Abstract
c-Myc (Myc) is a well known transcription factor that regulates many essential cellular processes; however, its role in modulating immunity is not known. Here, we showed different species of mycobacteria can induce Myc expression via ERK1/2 and JNK activation. Unexpectedly, the induced Myc is localized in the cytoplasm but not in the nucleus. This induced Myc expression is associated with the induction of TNF-α and IL-6 and with the suppression of intracellular mycobacterial growth. To delineate the underlying mechanisms, we demonstrated that Myc enhances IRAK1 degradation, leading to specific activations of ERK1/2 and p38 MAPK but not Akt, and reduces IκBα protein recovery upon degradation. Hence, our findings may provide insights into a potential role for Myc in regulating the antimicrobial responses.
Keywords: cytokine, macrophages
Tuberculosis remains a devastating disease around the world and also a major cause of death in AIDS patients (1). It is caused by infection with Mycobacterium tuberculosis (M. tuberculosis). Additionally, nontuberculous mycobacteria such as M. avium-intracellulare and M. kansasii are other common opportunistic pathogens causing disseminated infection in patients in the late stages of AIDS (2).
To control mycobacterial infections, the pathogens are recognized by specific host receptors including toll-like receptors (TLR), leading to activation of signaling cascades and consequent expression of cytokines for innate and adaptive immunity (3). For example, upon pathogen recognition, the activated TLR2 or TLR4 recruit MyD88, IRAK1, and IRAK4. This activates IκBα and mitogen-activated protein kinases (MAPK) including ERK1/2, p38 kinase and JNK (4, 5). Furthermore, we have recently demonstrated that mycobacteria activate ERK1/2 and p38 kinase via the dsRNA-activated protein kinase (PKR) and MAPK-phosphatase-1 (MKP1) (6, 7). Consequently, activation of these pathways triggers transcription factors including NFκB, CREB, and AP-1 to up-regulate the expression of cytokines such as TNF-α and IL-6 (4–7).
c-Myc (Myc) was first identified as the human homolog of v-myc, an oncogenic protein of avian myelocytomatosis virus (8). It possesses a N-terminal transactivation domain and a C-terminal basic helix–loop–helix (HLH) leucine zipper (LZ) domain for binding to a CACGTG E-box DNA sequence (9). Max, which also contains a HLH–LZ domain, dimerizes with Myc to bind to the E-box element of the promoter of target genes (9). This leads to the recruitment of histone acetyltransferase complexes for opening the chromatin for transcription of the target genes (10). Moreover, Myc interacts with RNA polymerase I, II, and III for transcription of different genes (10) and binds with cofactors of the prereplicative complex on DNA of the origin of replication for initiating the S phase of cell cycle (11). Through these mechanisms in the nucleus, Myc controls different cellular processes including cell cycle, cell growth and differentiation, metabolism, protein synthesis, cell adhesion and migration, angiogenesis, chromosomal instability, stem cell renewal, and apoptosis (9).
Myc is crucial to control the differentiation and self-renewal processes of hematopoietic stem cells, leading to the development of the progenitor cells (12, 13). These progenitor cells then differentiate into immune cells including monocytes and lymphocytes for mediating innate and adaptive immune responses. However, the precise roles of Myc in regulating cytokine induction and related immune responses against invading microbes have not been reported. Here, we showed that Myc plays a critical role in regulating innate immune responses against mycobacterial infection.
Results
Myc Expression Is Induced by Mycobacterial Infection.
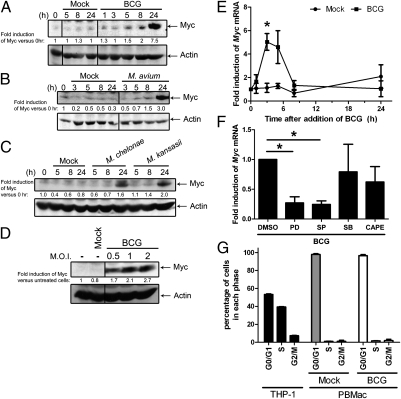
To examine whether mycobacteria induce Myc expression, primary human blood macrophages (PBMac) were treated with mycobacteria including Bacillus Calmette-Guérin (BCG), M. avium, M. chelonae, or M. kansasii. The results showed that the Myc protein levels were increased by these mycobacteria in a time-dependent manner (Fig. 1 A–C). Moreover, the level of Myc induction was dependent on the concentration of mycobacteria used (Fig. 1D). Apart from protein expression, Myc mRNA levels were significantly up-regulated by BCG in a time-dependent manner (Fig. 1E).
