Abstract
The 5′ cap structure (m7GpppX-) is an essential feature of eukaryotic mRNA required for mRNA stability and efficient translation. Influenza virus furnishes its mRNA with this structure by a cap-snatching mechanism, in which the viral polymerase cleaves host mRNA endonucleolytically 10–13 nucleotides from the 5′ end and utilizes the capped fragment as a primer to synthesize viral transcripts. Here we report a unique cap-snatching mechanism by which the yeast double-stranded RNA totivirus L-A furnishes its transcript with a cap structure derived from mRNA. Unlike influenza virus, L-A transfers only m7Gp from the cap donor to the 5′ end of the viral transcript, thus preserving the 5′ α- and β-phosphates of the transcript in the triphosphate linkage of the final product. This in vitro capping reaction requires His154 of the coat protein Gag, a residue essential for decapping of host mRNA and known to form m7Gp-His adduct. Furthermore, the synthesis of capped viral transcripts in vivo and their expression were greatly compromised by the Arg154 mutation, indicating the involvement of Gag in the cap-snatching reaction. The overall reaction and the structure around the catalytic site in Gag resemble those of guanylyltransferase, a key enzyme of cellular mRNA capping, suggesting convergent evolution. Given that Pol of L-A is confined inside the virion and unable to access host mRNA in the cytoplasm, the structural protein Gag rather than Pol catalyzing this unique cap-snatching reaction exemplifies the versatility as well as the adaptability of eukaryotic RNA viruses.
Keywords: transcription, Saccharomyces cerevisiae, killer toxin
Eukaryotic mRNA is capped at the 5′ end by a 7-methyl GMP moiety via an inverted 5′-5′ triphosphate linkage (m7GpppX) (1, 2). The cap structure is required for mRNA stability and efficient translation. Capping of the RNA 5′ end is accomplished in cells and for most viruses in three sequential catalytic reactions (3, 4): removal of the 5′ γ-phosphate by RNA triphosphatase, addition of GMP from GTP by guanylyltransferase, and methylation of the added GMP by methyltransferase. Influenza virus, however, employs a different mechanism (cap-snatching) to furnish its mRNA with the structure (5–7). The trimeric viral polymerase binds the cap of host mRNA, cleaves the RNA endonucleolytically 10–13 nucleotides downstream, and utilizes the capped fragment as a primer to synthesize its transcript. So far, only negative stranded RNA viruses and ambiviruses (the Orthomyxoviridae, Bunyaviridae, and Arenaviridae families) have been known to use this strategy to furnish their mRNAs.
The totivirus L-A, which infects the yeast Saccharomyces cerevisiae, has a nonsegmented dsRNA genome of 4.6 kb (8). Typical of fungal viruses, L-A has no extracellular transmission pathway. This virus is transmitted vertically from mother to daughter cells, or horizontally through mating. The L-A genome contains two overlapping genes, gag and pol, and the latter is expressed as a Gag-Pol fusion protein by a −1 ribosomal frameshift (9, 10). The genome is packed inside of a 39-nm icosahedral capsid consisting of 60 asymmetric Gag dimers, in which one or two Gag molecules are substituted by Gag-Pol. The N-terminal Pol region is necessary for genome packaging, although Gag alone is sufficient to form morphologically normal capsids (11). M1, a satellite RNA of L-A, has a dsRNA genome (1.6–1.8 kb) that encodes a protein toxin and immunity (12) but no proteins necessary for its own replication. It requires L-A for encapsidation and replication. Thus M1 can be maintained in the cell without the helper virus provided that L-A proteins are expressed from a vector (13). Two decades ago Blanc et al. (14) found that the L-A coat protein Gag covalently binds the cap structure of mRNA. The reaction was inhibited by the cap analogue m7GpppG but not by the nonmethylated GpppG. The subsequent study revealed that m7Gp decapped from mRNA covalently attached to His154 of Gag (15). Site-directed mutagenesis found His154 essential for decapping of mRNA, but its mutation (Arg154) did not affect transcription, replication, and encapsidation of viral RNA (15). L-A virions can synthesize positive strand transcripts in vitro in a conservative manner (16). Recently we found that L-A and M1 transcripts have diphosphate at their 5′ termini (5′-ppGAAAAAU…; L-A and M1 share the same 7-nt sequences at the 5′ ends) (17). More strikingly, when transcription was primed with GTP or GMP, the transcripts again bore the diphosphate at the 5′ ends. In the latter case, the 5′ β-phosphate was derived from the γ-phosphate of ATP present in the reaction. Therefore, L-A virus deliberately keeps its transcripts diphosphorylated at the 5′ ends.
