Abstract
GSK3/shaggy-like genes encode kinases that are involved in a variety of biological processes. By functional complementation of the yeast calcineurin mutant strain DHT22-1a with a NaCl stress-sensitive phenotype, we isolated the Arabidopsis cDNA AtGSK1, which encodes a GSK3/shaggy-like protein kinase. AtGSK1 rescued the yeast calcineurin mutant cells from the effects of high NaCl. Also, the AtGSK1 gene turned on the transcription of the NaCl stress-inducible PMR2A gene in the calcineurin mutant cells under NaCl stress. To further define the role of AtGSK1 in the yeast cells we introduced a deletion mutation at the MCK1 gene, a yeast homolog of GSK3, and examined the phenotype of the mutant. The mck1 mutant exhibited a NaCl stress-sensitive phenotype that was rescued by AtGSK1. Also, constitutive expression of MCK1 complemented the NaCl-sensitive phenotype of the calcineurin mutants. Therefore, these results suggest that Mck1p is involved in the NaCl stress signaling in yeast and that AtGSK1 may functionally replace Mck1p in the NaCl stress response in the calcineurin mutant. To investigate the biological function of AtGSK1 in Arabidopsis we examined the expression of AtGSK1. Northern-blot analysis revealed that the expression is differentially regulated in various tissues with a high level expression in flower tissues. In addition, the AtGSK1 expression was induced by NaCl and exogenously applied ABA but not by KCl. Taken together, these results suggest that AtGSK1 is involved in the osmotic stress response in Arabidopsis.
Salt stress, such as that caused by high concentrations of NaCl, causes hyperosmotic stress, imbalance in the cellular ion concentration, and general toxicity that adversely affects plant development. Numerous studies have been conducted to delineate the cellular changes that occur upon exposure to osmotic stress (Walton, 1980; DuPont, 1992; Bohnert et al., 1995; Niu et al., 1995). It has been noted that different plant species employ a variety of different mechanisms to cope with osmotic stress. Responses are especially well understood with regard to physiological and metabolic changes (LaRosa et al., 1987; Binzel et al., 1988; Skriver and Mundy, 1990; Delauney and Verma, 1993; Bohnert et al., 1995; Niu et al., 1995). However, not much has been learned about the mechanism of osmotic stress signaling in plants. In contrast, the signaling mechanism has been studied in detail in yeast. There it has been found that the NaCl stress or high osmotic stress signal is mediated by a MAP kinase pathway (Maeda et al., 1994, 1995). Osmosensor molecules, such as Sln1p and Sho1p, which are located on the plasma membrane, initiate the signaling pathway (Ota and Varshavsky, 1993). The signal then reaches various kinases such as Ssk1p, Pbs2p, and Hog1p (Maeda et al., 1994, 1995; Posas et al., 1996). In addition to the MAP kinase pathway, yeast has another signal transduction pathway that is specific for high NaCl stress. This pathway includes calcineurin, a phosphatase dependent on Ca2+, and calmodulin (Nakamura et al., 1993; Mendoza et al., 1994; Wieland et al., 1995). Therefore, it is possible that external high NaCl stress increases intracellular Ca2+ concentration, which then causes calmodulin to transmit signals to other, downstream components, such as calcineurin (Matheos et al., 1997). It is also very likely that protein kinases and protein phosphatases are involved in signal transduction in plants. In fact, many genes encoding protein kinases have been shown to be induced under high NaCl conditions and under exogenous ABA treatment (Anderberg and Walker-Simmons, 1992; Urao et al., 1994; Hwang and Goodman, 1995; Jonak et al., 1996; Mizoguchi et al., 1996). It has been suggested that protein kinases may be involved in osmotic stress signal transduction. Recently, the genes at the ABA-insensitive 1 and 2 loci of Arabidopsis have been cloned and shown to encode homologs of phosphatases (Leung et al., 1994, 1997; Meyer et al., 1994). These phosphatases have a Ca2+-binding domain, which implies that Ca2+ may play a role in the osmotic stress signal transduction in plants. Also, the SOS3 gene, the mutation of which renders Arabidopsis hypersensitive to a high NaCl concentration, encodes a Ca2+ sensor homolog, which further confirms the involvement of Ca2+ in NaCl stress signaling (Liu and Zhu, 1998). However, in most cases the evidence for the involvement of protein kinases in the NaCl stress or osmotic stress signal pathway is rather circumstantial, and to our knowledge no in vivo substrates for the kinases have been isolated. Also, the in vivo targets of the phosphatases remain to be defined.
Originally, GSK3 was identified as a kinase that phosphorylates glycogen synthase (Embi et al., 1980; Woodgett, 1990), but it has been recently shown that GSK3 is involved in developmental processes such as cell fate determination in Drosophila melanogaster (Ruel et al., 1993), Xenopus (Dominguez et al., 1995), and Dictyostelium (Harwood et al., 1995), and in insulin regulation and transcriptional activation of a variety of proteins in mammalian cells (Nikolakaki et al., 1993; Welsh and Proud, 1993). In yeast there are two GSK3 homologs, Mck1p and Mds1p (Neigeborn and Mitchell, 1991; Puziss et al., 1994). Mutation at the MCK1 gene showed a cold-sensitive phenotype, a temperature-sensitive phenotype, and loss of chromosomes during growth on benomyl. Based on these observations it has been suggested that Mck1p plays an important role in the regulation of kinetochore activity and entry into meiosis. In plants many genes encoding homologs of GSK3 have been identified by virtue of the conservation of the primary structure, i.e. the amino acid sequence, and in many cases it has been suggested that they are involved in developmental processes (Bianchi et al., 1993, 1994; Pay et al., 1993; Dercroocq-Ferrant et al., 1995; Jonak et al., 1995; Dornelas et al., 1998).