Fig. 1.
Mycobacteria induce Myc expression. PBMac were treated with (A) mock or BCG [multiplicity of infection (M.O.I.) = 1], (B) mock or M. avium (M.O.I. = 20), and (C) mock or M. chelonae (M.O.I. = 10) or M. kansasii (M.O.I. = 1) for the indicated time. (D) PBMac were treated with mock or BCG with the indicated M.O.I. for 24 h. (A–D) Cellular proteins were extracted for Western blots to analyze the levels of Myc and Actin. The data are representative results from cells isolated from three independent blood donors. The values under each lane represent the fold induction of Myc versus 0 h or the unteated cells after normalization to the level of Actin. (E) PBMac were treated with mock or BCG (M.O.I. = 1) for the indicated time. (F) PBMac were pretreated with DMSO, 10μM inhibitors antagonizing ERK1/2 (PD), JNK (SP) or p38 MAPK (SB), or 15μg/mL inhibitor against NFκB (CAPE) for 1 h, and followed by treatment with mock or BCG (M.O.I. = 1) for 3 h. (E and F) Total RNA was extracted for analyzing the mRNA expression levels by quantitative RT-PCR. (G) PBMac were treated with mock or BCG (M.O.I = 1) for 24 h. PBMac and THP-1 cells were fixed and stained with propidium iodide. The DNA contents of the cells were analyzed by flow cytometry and ModFit 3.2 software. (E–G) The data are presented as mean ± SEM from three independent blood donors. * denotes P < 0.05 as determined by Student's t test.
To delineate the signaling mechanisms underlying mycobacteria induction of Myc expression, PBMac were pretreated with specific inhibitors against various signaling molecules or kinases, and followed by BCG treatment. As measured by quantitative reverse-transcription PCR (RT-PCR), the levels of BCG-induced Myc mRNA were significantly reduced after pretreatment of the cells with inhibitors against ERK1/2 (PD98059) and JNK1/2 (SP600159), compared with those treated with DMSO (Fig. 1F). However, pretreatment with inhibitors against p38 kinases (SB203580) and NFκB (CAPE) did not affect the BCG-induced Myc mRNA transcription (Fig. 1F).
Previous studies have shown that Myc protein expression is associated with cell proliferation, regulates cell cycle, and possesses an oncogenic activity in cancer (9, 11). As mycobacteria induced Myc protein expression, we investigate whether the microbes may also affect the cell proliferation or cell cycle. To address this, PBMac were treated with mock or BCG, and analyzed for DNA content by flow cytometry. As a positive control, around 53%, 39%, and 8% of the THP-1 promonocytic cells are in the G0/G1, S, and G2/M phases, respectively, indicating that THP-1 cells are proliferating (Fig. 1G). In contrast, more than 90% and less than 4% and 5% of PBMac, which were treated with mock or BCG for 24 (Fig. 1G), 48, and 96 h (Fig. S1A), were in the G0/G1, S, and G2/M phases, respectively. In addition, all these percentages of cells in each phase did not significantly changed with the increasing duration of mycobacteria treatment (Fig. S1A). The results also showed that BCG treatment did not affect the cell number of PBMac (Fig. S1 C and D). These results suggest that unlike in other cellular system, mycobacteria neither affected the proliferation nor changed the G0/G1 phase of cell cycles of human macrophages.
Together, these results demonstrated that induction of Myc protein is a common pathway used by different species of mycobacteria and was mediated by the activation of ERK1/2 and JNK1/2 but not that of p38 kinases and NFκB in human macrophages with a G0/G1 phase.
Myc Is Localized in the Cytoplasm of Macrophages.