We speculated that, if m7Gp derived from host mRNA was transferred to the diphosphorylated 5′ end of the viral transcript, it would produce an authentic cap structure in yeast. We tested this hypothesis. Here we describe a unique cap-snatching mechanism in the dsRNA virus L-A. L-A only transfers the m7Gp moiety derived from mRNA to its transcript. We also demonstrate that this capping reaction is essential for efficient expression of the viral transcript.
Results
Cap Transfer Reaction.
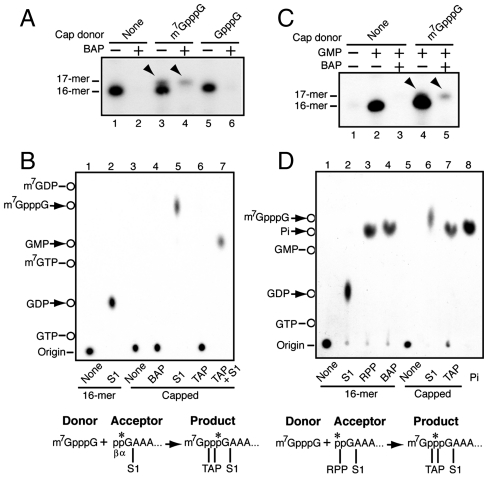
A 5-nt cap donor, 32p-labeled at the γ-phosphate of the triphosphate linkage, was incubated with L-A virions in a transcription mixture containing the 4 NTPs. The full-length L-A transcript incorporated the radioactivity (Fig. 1A, lane 1). To further analyze the reaction, CTP was omitted and GTP was replaced by GDP. Previously we observed that GDP is a better substrate to prime transcription than GTP (17). Because the first C appears at position 17 in the L-A transcript, the virions synthesize a 16-nt L-A fragment in the absence of CTP. L-A virions can also incorporate a cap analogue (m7GpppG) as a primer and produced a 17-nt transcript (17). In the presence of the cap donor, GDP-primed transcripts incorporated the label, which moved in a polyacrylamide gel slightly slower than the 16-nt transcript but as fast as a cap analogue (m7GpppG)-primed 17-nt transcript did (Fig. 1B, lanes 4–6). The omission of GDP abrogated the incorporation, indicating that the reaction is transcription-dependent and that the cap donor (or part of it) was not used as a primer (Fig. 1B, lane 3). The labeled product was isolated from the gel and analyzed by TLC. Bacterial alkaline phosphatase (BAP) treatment did not release radioactivity (Fig. 1C, lane 7), indicating that the product was internally labeled. Nuclease S1 treatment digested the RNA body but could not work on the triphosphate linkage, thus releasing the labeled cap analogue (m7GpppG) (Fig. 1C, lane 5). Tobacco acid pyrophosphatase (TAP) treatment released labeled m7Gp (Fig. 1C, lane 6). Because TAP can hydrolyze anhydrous bonds between α- and β-, and also β- and γ-phosphates of the triphosphate linkage, it indicates that the label was located at the γ-phosphate in the capped product. These results demonstrate that, at least, the m7Gp moiety of the cap donor was transferred to the 5′ end of the viral transcript and formed a new cap structure on it. The reaction required a high concentration of PEG 4000 (Fig. S1) whereas a nonmethylated capped molecule made in the absence of SAM did not function as cap donor (Fig. 1D, lane 4). PEG is known to have bulk structure in aqueous solution that causes crowding of macromolecules in the solution and has successfully been used to reconstitute the transcriptase activity of L-A virus in vitro (18). A 107-nt-capped molecule exhibited similar or better donor activity than the 5-nt donor (Fig. S2).