In an effort to isolate genes involved in the NaCl stress signal transduction pathway, we screened Arabidopsis cDNA clones for complementation of the yeast strain DHT22-1a, which has deletion mutations in both calcineurin genes. In this paper we report that Arabidopsis cDNA AtGSK1 encodes a protein highly homologous to the glycogen synthase kinase 3 of mammalian cells and to the shaggy gene of D. melanogaster, and that it can rescue the NaCl stress-sensitive phenotype of the yeast calcineurin and mck1 mutants. In addition, we show that the expression of the AtGSK1 gene is induced by NaCl and ABA treatments in Arabidopsis.
MATERIALS AND METHODS
Screening of cDNA Clones
To screen for cDNAs complementing a NaCl stress-sensitive strain of yeast (Saccharomyces cerevisiae), DHT22-1a (MATa trp1 ade2 ura3 can1-100 cmp::LEU2 cmp2::HIS3) (Nakamura et al., 1993), an Arabidopsis expression library constructed in pFL61, was introduced into competent cells of the mutant yeast by the LiCl method (Ito et al., 1983). Approximately one million colonies were obtained on SC-Ura plates and pooled to yield a transformed yeast library. The transformed yeast cells were then plated at a density of 107 cells per YPD plate (12 × 12 cm) supplemented with 0.9 m NaCl. Putative positive colonies were obtained 4 d after plating. The putative positive colonies were then rescreened on 0.9 m NaCl YPD plates. We selected one of them, clone 49, for further characterization. Plasmid DNA was isolated from the yeast cells and reintroduced into DHT22-1a cells to confirm the complementation (Ausubel et al., 1989). Finally, the inserts of clone 49 were subcloned into pBluescript and were sequenced with the dideoxy termination method using a dye-terminator cycle sequencing kit. The sequencing reaction was analyzed with the help of an automatic sequencing machine (ABI, Columbia, MD). To isolate a full-length cDNA clone, the insert of a positive clone was used as a hybridization probe in the screening of an Arabidopsis leaf cDNA library constructed in λZAPII. Positive clones were in vivo excised as pBluescript clones. The clone that appeared to contain a full-length cDNA was named AtGSK1. The whole sequence of the cDNA was obtained by sequencing serial deletion constructs.
Northern- and Southern-Blot Analyses
Total RNA was isolated from Arabidopsis seedlings as described previously (Ausubel et al., 1989). Fifteen micrograms of total RNA was separated in a 1.2% northern-blot gel and transferred onto nylon membranes (Biotrans, ICN). After transfer, RNA was UV cross-linked to the membrane and used in northern-blot analysis. For Southern-blot analysis genomic DNA was prepared according to a previously described protocol (Watson and Thompson, 1986), and 2 μg of the genomic DNA was digested with restriction endonucleases. To prepare a specific probe for hybridization two PCR primers were designed: 5′-AACTGTGCATGTCTGAAG-3′ and 5′-TACGTATCTGTCAGAATTG-3′. The PCR product was amplified from the cDNA clone, gel purified, and used for labeling with [α-32P]dATP by PCR. Hybridization and washings were carried out according to a published protocol (Church and Gilbert, 1984).
Complementation of the Yeast Mutation
The full-length cDNA designated AtGSK1 was subcloned into pVT-U, an expression vector with the ADH promoter and terminator and the URA3 selection marker. The resulting construct was introduced into DHT22-1a cells, which were plated on SC-Ura plates (Ausubel et al., 1989). The transformed cells were grown in YPD medium overnight, and 104 cells were spotted on YPD agar plates supplemented with 1.0 m NaCl. The growth was examined after 4 d at 30°C (Cunningham and Fink, 1996).
Assay of the PMR2A Gene Activation
To examine the transcriptional activation of the PMR2A gene in the mutant cells, the AtGSK1 cDNA was subcloned into an expression vector, pJG10 (a modified version of pJG4-5; the GAL1 promoter has been replaced by the ADH promoter) (Zervos et al., 1993). The construct was introduced into NaCl stress-sensitive cells, DHT22-1a (MATa trp1 ade2 ura3 can1-100 cmp::LEU2 cmp2::HIS3) cells, or mck1 (MATa trp1 ade2 ura3 can1-100 mck1:HIS3) cells, and transformants were selected on SC-Trp plates. Subsequently the reporter gene construct PMR2A:lacZ (Cunningham and Fink, 1996) was introduced into the cells harboring the AtGSK1 cDNA, and the cells were plated on SC-Ura plates. The transformants were rescreened on SC-Ura-Trp plates. Yeast cells harboring both genes were cultured at 30°C overnight and inoculated to an A600 of 0.1 into YPD medium supplemented with 0.5 m NaCl. Cells were harvested at various time points. Cell extracts were prepared, and β-galactosidase activity was measured according to published protocols (Cunningham and Fink, 1996).