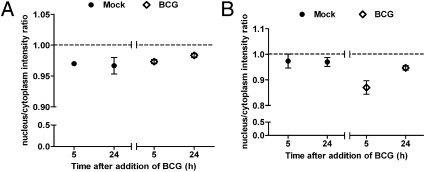
Because previous studies have reported that Myc is localized in the nucleus and functions as a transcription factor to regulate various cellular processes (9, 14–16), we investigated the subcellular location of Myc in PBMac following mycobacterial infection. As assayed by quantitative immunofluorescence microscopy using two different antibodies against Myc, we showed that the nucleus/cytoplasm intensity ratios of Myc protein were below one in macrophages treated with mock or BCG for 5 and 24 h (Fig. 2 A and B). In addition, there were no significant changes of the ratios before and after BCG treatment for either 5 or 24 h (Fig. 2 A and B). Consistent with Fig. 1A, the total signal intensity of Myc was significantly increased by BCG in a time-dependent manner (Fig. S2 A and B). These results suggest that Myc protein remained localized in the cytoplasm of PBMac, which were treated with mock or BCG.
Fig. 2.
Myc localizes in the cytoplasm. PBMac were treated with mock or BCG (M.O.I. = 1) for 5 and 24 hours. Cells were fixed and labeled with monoclonal primary antibody against Myc (4A6, Millipore; A) or polyclonal primary antibodies against Myc (N262, Santa Cruz Biotechnology; B), and followed by the corresponding secondary antibodies. Subsequently, cell nuclei were labeled with DAPI. Images of cells were analyzed by Cellomics ArrayScan HCS VTI System. The cytoplasmic localization of Myc was quantified and expressed as nuclear/cytoplasmic intensity ratio (ratio > 1 implies in nucleus, ratio < 1 implies in cytoplasm). The data are presented as mean ± SEM from three independent blood donors.
As verified by Western blot analyses, Myc proteins were found in the cytoplasm, but not in the nucleus, of mock or BCG cells (Fig. S2C). Consistent with Fig. 1A, the Myc protein level in BCG-treated cells was higher than that in mock-treated cells by twofold (Fig. S2C). The specific cytoplasm marker, α-tubulin, and the specific nucleus marker, lamin B, were only found in the cytoplasm and nucleus, respectively, but not vice versa (Fig. S2C). These results indicated the purity of the fractionated proteins. Hence, these results suggest that both constitutive and mycobacteria-induced Myc proteins resided in the cytoplasm but not in the nucleus of PBMac.
Myc Specifically Augments the Mycobacteria Induction of TNF-α and IL-6, but not IL-10 Protein.
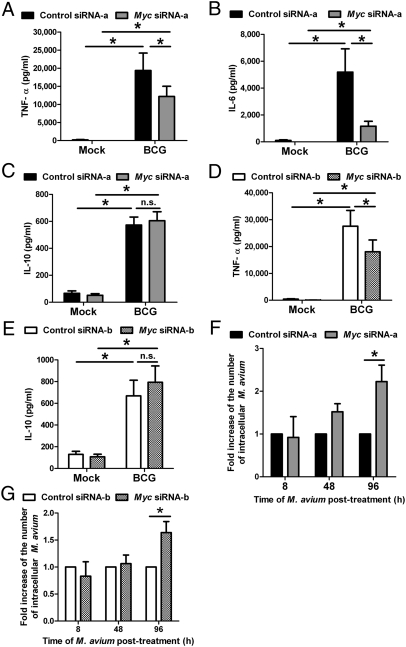
To investigate the role of Myc in regulating cytokine induction, PBMac were preincubated with control siRNA-a or Myc siRNA-a (Thermo Fisher Scientific), which targets the coding region of Myc mRNA, and then treated with BCG or M. avium. Myc siRNA-a significantly reduced the BCG-induced Myc protein levels compared with the control siRNA-a treated cells (Fig. S3A). With this condition, the culture media were harvested for measuring the cytokine levels by ELISA. The results showed that Myc siRNA-a significantly decreased the BCG- and M. avium-induced production of TNF-α and IL-6 protein (Fig. 3 A and B and Fig. S4 A and B). In contrast, Myc siRNA-a neither affected the basal nor the mycobacteria-induced IL-10 protein levels compared with the effects of the control siRNA-a in the treated cells (Fig. 3C). Likewise, Myc siRNA-a significantly down-regulated the mycobacteria-induced mRNA transcription of IL-6 and TNF-α (Fig. S4 C and D) but not that of IL-10 (Fig. S4E). In addition, Myc siRNA-a neither affected the mycobacterial survival nor the viability of PBMac at an early phase of infection despite its effects on inhibiting the mycobacteria-induction of TNF-α and IL-6 (Figs. S5 and S6). Furthermore, Myc siRNA-a neither significantly changed the G0/G1 phase nor the cell number of PBMac, indicating that Myc was not involved in regulating cell proliferation and cell cycle (Fig. S1 A and C).