Fig. 1.
Transfer of m7Gp from cap donor to the 5′ end of viral transcript. (A) Labeled 5-nt cap donor was incubated in a transcription reaction mixture with L-A virions (lane 1) or M1 virions with WT (lane 2) or mutant (Arg154, lane 3) Gag protein and the full-sized transcripts were separated in an agarose gel. Ethidium bromide staining and autoradiogram of the gel are shown. λ, lambda-HindIII markers. (B) L-A virions were incubated with 5-nt cap donor in a transcription mixture from which CTP and GTP were omitted. Transcription was done in the presence of GDP (lane 4) or in its absence (lane 3), and the products were analyzed on a 15% acrylamide gel. As mobility markers, GTP (lane 1), cap donor (lane 2), 16-mer (lane 5), and 17-mer primed with m7GpppG (lane 6) were run in parallel. The sequence of the cap donor and the position of 32P (indicated by the asterisk) are shown (Left). Under the panel the 5′ end sequence of L-A transcript is shown. The first C appears at position 17. (C) The labeled product (marked by the arrowhead in lane 4 of B) was isolated from the gel, treated with the enzymes as indicated, and analyzed on polyethyleneimine-cellulose (PEI-cellulose) with 0.3 M (NH4)2SO4. Cap donor was also processed in parallel as control. The mobility of nonlabeled nucleotides is indicated (Left). Deduced transfer reaction is shown under the panel along with TAP and S1 cleavage sites. (D) 7-methylated and nonmethylated cap donors were synthesized in the presence and in the absence of S-adenosyl methionine (SAM), respectively, and incubated with L-A virions as described in the legend to B. The product (17-mer) was separated in a 8-M urea/15% acrylamide gel.
Preservation of the 5′ α- and β-Phosphates of the Viral Transcripts in the Triphosphate Linkage During Cap Transfer.
The gel mobility of the capped transcript (Fig. 1B) strongly suggests that only the m7Gp moiety of the cap donor was transferred to the 5′ end of the viral transcript. Here we confirm this. We found that the cap analogue m7GpppG but not GpppG could be used as cap donor and produced the 17-nt-capped product (Fig. 2A). This allowed us to specifically label the 5′ end of the viral transcript (acceptor) and examine its fate during cap transfer. In the absence of CTP, L-A virion incorporates [α-32P] GTP at the 5′ end of the 16-nt transcript with the label located at the α-position. In the presence of m7GpppG, part of transcripts (ca. 20%) was converted to a BAP-resistant 17-mer (Fig. 2A, lane 4). This species was gel-purified and analyzed by TLC. S1 treatment released m7GpppG, thus confirming the formation of cap structure at the 5′ end (Fig. 2B, lane 5). By contrast the 16-nt transcript released GDP upon S1 digestion (Fig. 2B, lane 2) as observed previously (17). After TAP treatment the capped product remained at the origin (Fig. 2B, lane 6), indicating that the label was still associated with the RNA body of 16 nt. A further treatment with S1, however, converted it to GMP (Fig. 2B, lane 7). These results indicate that during cap transfer the α-phosphate at the 5′ end of the acceptor is preserved at the α-position in the triphosphate linkage of the capped product. When transcription is primed with GMP, the virion adds the β-phosphate at the 5′ end of the transcript at the expense of ATP (17). Thus the 5′ β-phosphate of the acceptor can be labeled specifically with [γ-32P] ATP, as shown in Fig. 2D. S1 treatment released GDP (Fig. 2D, lane 2), indicating that the 16-nt transcript had diphosphate at the 5′ end. Furthermore, the transcript released labeled Pi upon RNA 5′ polyphosphatase (RPP) treatment (Fig. 2D, lane 3). Because the enzyme does not work on the 5′ α-phosphate, it indicates that the label was located at the 5′ β-phosphate in the transcript. In the presence of m7GpppG, part of 16-mer again was converted to a BAP-resistant 17-mer (Fig. 2C, lane 5). The formation of cap structure at the 5′ end of the 17-mer was confirmed by S1 digestion (Fig. 2D, lane 6). This time, however, the 17-mer produced labeled Pi upon TAP treatment (Fig. 2D, lane 7), thus confirming that the marker was located at the β-position of the triphosphate linkage in the cap structure. These results altogether depict the L-A virus’ cap-snatching mechanism distinct from that of influenza virus: L-A virus transfers only m7Gp from mRNA to the diphosphorylated 5′ terminus of its transcript. In addition, although influenza virus requires a capped mRNA fragment as a primer for transcription (19), L-A virus can synthesize its transcript in the absence of cap transfer.