Construction of the mck1 Deletion Mutant
The MCK1 gene was isolated from yeast genomic DNA by PCR using two primers: 5′-GGAGTTAAGCCCAAGAC-3′ and 5′-ACAGCGGATCAAAGGTG-3′. The PCR product was subcloned into pBluescript and confirmed by partial sequencing of both ends. The His3 gene fragment was inserted into the NcoI site of the MCK1 gene. A DNA fragment containing the mck1:His3 gene was then introduced into DHT22-1b (MATa trp1 leu2 ade2 ura3 his3 can1-100) to generate a mck1 mutant strain, and colonies were selected on SC-His plates. The insertion of the His3 gene was confirmed by PCR amplification using the MCK1-specific primers and genomic DNA obtained from the His3+ cells. To assay the NaCl stress sensitivity, mck1 mutant cells were plated on the YPD plate containing 1.0 m NaCl.
RESULTS
Isolation of the AtGSK1 cDNA
To clone genes involved in the NaCl stress signaling pathway in plants, we attempted to isolate cDNAs that complement a yeast NaCl stress-sensitive strain. We decided to use the yeast strain DHT22-1a, which has deletion mutations in both genes encoding catalytic subunits of yeast calcineurin (Nakamura et al., 1993). An Arabidopsis leaf cDNA library constructed in pFL61 was introduced into the yeast cells, and approximately 1 × 106 yeast transformants were obtained. To screen for complementing cDNA clones, the transformed yeast cells were plated on YPD plates supplemented with 0.9 m NaCl. The screening resulted in five positive clones from which we chose clone 49 to be characterized in detail. The insert of the cDNA clone was transferred into pBluescript for sequencing. The sequence of the 5′ end revealed that the cDNA encodes aprotein with a high degree of amino acid sequence homology to GSK3 of mammalian cells (Woodgett, 1990) and to the shaggy gene of Drosophila malanogaster (Ruel et al., 1993). However, the cDNA appeared to be missing approximately 25 amino acid residues from the N terminus when the deduced amino acid sequence of the cDNA was compared with other homologs in the public database. Therefore, to obtain a full-length cDNA, an Arabidopsis λZAPII leaf cDNA library was screened again with the insert of clone 49 as the hybridization probe. The screening resulted in the isolation of 10 positive λ clones. pBluescript clones were obtained from the λ clones by in vivo excision, and the 5′ ends of the these pBluescript clones were sequenced to confirm the identity. These cDNA clones appeared to be identical with a minor difference in size. Therefore, the clone that contained the largest insert was named AtGSK1 (Arabidopsis thaliana GSK3 1). The full sequence of the cloned cDNA was obtained by the dideoxy termination method using a dye-terminator cycle sequencing kit. The cDNA insert consists of 1572 bp with a putative initiation codon at nucleotide position 26, which is followed by 1224 bp of the open reading frame and 323 bp of the 3′-untranslated region.
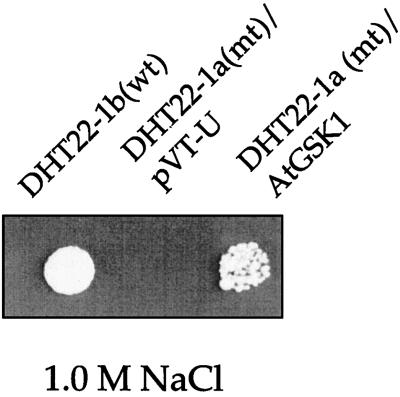
Since the original cDNA clone was missing 25 amino acid residues from the N terminus, we introduced into the mutant yeast strain the full-length cDNA under the control of the ADH promoter. The yeast transformant was reexamined on a 1.0 m NaCl plate. As shown in Figure 1, AtGSK1 rescued the NaCl stress-sensitive phenotype of the DHT22-1a cells, which confirmed the original finding. This result suggests that the missing 25 amino acid residues may not affect the ability of AtGSK1 to complement the yeast calcineurin mutant.
Figure 1.
Complementation of DHT22-1a mutant (mt) cells with AtGSK1. The DHT22-1a (MATa trp1 ade2 ura3 can1-100 cmp::LEU2 cmp2::HIS3) mutant cells were transformed with the full-length AtGSK1 under the ADH1 promoter. The transformed cells were plated on a YPD plate containing 1.0 m NaCl and incubated at 30°C for 4 d. wt, Wild type.
Sequence Analysis of AtGSK1
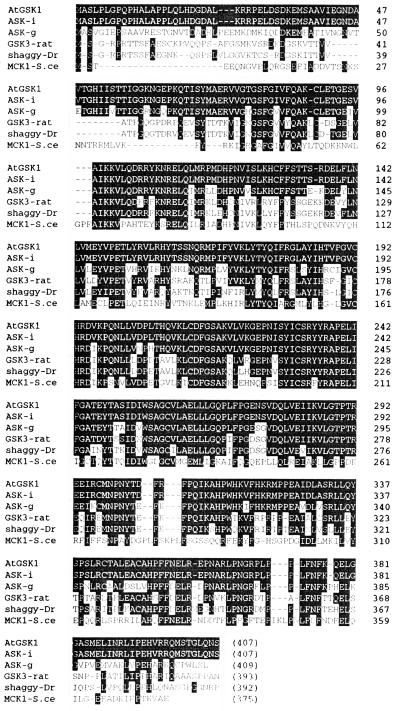
The amino acid sequence of the AtGSK1 cDNA (accession no. AF019927) was highly homologous to seven ASK sequences in the database. AtGSK1 was nearly identical to ASK iota at the level of nucleotide sequence with only minor differences such as the sizes of the 5′-untranslated region and the poly(A+) tail, suggesting that the AtGSK1 cDNA may be identical to ASK iota (Dornelas et al., 1998). Conceptual translation of the AtGSK1 cDNA results in a protein with 407 amino acid residues and with a molecular mass of 46 kD. Also, there are six additional Arabidopsis sequences with high degree of homology in the public databases. The amino acid sequence of AtGSK1 was compared with protein sequences obtained from the public databases using the BLAST program (Altshul et al., 1990). As shown in Figure 2, AtGSK1 exhibits a high degree of amino acid sequence homology with known GSK3 homologs in a variety of organisms, such as mammals, D. melanogaster, and Arabidopsis. AtGSK1 shares 100% amino acid sequence similarity with ASK-iota, 65% with ASK-gamma, 56% with D. melanogaster shaggy, 38% with rat GSK3β, and 36% with yeast Mck1p.
