mm and by C.N.S. using digital calipers accurate to 0.01 mm. Measurements taken by different observers and instruments were highly repeatable (43) when remeasured by C.N.S. blind to previous measurements by J.F.R.C.-R. (length: repeatability = 0.999, F19,20 = 2,706, P < 0.001; breadth: repeatability = 0.999, F19,20 = 1,533, P < 0.001). Egg volume was estimated using a general equation (44) (volume = 0.51 × length × breadth2) and log10-transformed. Estimated volume was highly correlated to fresh egg mass where this was available (untransformed variables: Greater Honeyguides: R2 = 0.948, slope ± SE = 0.900 ± 0.036, F1,35 = 632.0, P < 0.001; hosts: R2 = 0.964, slope ± SE = 0.911 ± 0.028, F1,40 = 1,065.2, P < 0.001). Egg shape was taken as the log10-transformed ratio of egg breadth to egg length.
Genetic Data.
We confirmed the identification of honeyguide eggs to species and tested for mitochondrial DNA divergence between honeyguides parasitizing different host species using samples of known host origin. DNA was extracted from eggshell membranes (45), toepads and feathers from nestling skins, fresh eggshell membranes from deserted or damaged eggs, and blood from nestlings, all originating from Zambia and primarily from a ca. 50-km2 region within the Choma District. Eggshell membranes and museum skins were from J.F.R.C.-R.'s collection (bequeathed to the Natural History Museum, London) and collected during 1978–2000; blood and fresh eggshell membranes were collected in the field by C.N.S. during 2008–2009. Samples for other honeyguide species and for Greater Honeyguides from other locations in Africa were obtained from museum tissue collections as detailed in Table S2.
For egg samples, we amplified 294 bp of the mitochondrial small subunit ribosomal RNA (12S) using conserved avian primers. For three egg-membrane samples that initially failed, a shorter, overlapping fragment was amplified. Either fragment was sufficient to unambiguously assign Greater Honeyguide samples to one of two divergent clades. For a subset of individuals with sufficiently high-quality DNA samples (mostly represented by nestling tissue or adult skins; Table S2), we also amplified and sequenced five nuclear introns to test for possible divergence between Greater Honeyguide samples representing the two divergent mtDNA lineages. Samples were assigned to one of two groups on the basis of the mtDNA results as host association was not known for all samples (e.g., skins from adult birds), but ΦST was calculated only for individuals collected near Choma. We also sequenced both mtDNA and nuclear loci for other honeyguide species and for Greater Honeyguide samples from elsewhere in Africa. Finally, we confirmed the sex of Greater Honeyguide museum skins by amplifying and sequencing an intron of the sex-linked CHD-1 gene; we designed Z- and W-chromosome–specific primers for honeyguides on the basis of the initial sequences obtained using published primers (46).
All primer sequences are provided in Table S3. PCR reactions, purification of PCR products, and DNA sequencing followed standard protocols (47). Nuclear alleles were computationally inferred using Phase v. 2.1.1 (48). Phylogenetic analyses and calculation of genetic distances were completed in Paup* v. 4.0b10 (49). Nuclear differentiation, measured as ΦST, and its statistical significance were calculated using Arlequin (50).
Statistical Analyses.
We used parasitized clutches as the unit of analysis, taking the mean of all host eggs in the clutch. We used one-way ANOVAs to test for differences in egg traits between host species and between honeyguide eggs found in the nests of different host species. We used linear models to test for the correlation between the egg traits of honeyguides and those of their hosts. Because sample sizes varied widely among host species, and because our intent was to investigate a possible relationship across host species, we did not treat individual host nests as independent data points. Instead, we used generalized estimating equations (GEEs) (51), which take into account similarity within groups and thus avoid bias from nonindependence within groups. In these analyses, “groups” are host species and individual data points are nest-level egg measures (mean of host eggs; dimensions of honeyguide egg). We used GEEs rather than other mixed-model approaches because GEEs estimate an overall slope across groups, consistent with our goal of testing for a relationship across host species and not within. We used the gee package of R (52), with exchangeable correlation structure and Gaussian error distribution.
Supplementary Material
Acknowledgments
We thank the Zambia Wildlife Authority for research permits; the Bruce-Miller, Counsell, Danckwerts, Duckett, and Green families for their hospitality in Zambia; and many people for finding nests, especially K. Banda, C. Banda, G. Banda L. Hamusikili, K. Moto, C. Moya, A. Munkombwe, C. Munkombwe, R. Munkombwe, S. Munkombwe, G. Siabulanda, F. Sinoya, and D. Siyapolo. We are grateful to T. R. Birkhead, N. B. Davies, and three referees for comments. K. M. Sefc and R. B. Payne shared phylogenetic data for honeyguides. For access to egg and tissue collections, we thank the Natural History Museum, London; the American Museum of Natural History; Louisiana State University Museum of Natural History; the Field Museum of Natural History, Chicago; and the Marjorie Barrick Museum of Natural History, Las Vegas. C.N.S. was supported by a Royal Society Dorothy Hodgkin Research Fellowship and Sidney Sussex and Newnham Colleges, Cambridge, and S.Q. was supported by a Marie Curie fellowship of the European Commission. Genetic analyses were facilitated by a National Science Foundation grant to M.D.S. and the Department of Science and Technology/National Research Foundation Centre of Excellence at the Percy FitzPatrick Institute, University of Cape Town.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109630108/-/DCSupplemental.