Fig. 2.
Preservation of 5′ diphosphate of L-A transcript in the triphosphate linkage of the capped product during cap snatching. (A) L-A virions were incubated in a CTP-omitted transcription mixture in the presence or absence of methylated or nonmethylated cap analogue (0.5 mM). Transcription was primed with [α-32P] GTP. BAP-treated or nontreated products were separated in 8 M urea/15% acrylamide gel. The arrowheads indicate BAP-resistant capped transcript (17-mer). (B) BAP-resistant 17-mer (capped) shown in A was isolated from the gel, treated with the enzymes singularly or sequentially as indicated, and analyzed by TLC using 0.3 M (NH4)2SO4 as solvent. Noncapped transcript (16-mer) was also processed in parallel as control. Deduced reaction scheme is drawn under the panel along with TAP and S1 cleavage sites. (C) L-A virions were incubated in a GTP- and CTP-omitted transcription mixture in the presence or absence of m7GpppG (0.5 mM). Transcription was primed with GMP in the presence of [γ-32P] ATP. BAP-treated or nontreated products were separated in an 8 M urea/15% acrylamide gel. The arrowheads indicate BAP-resistant capped transcripts (17-mer). (D) BAP-resistant 17-mer (capped) shown in C was isolated and processed as described in the legend to Fig. 1C except that TLC was carried out with 1 M LiCl as solvent. Noncapped transcript (16-mer) was also processed in parallel as control. RPP: RNA 5′ polyphosphatase that hydrolyses β- and γ- phosphates at the 5′ terminus of RNA. Lane 8: Pi standard. Deduced cap transfer reaction with some enzymatic cleavage sites is drawn under the panel. The asterisks show the positions of the 32P label.
Involvement of Gag in the Cap-Snatching Reaction.
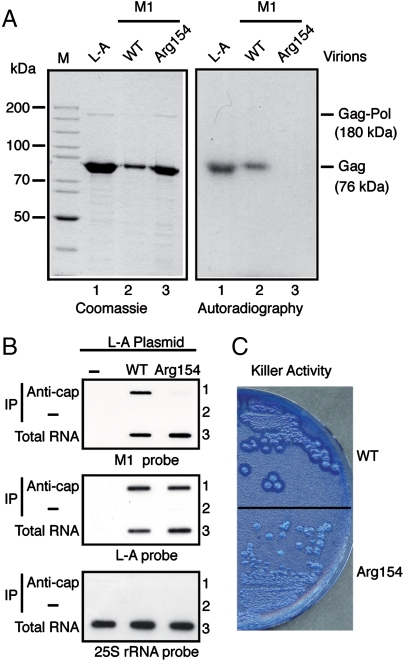
M1 requires L-A for encapsidation and replication. M1 can be maintained in the cell in the absence of the helper virus provided L-A proteins are expressed from a vector (13). Like L-A, M1 virions formed with wild-type L-A proteins have decapping activity and formed m7Gp-Gag adduct when incubated with the 5-nt cap donor (Fig. 3A, lanes 1 and 2). By contrast, M1 virions formed with a mutant Gag (Arg154) failed to form the adduct as demonstrated previously (15) (Fig. 3A, lane 3). This mutant Gag can support M1; therefore, transcription, encapsidation, and replication of M1 are not impaired by the mutation. When M1 virions were incubated in a transcription mixture with the 5-nt cap donor, the virions with the wild-type L-A proteins transferred m7Gp from the donor to the transcript (Fig. 1A, lane 2), whereas the virions with the mutant Gag failed to do so (Fig. 1A, lane 3). Because mutant M1 virions synthesized transcripts with 5′ diphosphate (Fig. S3) as wild-type virions did (17), these results indicate that the Arg154 mutation specifically impaired the cap-snatching reaction, thus signifying the involvement of the decapping reaction in cap snatching.