Figure 2.
Alignment of the amino acid sequence of AtGSK1 with other GSK3 homologs. The deduced amino acid sequence AtGSK1 is aligned with sequences obtained from public databases. ASK-i, ASK-g, GSK3-rat, shaggy-Dr, and MCK1-S.ce indicate the following: ASK-iota (accession no. 1480078), ASK-gamma (accession no. 1170714), rat GSK3β (accession no. 125374), D. melangaster shaggy (accession no. 125701), and yeast Mck1p, respectively. The gaps were introduced to maximize the alignment. The identical amino acid residues between AtGSK1 and other homologs are shaded.
Genomic Structure of the AtGSK1 Gene
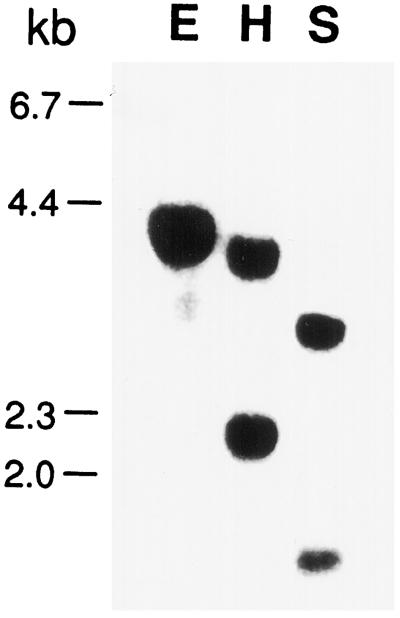
To investigate the copy number of the AtGSK1 gene in the Arabidopsis genome, Southern-blot analysis was carried out using the whole insert as a hybridization probe. Hybridization was carried out at a high stringency condition (Church and Gilbert, 1984). As shown in Figure 3, there are two types of bands: a strongly hybridizing band and weakly hybridizing bands. Thus, the Southern-blot result suggested that the AtGSK1 gene is a member of multicopy gene family. In fact, a BLAST search using the nucleotide sequence of the AtGSK1 gene revealed that there are other highly homologous genes, such as shaggy-like kinase-etha (accession no. 2129739). Therefore, it is also possible that the AtGSK1 cDNA hybridized with homologs of these genes at the high stringency condition we used for Southern-blot analysis.
Figure 3.
Southern-blot analysis of the AtGSK1 gene. Genomic DNA was digested with restriction enzymes and size separated onto a 0.8% agarose gel. The DNA was then transferred onto a nylon membrane and UV cross-linked. Hybridization was carried out with the AtGSK1 cDNA at 65°C overnight. E, H, and S indicate EcoRI, HindIII, and SacI, respectively.
Expression of the AtGSK1 Gene
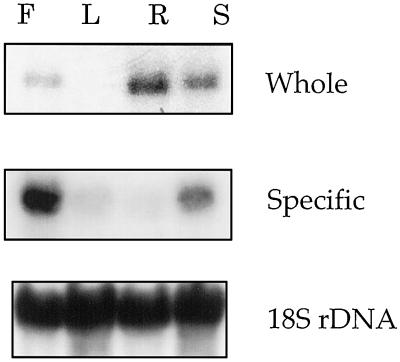
To gain insight into the AtGSK1 gene expression, total RNA was isolated from various tissues and used in northern-blot analyses. We first used the whole insert as a hybridization probe. As shown in Figure 4, the AtGSK1 transcript was most abundantly present in root and silique tissue and at a slightly lower level in flower tissue, whereas it was nearly undetectable in leaf tissue. However, when an AtGSK1-specific probe was used as a hybridization probe, the expression pattern was different. As seen in Figure 4, the transcript level was highest in the flower tissue followed by siliques but was quite low in leaf and root tissues. The results suggest that, depending on the tissue, the AtGSK1 gene is subject to differential regulation and in addition that there may be other closely related genes that are highly expressed in root tissue.
Figure 4.
Tissue-specific expression of the AtGSK1 gene. Total RNA (15 μg) isolated from various tissues was size separated onto a 1.2% formaldehyde/agarose gel and transferred onto a nylon membrane. The northern-blot gel was stained with EtBr, and the gel photograph was taken before transfer to examine the loading of the RNA samples. The whole cDNA (Whole) or the specific probe (Specific) of the AtGSK1 cDNA (nucleotide positions 1149–1513) was used as the hybridization probe, respectively. Hybridizations were carried out in a church buffer at 65°C overnight. The blots were washed with 2× SSC once at room temperature and twice with 0.1× SSC, 0.1% SDS at 60°C for 20 min. F, L, R, and S indicate flower, leaf, root, and silique, respectively.