References
- 1.Ehrlich PR, Raven PH. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 2.Yoder JB, Nuismer SL. When does coevolution promote diversification? Am Nat. 2010;176:802–817. doi: 10.1086/657048. [DOI] [PubMed] [Google Scholar]
- 3.Sorenson MD, Sefc KM, Payne RB. Speciation by host switch in brood parasitic indigobirds. Nature. 2003;424:928–931. doi: 10.1038/nature01863. [DOI] [PubMed] [Google Scholar]
- 4.Malenke JR, Johnson KP, Clayton DH. Host specialization differentiates cryptic species of feather-feeding lice. Evolution. 2009;63:1427–1438. doi: 10.1111/j.1558-5646.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 5.Forbes AA, Powell THQ, Stelinski LL, Smith JJ, Feder JL. Sequential sympatric speciation across trophic levels. Science. 2009;323:776–779. doi: 10.1126/science.1166981. [DOI] [PubMed] [Google Scholar]
- 6.Baldamus E. A new contribution to the reproductive history of cuckoos, Cuculus canorus. Naumannia. 1853;3:307–325. [Google Scholar]
- 7.Brooke MdL, Davies NB. Egg mimicry by cuckoos Cuculus canorus in relation to discrimination by hosts. Nature. 1988;335:630–632. [Google Scholar]
- 8.Newton A. A Dictionary of Birds. London: Adam & Charles Black; 1896. [Google Scholar]
- 9.Wynne-Edwards V. Inheritance of egg-colour in the parasitic cuckoo. Nature. 1933;132:822. [Google Scholar]
- 10.Punnett RC. Inheritance of egg-colour in the parasitic cuckoos. Nature. 1933;132:892–893. [Google Scholar]
- 11.Pycraft WP. A History of Birds. London: Methuen and Co.; 1910. [Google Scholar]
- 12.Marchetti K, Nakamura H, Gibbs HL. Host-race formation in the common cuckoo. Science. 1998;282:471–472. doi: 10.1126/science.282.5388.471. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs HL, et al. Genetic evidence for female host-specific races of the common cuckoo. Nature. 2000;407:183–186. doi: 10.1038/35025058. [DOI] [PubMed] [Google Scholar]
- 14.Mahler B, Confalonieri VA, Lovette IJ, Reboreda JC. Partial host fidelity in nest selection by the shiny cowbird (Molothrus bonariensis), a highly generalist avian brood parasite. J Evol Biol. 2007;20:1918–1923. doi: 10.1111/j.1420-9101.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- 15.de la Colina MA, Mahler B, Reboreda JC. Differences in morphology and colour pattern of shiny cowbird (Molothrus bonariensis) eggs found in nests of two hosts. Biol J Linn Soc Lond. 2011;102:838–845. [Google Scholar]
- 16.Jensen RAC. Genetics of cuckoo egg polymorphism. Nature. 1966;209:827. [Google Scholar]
- 17.Fossøy F, et al. Genetic differentiation among sympatric cuckoo host races: Males matter. Proc Biol Sci. 2010;278:1639–1645. doi: 10.1098/rspb.2010.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honza M, et al. Behaviour of female common cuckoos, Cuculus canorus, in the vicinity of host nests before and during egg laying: A radiotelemetry study. Anim Behav. 2002;64:861–868. [Google Scholar]
- 19.Roy MS, Sponer R, Fjeldså J. Molecular systematics and evolutionary history of akalats (genus Sheppardia): A pre-Pleistocene radiation in a group of African forest birds. Mol Phylogenet Evol. 2001;18:74–83. doi: 10.1006/mpev.2000.0862. [DOI] [PubMed] [Google Scholar]
- 20.Njabo KY, Bowie RCK, Sorenson MD. Phylogeny, biogeography and taxonomy of the African wattle-eyes (Aves: Passeriformes: Platysteiridae) Mol Phylogenet Evol. 2008;48:136–149. doi: 10.1016/j.ympev.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Dobzhansky T. Evolution in the tropics. Am Sci. 1950;38:209–221. [Google Scholar]
- 22.Rothstein SI. Brood parasitism: The importance of experiments and host defences of avifaunas on different continents. Proceedings of the VII Pan-African Ornithological Congress. 1992:521–535. [Google Scholar]
- 23.Davies NB. Cuckoos, Cowbirds and Other Cheats. London: T & A D Poyser; 2000. [Google Scholar]
- 24.Isack HA, Reyer HU. Honeyguides and honey gatherers: Interspecific communication in a symbiotic relationship. Science. 1989;243:1343–1346. doi: 10.1126/science.243.4896.1343. [DOI] [PubMed] [Google Scholar]
- 25.Friedmann H. The honey-guides. Bull US Natl Mus. 1955;208:1–292. [Google Scholar]