Fig. 3.
Cap snatching is essential for viral expression. (A) Decapping activity of Gag to form m7Gp-Gag adduct. L-A virions (lane 1) or M1 virions with WT (lane 2) or mutant (Arg154, lane 3) Gag protein were incubated for decapping with 5-nt cap donor labeled at the γ-phosphate with 32P. Then proteins were separated in an SDS gel and detected by Coomassie blue staining (Left) or by autoradiography (Right). M; protein standards for mobility. (B) Detection of capped viral transcripts in vivo. RNA was extracted from cells containing M1 supported by L-A plasmid expressing WT or mutant Gag (Arg154) or from L-A plasmid-cured cells (−) as control. Capped RNA was immunoprecipitated with (Anti-cap) or without (−) anti-cap antibody and hybridized with M1 (+) strand-specific (Upper), L-A (+) strand-specific (Middle), or 25S rRNA-specific (Lower) probe. As control, total RNA was processed in parallel (Total RNA). (C) Killer assays.
Cap Snatching Is Essential for Efficient Expression of Viral Transcripts.
We have demonstrated that L-A and M1 virions can synthesize capped transcripts in vitro. However, capped viral transcripts in vivo have not been detected yet. We tried to prove their existence in the cell. RNA was extracted from cells containing M1 virions with wild-type or Arg154 mutant Gag protein. Capped RNA was then isolated using a monoclonal anticap antibody. As shown in Fig. 3B, Upper, lane 1, we could detect capped M1 transcripts from virions with wild-type L-A protein, whereas capped RNA from M1 virions with mutant Gag was barely detectable. Because M1 in these strains was supported by plasmid-encoded L-A proteins, capped L-A RNA transcribed by PolII was used as a positive control. There was no difference in the amount of capped L-A RNA between these strains (Fig. 3B, Middle, lane 1). The results therefore demonstrate that the Gag mutation Arg154 specifically affects capping of virion-derived transcripts. The strain carrying M1 with wild-type L-A proteins produces protein toxin and kills sensitive cells. By contrast the strain harboring the mutant Gag barely produces the toxin as described (15) (Fig. 3C). From these in vivo results we conclude that the cap-snatching mechanism operates in the cell and does produce capped viral transcripts, and that the capping reaction is essential for efficient expression of the viral genome.
Discussion
We have demonstrated a unique mechanism of cap snatching in the yeast dsRNA virus L-A. The virus transfers the m7Gp moiety from host mRNA to the diphosphorylated 5′ end of viral transcript, thus generating an authentic cap structure (referred to as cap0) in the budding yeast. The mechanism is distinct from that of influenza virus. Influenza virus endonucleolytically cleaves host mRNA 10–13 nt downstream from the 5′ end and utilizes the resulting capped oligo fragment as a primer to synthesize its transcript. The L-A cap-snatching mechanism is essential for efficient expression of viral transcripts and, strikingly, His154 of Gag is involved in this process. In influenza virus the trimeric polymerase performs cap snatching. It is surprising to find a unique cap-snatching activity in the structural coat protein Gag. However, considering that the Pol domain of the Gag-Pol fusion protein is confined inside the virion and unable to access mRNA in the cytoplasm, it is quite reasonable that Gag but not Pol performs the reaction. Thus these findings exemplify the versatility and adaptability of eukaryotic RNA viruses.