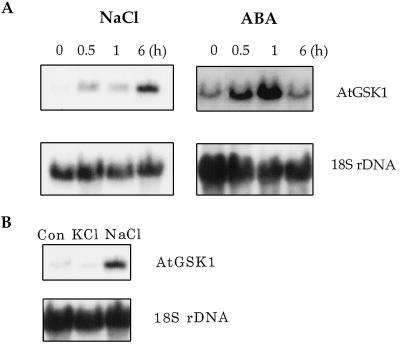
Since the AtGSK1 gene was isolated by rescue of a NaCl stress-sensitive yeast mutant, we examined the possibility of induction of the gene's expression under NaCl stress conditions. To apply NaCl stress to Arabidopsis plants, we treated Arabidopsis seedlings grown for 1 week in liquid culture with 0.15 m NaCl or 100 μm ABA. The Arabidopsis seedlings were harvested at various times, and total RNA was prepared to examine the AtGSK1 transcript level. As shown in Figure 5, the AtGSK1 transcript level accumulated to high levels with time upon NaCl treatment. Also, when the Arabidopsis seedlings were treated with exogenous ABA, the transcript level was increased. However, the pattern of induction was different from that of the NaCl treatment. The transcript level was increased within 30 min, reached the maximum at 1 h, and decreased again at 6 h after the ABA treatment. Next, we addressed whether the induction is specific to NaCl stress. We treated Arabidopsis seedlings with 200 mm KCl in a similar manner and examined the level of AtGSK1 transcripts. As shown in Figure 5, the treatment of Arabidopsis seedlings with KCl did not affect the expression of AtGSK1. The results suggested that the induction of AtGSK1 expression is specific to high NaCl stress.
Figure 5.
Induction of the AtGSK1 gene expression. A, Arabidopsis seedlings grown in Murashige and Skoog liquid medium were treated with 150 mm NaCl and 100 μm ABA for the indicated times. B, Arabidopsis seedlings were treated with 150 mm and 200 mm KCl for 6 h. Total RNA (15 μg) was analyzed by northern-blot analysis using the AtGSK1-specific probe. To examine the loading of the RNA samples, the northern blot was hybridized with 18S rDNA. Con, No treatment.
AtGSK1 Activates the NaCl Stress Signal Pathway in Yeast
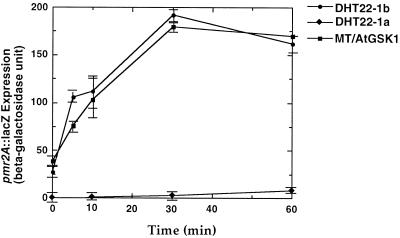
Since AtGSK1 is a protein kinase homolog, it is likely that AtGSK1 activates the NaCl stress signal transduction pathway in yeast that turns on the transcription of genes encoding proteins involved in the regulation of the internal concentrations of Na+. To investigate this further, we examined whether AtGSK1 can activate NaCl stress-inducible genes in yeast. It has been shown that the PMR2A gene, which encodes Na+-ATPase involved in Na+ transport, is induced by NaCl stress (Wieland et al., 1995). We therefore introduced a reporter gene, PMR2A:lacZ, together with the AtGSK1 cDNA into DHT22-1a cells. The overnight culture of the doubly transformed yeast cells was then inoculated into YPD medium supplemented with 0.5 m NaCl, and the cultures were harvested at various times. β-Galactosidase activity was measured in the cell extracts prepared from the doubly transformed cells. As shown in Figure 6, cells expressing the AtGSK1 cDNA exhibited increasing β-galactosidase activity with time of NaCl treatment, similar to what happens when the PMR2A:lacZ gene is expressed in the isogenic wild-type cells, DHT22-1b. In contrast, mutant cells harboring a control plasmid did not show any measurable β-galactosidase activity until 1 h. However, the mutant cells eventually expressed the β-galactosidase encoded by PMR2A:lacZ because the PMR2A gene can also be induced by other signaling pathways (data not shown) (Cunningham and Fink, 1996). This result clearly shows that AtGSK1 can function in the NaCl stress signal transduction pathway of yeast.
Figure 6.
Activation of the PMR2A gene in calcineurin mutant cells by AtGSK1. The AtGSK1 cDNA was ligated into an expression vector pJG10. The resulting construct was introduced into DHT22-1a cells, and the cells were screened on SC-Trp plates. The transformed yeast cells were then transformed with a reporter construct, PMR2A:lacZ, and the cells were plated on SC-Ura plates. The double transformants were grown in YPD medium at 30°C overnight, and the overnight culture was inoculated to an A600 of 0.1 into YPD medium. The culture was further incubated until mid-log phase, and NaCl was added to 0.5 m NaCl. Cells were harvested at various time points. Cell extracts were prepared and assayed for β-galactosidase activity. Three independent experiments were carried out and similar results were obtained. •, ♦, and ▪ indicate the wild type with PMR2A:lacZ, the mutant with pJG10 and PMR2A:lacZ, and the mutant with AtGSK1 and PMR2A:lacZ, respectively.