- 26.Spottiswoode CN, Koorevaar J. A stab in the dark: Chick killing by brood parasitic honeyguides. Biol Lett. 2011 doi: 10.1098/rsbl.2011.0739. 10.1098/rsbl.2011.0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason P, Rothstein SI. Coevolution and avian brood parasitism: Cowbird eggs show evolutionary response to host discrimination. Evolution. 1986;40:1207–1214. doi: 10.1111/j.1558-5646.1986.tb05745.x. [DOI] [PubMed] [Google Scholar]
- 28.Marchetti K. Egg rejection in a passerine bird: Size does matter. Anim Behav. 2000;59:877–883. doi: 10.1006/anbe.1999.1388. [DOI] [PubMed] [Google Scholar]
- 29.Langmore NE, Hunt S, Kilner RM. Escalation of a coevolutionary arms race through host rejection of brood parasitic young. Nature. 2003;422:157–160. doi: 10.1038/nature01460. [DOI] [PubMed] [Google Scholar]
- 30.Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- 31.Webb WC, Marzluff JM, Omland KE. Random interbreeding between cryptic lineages of the Common Raven: evidence for speciation in reverse. Mol Ecol. 2011;20:2390–2402. doi: 10.1111/j.1365-294X.2011.05095.x. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian S, et al. High mitogenomic evolutionary rates and time dependency. Trends Genet. 2009;25:482–486. doi: 10.1016/j.tig.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- 34.Moyle RG, Filardi CE, Smith CE, Diamond J. Explosive Pleistocene diversification and hemispheric expansion of a “great speciator.”. Proc Natl Acad Sci USA. 2009;106:1863–1868. doi: 10.1073/pnas.0809861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren BH, Bermingham E, Bowie RCK, Prys-Jones RP, Thébaud C. Molecular phylogeography reveals island colonization history and diversification of western Indian Ocean sunbirds (Nectarinia: Nectariniidae) Mol Phylogenet Evol. 2003;29:67–85. doi: 10.1016/s1055-7903(03)00063-0. [DOI] [PubMed] [Google Scholar]
- 36.Schemske DW. Biotic interactions and speciation in the tropics. In: Butlin R, Bridle J, Schluter D, editors. Speciation and Patterns of Diversity. Cambridge, UK: Cambridge University Press; 2009. pp. 219–239. [Google Scholar]
- 37.Moksnes A, Røskaft E. Egg-morphs and host preferences in the common cuckoo. J Zool (Lond) 1995;236:625–648. [Google Scholar]
- 38.Rahn H, Paganelli CV. Length, breadth and elongation of avian eggs from the tables of Schönwetter. J Ornithol. 1988;129:366–369. [Google Scholar]
- 39.Avise JC. Molecular Markers, Natural History, and Evolution. 2nd Ed. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 40.Spottiswoode CN, Colebrook-Robjent JFR. Eggshell puncturing by the brood parasitic Greater Honeyguide and potential host counteradaptations. Behav Ecol. 2007;18:792–799. [Google Scholar]
- 41.Payne RB. Clutch size, laying periodicity and behaviour in the honeyguides. Proceedings VII Pan-African Ornithological Congress. 1992:537–547. [Google Scholar]
- 42.Short LL, Horne JFM. Toucans, Barbets and Honeyguides. Oxford: Oxford University Press; 2001. [Google Scholar]
- 43.Lessells C, Boag P. Unrepeatable repeatabilities: A common mistake. Auk. 1987;104:116–121. [Google Scholar]
- 44.Hoyt DF. Practical methods of estimating volume and fresh weight of bird eggs. Auk. 1979;96:73–77. [Google Scholar]
- 45.Chilton G, Sorenson MD. Genetic identification of eggs purportedly from the extinct Labrador duck. Auk. 2007;124:962–968. [Google Scholar]
- 46.Kahn NW, St John J, Quinn TW. Chromosome-specific intron size differences in the avian CHD gene provide an efficient method for sex identification in birds. Auk. 1998;115:1074–1078. [Google Scholar]
- 47.Sorenson MD, Payne RB. A single ancient origin of brood parasitism in African finches: implications for host-parasite coevolution. Evolution. 2001;55:2550–2567. doi: 10.1111/j.0014-3820.2001.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 48.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0b10. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 50.Schneider S, Roessli D, Excoffier L. ARLEQUIN Version 2.0: A Software for Genetics Data Analysis. Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- 51.Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 52.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.