Fig. 3.
Myc specifically mediates mycobacteria-induction of cytokines and suppresses the intracellular growth of mycobacteria. (A–E) PBMac were transfected with control or Myc siRNA-a (A–C) or control or Myc siRNA-b (D and E) for 48 h, and followed by treatment with mock or BCG (M.O.I. = 1) for 24 h. Supernatants were harvested for determining the levels of the indicated cytokines by ELISA. (F and G) PBMac were treated with control or Myc siRNA-a (F) or control or Myc siRNA-b (G) for 48 h and followed by treatment with M. avium (M.O.I. = 20) for 8 h. Cells were either lysed for determining the number of intracellular bacteria or were washed with PBS. After washings and treatment with trypsin/EDTA, the cells were incubated with fresh culture medium for another 40 or 88 h. At the indicated time, cells were then lysed for determining the number of intracellular bacteria. The numbers of intracellular M. avium bacteria in cells transfected with Myc siRNA-a or Myc siRNA-b cells were expressed as fold induction over those transfected with control siRNA-a or control siRNA-b, respectively. The data are expressed as mean ± SEM from 8 (A), 7 (B), 3 (C), 4 (D), 4 (E), 3 (F), and 10 (G) independent blood donors. For A–E, * and n.s. denote P < 0.05 and P > 0.05, respectively, as determined by Student's t test. For F and G, * denotes P < 0.05 as determined by two-way ANOVA.
To further confirm these results, PBMac were transfected with control siRNA-b or Myc siRNA-b (Applied Biosystems), which targets the 3′ UTR of Myc mRNA, and then treated with BCG. Myc siRNA-b significantly decreased the BCG-induced Myc protein levels after BCG addition, compared with the control siRNA-b-treated cells (Fig. S3B). Consistent with the results obtained by Myc siRNA-a, Myc siRNA-b specifically reduced the mycobacteria-production of TNF-α protein (Fig. 3D) but not that of IL-10 protein (Fig. 3E). Similar to Myc siRNA-a, Myc siRNA-b neither significantly changed the G0/G1 phase nor the cell number of PBMac (Fig. S1 B and D). Therefore, these results suggest that Myc was required for specifically mediating mycobacteria-induction of TNF-α and IL-6 but not that of IL-10.
Myc Mediates the Suppression of the Intracellular Growth of Mycobacteria.
To address whether Myc affects the intracellular growth of mycobacteria, PBMac were transfected with control or Myc siRNA-a or with control or Myc siRNA-b, and then treated with M. avium for up to 96 h. The number of intracellular mycobacteria was quantified. After 8- and 48-h of M. avium treatment, the numbers of intracellular mycobacteria in cells transfected with control and Myc siRNA-a-transfected cells, and with control and Myc siRNA-b were comparable (Fig. 3 F and G). However, after 96 h of M. avium treatment, the numbers of intracellular mycobacteria in cells transfected with Myc siRNA-a and Myc siRNA-b, were significantly higher than that of the cells transfected with control siRNA-a and control siRNA-b, respectively (Fig. 3 F and G). In addition, this effect of Myc was not resulted from the oncogene-induced protection from cell death as shown by the nonsignificant differences of viabilities between the control and Myc siRNA-a-transfected cells after 96 h of mycobacteria treatment (Fig. S7). Moreover, as a previous study showed that Myc represses the transcription of NRAMP1 gene in which its encoded protein is responsible for inhibiting the intracellular mycobacterial growth (17), the expression level of NRAMP1 mRNA was examined. The results showed that the NRAMP1 mRNA expressions were not affected by either mycobacteria or Myc (Fig. S8 A and B), indicating that NRAMP1 was not required for the Myc-inhibition of intracellular mycobacterial growth. These results suggest that Myc was crucial for limiting the intracellular growth of mycobacteria in macrophages at a later phase of infection.