AtGSK1 Complements mck1 Mutant Cells in Yeast
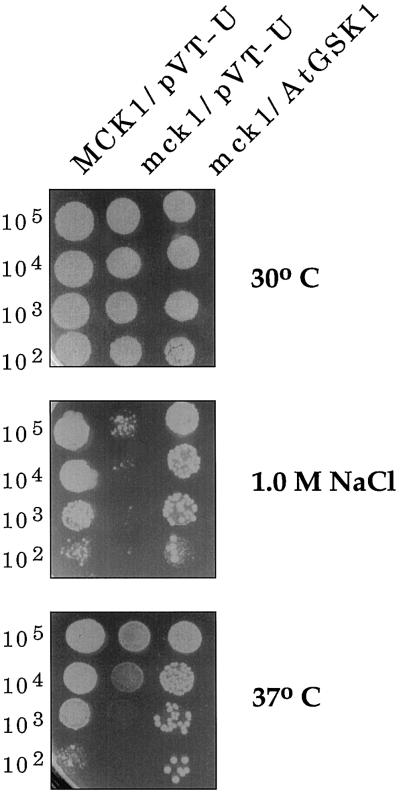
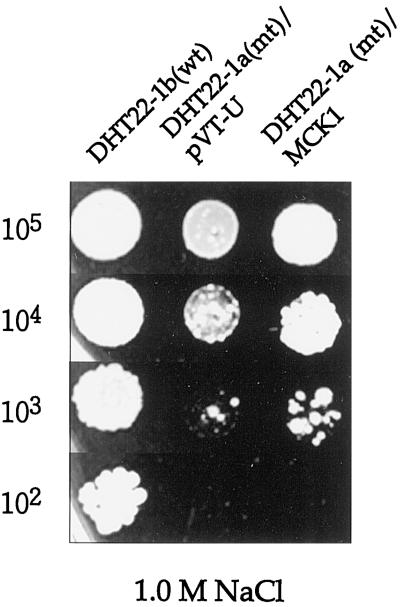
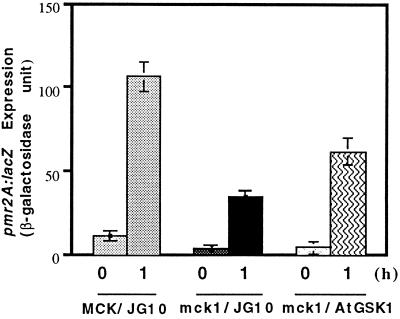
Two GSK3/shaggy-like protein kinase genes, MCK1 (Neigeborn and Mitchell, 1991) and MDS1 (Puziss et al., 1994), have been identified in yeast. Therefore, we investigated a possibility that AtGSK1 may functionally mimic any of the yeast GSK3/shaggy homologs in the NaCl stress signaling in the calcineurin mutant. However, it was not known whether either Mck1p or Mds1p is involved in the NaCl stress signal transduction pathway. Therefore, first we generated mck1:His3 and mds1:His3 mutants and examined whether these mutant cells show NaCl stress sensitivity. As shown in Figure 7, the mck1 mutant cells showed sensitivity to high NaCl concentration. However, the mds1 mutant cells did not show NaCl stress sensitivity (data not shown). Next, we investigated whether AtGSK1 gene can complement the NaCl stress-sensitive phenotype of the mck1 mutant cells. As shown in Figure 7, AtGSK1 rescued the NaCl sensitivity of the mutant. Thus, this result clearly suggests that AtGSK1 can functionally replace Mck1p in the yeast cells. Next, we addressed whether MCK1 can rescue the calcineurin mutant from the NaCl-sensitive phenotype. We introduced the ADH1:MCK1 hybrid gene into calcineurin mutant cells and then examined the phenotype of the mutant cells. As shown in Figure 8, MCK1 complemented the NaCl-sensitive phenotype of DHT22-1a cells. To further characterize the mutant we introduced the chimeric PMR2A:lacZ gene into mck1 mutant cells and assayed for the induction of the PMR2A:lacZ gene in the mutant by 0.5 m NaCl stress for 1 h. As shown in Figure 9, the level of β-galactosidase activity was markedly reduced in the mck1 mutant cells. Next, we examined whether AtGSK1 can induce the PMR2A gene under the NaCl stress conditions. As shown in Figure 9, AtGSK1 partially restored the inducibility of the PMR2A:lacZ gene in the mutant. Therefore, these results suggest that Mck1p plays a role in the NaCl stress signal transduction pathway and that AtGSK1 can functionally replace Mck1p in NaCl stress signaling in the yeast cells.
Figure 7.
Complementation of the mck1 mutant by the AtGSK1 gene. The AtGSK1 gene was introduced into mck1 mutant cells, and the transformed cells were examined for the complementation of temperature sensitivity and NaCl sensitivity of the mutants. The transformed cells were grown liquid SC-Ura− medium at 30°C overnight. The overnight cultures were diluted in YPD medium, plated on YPD, and incubated at 37°C for 3 d to examine temperature sensitivity. To examine NaCl sensitivity the cells were plated in a serial dilution on YPD supplemented with 1.0 m NaCl and incubated at 30°C for 3 d. The numbers on the left side indicate the density of yeast cells.
Figure 8.
Complementation of the calcineurin mutant by the MCK1 gene. The MCK1 gene was introduced into calcineurin mutant (mt) cells, and the transformed cells were examined for the complementation of NaCl sensitivity of the mutants. The transformed cells were grown in liquid SC-Ura− medium at 30°C overnight. The overnight cultures were diluted in YPD medium, plated in a serial dilution on YPD supplemented with 1.0 m NaCl, and incubated at 30°C for 3 d. The numbers on the left side indicate the density of yeast cells. wt, Wild type.
Figure 9.