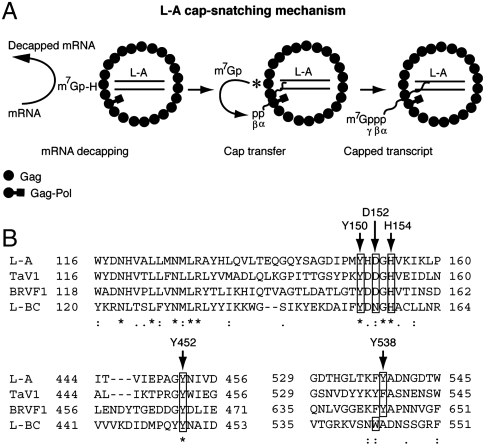
The cap-snatching reaction of L-A virus requires a high concentration of PEG 4000 and 7-methylation of cap is essential for cap-donor activity. Interestingly L-A virions used m7GpppG in vitro as a cap donor. Thus the virus can potentially utilize not only host mRNA but also m7GpppG or capped oligo fragments generated from the 3′-5′ mRNA degradation pathway carried out by the exosome (20, 21), as cap donors. A substantial amount (ca. 20%) of transcripts (16-mer) can acquire the cap structure in vitro. The capped species preserved the α- and β-phosphates of the transcript in the triphosphate linkage. Current data show that 5′ monophosphorylated or triphosphorylated transcripts do not function as cap acceptors in the reaction, thus suggesting that the diphosphorylated 5′ end status is established at the very early stage of transcription. His154 is essential for cap snatching in vitro and in vivo. The model depicted in Fig. 4A postulates that the m7Gp-Gag adduct generated by the decapping reaction is an intermediate of m7Gp transfer reaction.
Fig. 4.
(A) Schematic diagram of L-A cap-snatching mechanism. In the middle virion, m7Gp-His154 adduct is depicted by the asterisk. It is not known which Gag protein in the virion participates in the cap transfer reaction. mRNA decapping and cap transfer may be coupled with transcription (see text). (B) Comparison of selected L-A Gag sequences with those of other fungal totiviruses. His154 and crucial residues for cap recognition are indicated by the arrows. GenBank accession numbers are L-A (AAA50320.1), TaV1 (ADQ54105.1), BRVF (YP 001497150.1), and L-BC (NP 042580.1).
Crystallographic studies of the L-A virion have identified a trench on the outer surface of Gag that includes His154 (22, 23). L-A transcripts are made inside the virion and presumably released to the cytoplasm through one of the pores located at the icosahedral 5-fold symmetric axes (22). The trenches are positioned in the Gag asymmetric dimer close to the 2-fold and 3-fold axes, far from the pores. It is likely that the high concentration of PEG 4000 required in the reaction increases the local concentration of the 5′ end of the viral transcript (or the host mRNA cap donor) in the trench, presumably mimicking the cytoplasmic environment overcrowded with macromolecules. The physical separation of the cap-snatching site from the transcription site may ensure that not all transcripts are capped. If capped transcripts were destined for translation and the noncapped ones for encapsidation, then the virus might actively participate in this differentiation process by changing the ratio of capping. Our current data suggest that only virions actively engaging in polymerization have capping activity, implying that the two sites are coordinated. Previously Masison et al. (24) failed to detect the cap-snatching activity of L-A virus, presumably because they did not include PEG in the reaction. The authors found that a ski1 mutation suppresses the effect of the mutation of His154 on expression of M1 toxin and proposed an alternative model, a decoy hypothesis in which L-A virus utilizes cellular mRNAs decapped by the virus as a decoy to protect its noncapped transcripts from degradation by the XRN1/SKI1 5′-3′ exonuclease (24).