Myc Regulates the Activation of Mycobacteria-Induced Signaling Molecules.
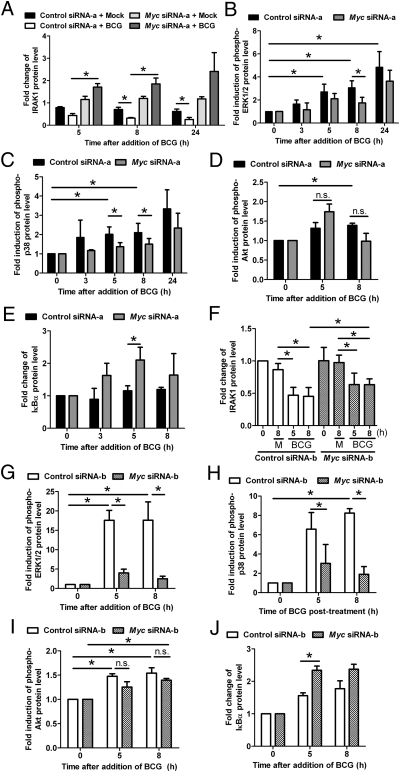
To delineate the mycobacteria-triggered pathways that are regulated by Myc, PBMac were transfected with control or Myc siRNA-a before BCG treatment, and followed by additional Western analysis. The results demonstrated that BCG induced the degradation of IRAK1 in a time-dependent manner (Fig. 4A). However, Myc siRNA-a substantially inhibited this IRAK1 degradation (Fig. 4A). Furthermore, Myc siRNA-a decreased the phosphorylation levels of ERK1/2 and p38 kinases induced by BCG treatment (Fig. 4 B and C). In contrast, while the Akt phosphorylation levels increased at eight hours after BCG addition, they were not affected by the Myc siRNA-a (Fig. 4D). Additionally, similar results were observed when transfection with Myc siRNA-a was replaced by that with Myc siRNA-b (Fig. 4 F–I). These results indicate that Myc specifically enhanced mycobacteria-activation of IRAK1 as well as the downstream kinases including ERK1/2 and p38 kinases but not that of Akt.
Fig. 4.
Myc augments mycobacteria-activation of IRAK1, MAPK and NFκB. PBMac were transfected with control or Myc siRNA-a as in Fig. 3 A–C for A–E, and with control or Myc siRNA-b as in Fig. 3 D and E for F–J before BCG (M.O.I. = 1) treatment for the indicated time. Cytoplasmic proteins were analyzed by Western blot. (A and F) The protein level of IRAK1 was quantified, normalized with that of Actin, and expressed as fold change over the treatment with control siRNA-a or siRNA-b and mock. (B–D and G–I) The levels of phosphorylated proteins of EKR1/2, p38 kinase and Akt at each time point were normalized with the level of their respective total proteins and expressed as fold induction over their basal levels at 0 h. (E and J) The protein level of IκBα was normalized with that of Actin at each time point and expressed as fold change over its basal level at 0 h. The data are expressed as mean ± SEM from three (A), five (B), four (C), three (D), four (E), four (F), four (G), three (H), three (I), and three (J) independent blood donors. M denotes mock. * denotes P < 0.05 as determined by Student's t test.
Moreover, the IκBα protein levels were significantly up-regulated by Myc siRNA-a and by Myc siRNA-b at five hours (Fig. 4 E and J). These results suggest that Myc reduced the mycobacteria-induced IκBα protein levels. Additionally, as assayed by immunocytochemistry, our results showed that BCG induced the NFκB p65 nuclear localization with an increase of the nucleus/cytoplasm intensity ratio, but Myc siRNA-a reduced this nuclear localization with a significant decrease of the ratio (Fig. S9). These results suggest that Myc augmented the mycobacteria-induced NFκB pathway.