Induction of PMR2A:lacZ in the mck1 mutant cells. The PMR2A:lacZ and AtGSK1 genes were introduced into the mck1 mutant cells. The β-galactosidase activity was assayed at 1 h, as described in the Figure 6 legend. Three independent assays were carried out and showed nearly identical results.
DISCUSSION
To understand a mechanism of signal transduction, it is important to identify the components involved in the pathway; however, it is difficult to isolate such components. In this study we exploited the yeast mutant strain DHT22-1a, which has deletions at both genes of the calcineurin catalytic subunit (Nakamura et al., 1993), thus leaving it sensitive to NaCl stress. The Arabidopsis cDNA AtGSK1, which can rescue this yeast calcineurin mutant from demise under high NaCl conditions, encodes a protein highly homologous to GSK3 found in rat and other GSK3-like proteins in other organisms, including Arabidopsis (Woodgett, 1990; Ruel et al., 1993; Blair, 1994; Dornelas et al., 1998). In Arabidopsis alone, more than five genes encoding proteins highly homologous to GSK3/shaggy have been identified (Jonak et al., 1995; Dornelas et al., 1998). However, in most plants it is not known in which biological processes the encoded proteins function. Also, it has not been previously shown that GSK3 homologs are involved in the NaCl stress or osmotic stress signal transduction pathways. The complementation of the NaCl-sensitive phenotype by AtGSK1 in the calcineurin mutant raised a possibility that AtGSK1 is involved in the NaCl stress signal transduction pathway in plants. This notion was further supported by the fact that AtGSK1 can turn on the transcription of the PMR2A gene in yeast.
In yeast two genes, MCK1 and MDS1, encode homologs of GSK3 (Neigeborn and Michell, 1991; Puziss et al., 1994). It is possible that AtGSK1 may functionally replace Mck1p and Mds1p in the NaCl stress signal transduction in the transformed yeast cells. However, previous studies have shown that these proteins are involved in processes such as meiosis and temperature sensitivity in yeast, but it is not known whether these proteins are also involved in the NaCl stress responses. Therefore, in this study we first addressed whether Mck1p and Mds1p play a role in the NaCl stress signal transduction pathway. The mck1 mutant strain, which has an insertion mutation at the MCK1 gene, exhibited an increased sensitivity to a high NaCl condition in addition to the temperature sensitivity. However, the mds1:His3 mutant did not show any noticeable phenotype. Also, the induction level of the PMR2A gene was greatly reduced in the mck1 mutant cells. When AtGSK1 was introduced into the mck1 mutant, AtGSK1 rescued the NaCl stress sensitivity and partially restored the inducibility of the PMR2A gene in the mutant. Therefore, these results suggest that AtGSK1 can functionally replace Mck1p in the NaCl stress response in yeast. Here we did not address the detailed mechanism of the Mck1p in the NaCl stress signal transduction pathway in yeast. Recently, Matheos et al. (1997) have shown that calcineurin directly interacts with Tcn1p to activate the PMR2A gene in yeast, indicating that no additional components are necessary for the activation. However, calcineurin also plays a role in the activation of PMR2A and the NaCl stress response by a Tcn1p-independent pathway. Therefore, it is possible that Mck1p may be involved in this Tcn1p-independent pathway for the activation of the PMR2A gene. Since expression of the PMR2A gene is regulated by various mechanisms, it is equally possible that Mck1p may activate the expression of the PMR2A gene by other pathways, such as the Hog pathway, or by influencing the carbon metabolism in yeast.
Previously, it has been shown that other homologous genes, such as AtK-1 and MsK, are expressed at high levels in floral tissue (Pay et al., 1993; Jonak et al., 1995). Northern-blot analysis suggests that AtK-1 may be involved in developmental processes, as is the case with homologous genes in animals. However, it is more likely that plant GSK3 homologs are involved in several diverse biological processes, an example being the NaCl stress signal transduction pathway. The data presented here strongly suggest that AtGSK1 is involved in the NaCl stress signal transduction pathway in Arabidopsis. Recently, Pardo et al. (1998) have shown that constitutively activated yeast calcineurin in tobacco plants can substantially increase tolerance to high NaCl stress. Therefore, these results suggest that a calcineurin homolog functions in the NaCl-adaptation process in plants. If this is the case in plants, it is possible that AtGSK1 may be involved in a calcineurin signal transduction pathway in plants. The notion that AtGSK1 may be involved in the NaCl stress signal transduction pathway was further supported by induction of AtGSK1 expression upon NaCl and ABA treatments. However, additional studies are needed to fully elucidate the detailed mechanism by which AtGSK1 plays a role in the NaCl stress response in plants.
The accession number for the nucleotide sequence of the AtGSK1 cDNA reported in this article is AF019927.
ACKNOWLEDGMENTS
We would like to thank Dr. T. Miyakawa (Hiroshima University, Japan) for the yeast strains DHT22-1a and DHT22-1b, and Dr. G.R. Fink (Massachusetts Institute of Technology, Cambridge) for the reporter construct PMR2A:lacZ.
Abbreviations:
- SC
synthetic complete
- Ura
uracil
- YPD
yeast peptone dextrose
Footnotes
This work was supported in part by grants from the Korea Ministry of Science and Technology and from Korea Science and Engineering Foundation to the Plant Molecular Biology and Biotechnology Research Center, Gyeongsang National University.