His154 is located at the tip of a loop (residues 144–163) that is part of the upper rim of the trench. Upon ligand binding (m7GDP) the rim moves inwardly and forms a closed conformation (23). Guanylyltransferase also contains a trench that can adopt either open or closed conformations during the mRNA capping reaction (25, 26). Furthermore, it has been noticed that secondary structure elements around the trench of Gag resemble those in guanylyltransferase (22). This enzyme forms a Gp-enzyme intermediate with GTP and transfers Gp to the diphosphorylated 5′ termini of PolII transcripts to generate a nonmethylated cap structure. Thus, the cap-snatching reaction of L-A (Fig. 4A) resembles that of guanylyltransferase. These similarities suggest convergent evolution. In the standard capping reaction, guanylyltransferase forms a covalent bond with Gp through Lys but not His. However, it has been demonstrated that in the capping reaction of vesicular stomatitis virus, L protein covalently binds the 5′ monophosphorylated pre-mRNA through His and transfers the bound pre-mRNA to GDP (27). Members of the alphavirus-like superfamily methylate GTP first, then seem to utilize His to form a phosphoamide bond with m7Gp and transfer the m7Gp moiety to the 5′ diphosphate end of a viral transcript during the capping reaction (28–30).
In the trench, Tyr150, Asp152, Tyr452, and Tyr538 are located close to the bound m7GDP, suggesting their involvement in cap recognition (23). Mutagenesis studies indicate that these residues are crucial for decapping activity. Tyr452 and Tyr538 sandwich m7GDP by a potential stacking (23). Cap-binding proteins normally use two aromatic rings of Tyr, Phe, or Trp in cap recognition to sandwich the m7G moiety (31, 32). Fungal totiviruses closely related to L-A share these residues (or similar ones) as well as His154 at comparative positions in their coat proteins (Fig. 4B). Decapping activity has also been demonstrated in Gag of L-BC virus (14). Therefore, it is likely that the cap-snatching mechanism of L-A virus is widespread among totiviruses of fungi. Standard mRNA capping requires three sequential enzymatic reactions. L-A virus, however, obtains these caps from host mRNA, thus reducing the burden of encoding its own capping machinery in a small dsRNA genome (4.6 kb). Typical of a fungal virus, L-A has no extracellular transmission pathway. This further eliminates the necessity to encode proteins for exit and reentry to a new cell. These features have apparently contributed to reduce the viral genome to a single dsRNA segment. By contrast, infectious dsRNA viruses (Reoviridae) found in higher eukaryotes possess their own capping machinery (33) and have genomes with 10 or more segments.
Materials and Methods
Preparation of the 5-nt Cap Donor.
Nonlabeled 106-nt RNA fragment containing the polylinker sequence from Bluescript KS+ vector was made by T7 runoff transcription. The cap structure was installed on it using [α-32P] GTP with the aid of mScript mRNA production system (Epicentre) following the instructions provided by the manufacturer. The 5-nt cap donor was generated by digesting it with RNase A (14) and purified through a 15% acrylamide gel.
Cap Transfer Reaction.
The standard cap transfer reaction contained 50 mM Tris HCl pH 7.5, 5 mM MgCl2, 0.1 mM EDTA, 20 mM NaCl, 5 mM KCl, 0.5 mM each of ATP, CTP, GTP, and UTP, 20% PEG 4000, and labeled 5-nt cap donor (2–5,000 cpm). The reaction was started by the addition of L-A or M1 virions and kept at 30 °C for 1 h. Phenol-extracted products were analyzed on either agarose or acrylamide gels. TLC using PEI-cellulose was done as described (17). Decapping reaction was done as described (14).
Immunoprecipitation with Anticap Monoclonal Antibody.
Freshly growing cells (109 cells) were harvested and broken by vortex mixing (15 s, 10 times) with glass beads in a buffer containing 20 mM Tris HCl, pH 8.0 and 100 mM NaCl. After removing cell debris, RNA was extracted with phenol from the supernatant and precipitated with ethanol. RNA was incubated for 30 min at 4 °C with 30 μL of protein G-Sepharose (GE Healthcare) prebound to anti-m3G/m7G-cap monoclonal antibody (Synaptic Systems). RNA bound to the Sepharose was extracted, blotted to a nylon membrane, and hybridized with an L-A-, M1-, or 25S rRNA-specific probe as described previously (34). The L-A and M1 probes recognize the positive strand sequences of L-A (nt 1,323–1,786) and M1 (nt 14–500), respectively. The 25S rRNA probe recognizes nt 139–608 of 25S rRNA.