LITERATURE CITED
- Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1989) Current Protocol in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York
- Bianchi MW, Guivarch D, Thomas M, Woodgett JR, Kreis M. Arabidopsis homologs of the shaggy and GSK-3 protein kinases: molecular cloning and functional expression in Escherichia coli. Mol Gen Genet. 1994;242:337–345. doi: 10.1007/BF00280424. [DOI] [PubMed] [Google Scholar]
- Bianchi MW, Plyte SE, Kreis M, Woodgett JR. A Saccharomyces cerevisiae protein-serine kinase related to mammalian glycogen synthase kinase-3 and the Drosophila melanogaster gene shaggy product. Gene. 1993;134:51–56. doi: 10.1016/0378-1119(93)90173-z. [DOI] [PubMed] [Google Scholar]
- Binzel ML, Hess FD, Bressan RA, Hassegawa PM. Intracellular compartmentation of ions in salt adapted tobacco cells. Plant Physiol. 1988;86:607–614. doi: 10.1104/pp.86.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair SS. A role for the segment polarity gene shaggy-zeste white 3 in the specification of regional identity in the developing wing of Drosophila. Dev Biol. 1994;162:229–244. doi: 10.1006/dbio.1994.1081. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq-Ferrant V, Van Went J, Bianchi MW, de Vries SC, Kreis M. Petunia hybrid homologues of shaggy/zeste-white 3 expressed in female and male reproductive organs. Plant J. 1995;7:897–911. doi: 10.1046/j.1365-313x.1995.07060897.x. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma D.P.S (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4: 215–223
- Dominguez I, Itoh K, Sokol SY. Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc Natl Acad Sci USA. 1995;92:8498–8502. doi: 10.1073/pnas.92.18.8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas MC, Lejeune B, Dron M, Kreis M. The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: structure, organization and evolution. Gene. 1998;212:249–257. doi: 10.1016/s0378-1119(98)00147-4. [DOI] [PubMed] [Google Scholar]
- DuPont FM (1992) Salt-induced changes in ion transport: regulation of primary pumps and secondary transporters. In DT Cooke, DT Clarkson, eds, Transport and Receptor Proteins of Plant Membranes. Plenum Press, New York, pp 91–100
- Embi N, Rylatt DB, Cohen P. Glycogen synthase kinase-3 from rabbit skeletal muscle: separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur J Biochem. 1980;107:519–527. [PubMed] [Google Scholar]
- Harwood AJ, Plyte SE, Woodgett J, Strutt H, Kay RR. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- Hwang I, Goodman HM. An Arabidopsis thaliana root-specific kinase homolog is induced by dehydration, ABA, and NaCl. Plant J. 1995;8:37–43. doi: 10.1046/j.1365-313x.1995.08010037.x. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Heberle-Bors E, Hirt H. Inflorescence-specific expression of AtK-1, a novel Arabidopsis thaliana homologue of shaggy/glycogen synthase kinase-3. Plant Mol Biol. 1995;27:217–221. doi: 10.1007/BF00019194. [DOI] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H. Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa PC, Hasegawas PM, Rhodes D, Clithero JM, Watad A-EA, Bressan RA. Abscisic acid stimulated osmotic adjustment and its involvement in adaptation of tobacco cells to NaCl. Plant Physiol. 1987;85:174–181. doi: 10.1104/pp.85.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE 2 (ABI2) and ABI1 genes encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Matheos DP, Kingbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Gene Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S-I, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Nikolakaki E, Coffer PJ, Hemelsoet R, Woodgett JR, Defize LH. Glycogen synthase kinase 3 phosphorylates Jun family members in vitro and negatively regulates their transactivating potential in intact cells. Oncogene. 1993;8:833–840. [PubMed] [Google Scholar]
- Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota IM, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Pardo JM, Reddy MP, Yang S, Maggio A, Huh G-H, Matsumoto T, Coca MA, Paino-D'Urzo M, Koiwa H, Yun D-J and others. Stress signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proc Natl Acad Sci USA. 1998;95:9681–9686. doi: 10.1073/pnas.95.16.9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pay A, Jonak C, Bogre L, Meskiene I, Mairinger T, Szalay A, Heberle-Bors E, Hirt H. The MsK family of alfalfa protein kinase genes encodes homologues of shaggy/glycogen synthase kinase-3 and shows differential expression patterns in plant organs and development. Plant J. 1993;3:847–856. doi: 10.1111/j.1365-313x.1993.00847.x. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD-SSK1 “two-component“ osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Puziss JW, Hardy TA, Johnson RB, Roach PJ, Hieter P. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 1994;14:831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Bourouis M, Heitzler P, Pantesco V, Simpson P. Drosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch. Nature. 1993;362:557–560. doi: 10.1038/362557a0. [DOI] [PubMed] [Google Scholar]
- Skriver K, Mundy J. Gene expression in response to abscisic acid and osmotic stress. Plant Cell. 1990;2:503–512. doi: 10.1105/tpc.2.6.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hyashida N, Shinozaki K. Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet. 1994;244:331–340. doi: 10.1007/BF00286684. [DOI] [PubMed] [Google Scholar]
- Walton DC. Biochemistry and physiology of abscisic acid. Annu Rev Plant Physiol. 1980;31:453–489. [Google Scholar]
- Watson JC, Thompson WF. Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol. 1986;118:57–75. [Google Scholar]
- Welsh GI, Proud CG. Glycogen synthase kinase-3 is rapidly inactivated in response to insulin and phosphorylates eukaryotic initiation factor eIF-2B. Biochem J. 1993;294:625–629. doi: 10.1042/bj2940625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]