Supplementary Material
Acknowledgments.
We thank Reed B. Wickner (National Institutes of Health, Bethesda, MD) for providing the L-A-free killer strains. This work was supported by Grants BFU2007–60057 and BFU2010–15768 from the Spanish Ministry of Education and Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111900108/-/DCSupplemental.
References
- 1.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y, LaFiandra A, Shatkin AJ. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 3.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 4.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 5.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 6.Guilligay D, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 7.Boivin S, Cusack S, Ruigrok RW, Hart DJ. Influenza A virus polymerase: Structural insights into replication and host adaptation mechanisms. J Biol Chem. 2010;285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner RB. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Willians and Wilkins; 2007. pp. 737–768. [Google Scholar]
- 9.Icho T, Wickner RB. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- 10.Dinman JD, Icho T, Wickner RB. A-1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimura T, Ribas JC, Makhov AM, Wickner RB. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature. 1992;359:746–749. doi: 10.1038/359746a0. [DOI] [PubMed] [Google Scholar]
- 12.Bostian KA, et al. Sequence of the preprotoxin dsRNA gene of type I killer yeast: Multiple processing events produce a two-component toxin. Cell. 1984;36:741–751. doi: 10.1016/0092-8674(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 13.Wickner RB, Icho T, Fujimura T, Widner WR. Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: A ski- phenocopy. J Virol. 1991;65:155–161. doi: 10.1128/jvi.65.1.155-161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanc A, Goyer C, Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol Cell Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanc A, Ribas JC, Wickner RB, Sonenberg N. His-154 is involved in the linkage of the Saccharomyces cerevisiae L-A double-stranded RNA virus Gag protein to the Cap structure of mRNAs and is essential for M1 satellite virus expression. Mol Cell Biol. 1994;14:2664–2674. doi: 10.1128/mcb.14.4.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura T, Esteban R, Wickner RB. In vitro L-A double-stranded RNA synthesis in virus-like particles from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:4433–4437. doi: 10.1073/pnas.83.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimura T, Esteban R. Yeast double-stranded RNA virus L-A deliberately synthesizes RNA transcripts with 5′-diphosphate. J Biol Chem. 2010;285:22911–22918. doi: 10.1074/jbc.M110.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimura T, Wickner RB. Reconstitution of template-dependent in vitro transcriptase activity of a yeast double-stranded RNA virus. J Biol Chem. 1989;264:10872–10877. [PubMed] [Google Scholar]
- 19.Yuan P, et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 21.Schaeffer D, Clark A, Klauer AA, Tsanova B, van Hoof A. Functions of the cytoplasmic exosome. Adv Exp Med Biol. 2011;702:79–90. doi: 10.1007/978-1-4419-7841-7_7. [DOI] [PubMed] [Google Scholar]
- 22.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, et al. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral cased protein: A unique viral response to host defense. J Mol Recognit. 2005;18:158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- 24.Masison DC, et al. Decoying the cap-mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.HÅkansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 26.Chu C, et al. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc Natl Acad Sci USA. 2011;108:10104–10108. doi: 10.1073/pnas.1106610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino T, Yadav SP, Banerjee AK. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YL, Hsu YH, Han YT, Meng M. mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of bamboo mosaic virus. J Biol Chem. 2005;280:13153–13162. doi: 10.1074/jbc.M412619200. [DOI] [PubMed] [Google Scholar]
- 31.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 32.Fechter P, Brownlee GG. Recognition of mRNA cap structures by viral and cellular proteins. J Gen Virol. 2005;86:1239–1249. doi: 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
- 33.Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda2. J Biol Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- 34.Fujimura T, Esteban R. Interactions of the RNA polymerase with the viral genome at the 5′- and 3′-ends contribute to 20S RNA narnavirus persistence in yeast. J Biol Chem. 2007;282:19011–19019. doi: 10.1074/jbc.M702432200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.