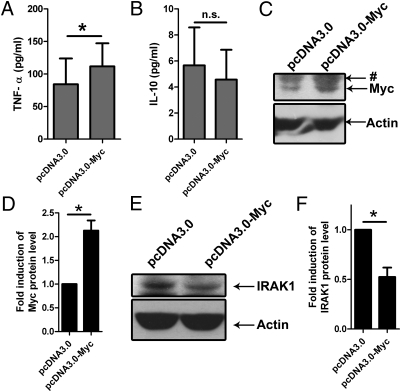
Myc Is Sufficient for TNF-α Induction and IRAK1 Activation.
To confirm the results obtained by the knockdown experiments using siRNA, we investigated whether Myc is sufficient to induce TNF-α. PBMac were electroporated with pcDNA3.0-Myc, a Myc protein-expression plasmid. As assayed by Western blot, the Myc protein level increased in cells electroporated with pcDNA3.0-Myc, compared with those electroporated with the control pcDNA3.0 plasmid (Fig. 5 C and D), indicating that Myc can be overexpressed in the transfected PBMac. Next, the cytokine protein levels of the Myc-overexpressing cells were determined by ELISA. The results showed that TNF-α protein production was significantly elevated by Myc overexpression compared with the control cells (Fig. 5A). However, IL-10 production of the pcDNA3.0-Myc-transfected cells was not affected by the Myc overexpression (Fig. 5B). These results indicate that Myc was sufficient for the specific induction of TNF-α but not for that of IL-10 production.
Fig. 5.
Overexpression of Myc is sufficient for TNF-α induction and IRAK1 activation. PBMac were electroporated with the indicated expression plasmid and incubated with culture medium for 48 h. Supernatants were discarded and the cells were incubated with fresh culture medium for 24 h. (A and B) Supernatants were harvested for determining the levels of the indicated cytokines by ELISA. The data are expressed as mean ± SEM from four (A) and three (B) independent blood donors. (C and E) Total proteins were harvested for analysis by Western blot. The data are representative results from cells isolated from three independent blood donors. (D and F) Protein levels of Myc (D) and IRAK1 (F) were normalized with that of Actin and expressed as fold induction relative to the cells electroporated with pcDNA3.0 plasmid. The data are expressed as mean ± SEM from three independent blood donors. The * and n.s. denote P < 0.05 and P > 0.05, respectively, as determined by Student's t test. The # denotes nonspecific bands.
To verify whether Myc induces IRAK1 activation, we also examined the protein levels of IRAK1 by Western Blot. The results demonstrated that the protein level of IRAK1 decreased significantly in cells with overexpression of Myc compared with the control cells (Fig. 5 E and F). These results suggest that Myc was adequate for reducing the expression of IRAK1 protein.
Discussion
In this report, we demonstrated that several species of mycobacteria with different pathogenicity including M. bovis (BCG), M. avium, M. kansasii, and M. chelonae stimulated the expression of Myc protein in a time- and dose-dependent manner, due to transcriptional activation of Myc. These results further verified previous uncharacterized microarray data obtained from cells infected with M. avium (18, 19). Indeed, the levels and kinetics of Myc expression vary among different experiments. This may be due to the individual variations among primary cells from different blood donors. In fact, other pathogens, including Helicobacter pylori and Salmonella typhimurium, had also been shown to trigger Myc protein expression in mouse macrophages (20, 21). Moreover, Myc has been shown to act as a transcription factor to induce the mRNA expression of cytokines including IL-2, IL-13, and IL-17C in cancer cell lines (22, 23). However, the mechanisms including how Myc is induced by these pathogens and the functional consequences of induced Myc in primary immune cells for triggering innate immune responses have not been reported. Here, we demonstrated that mycobacteria induced Myc transcription via the activation of ERK1/2 and JNK1/2 and the induced Myc plays a role in triggering anti-mycobacterial responses without affecting the cell proliferation and changing the G0/G1 phase of cell cycle of macrophages.
Although a previous report showed that the Myc protein level increases in the nucleus upon H. pylori infection in a mouse macrophage cell line (21), our finding is consistent with several other studies showing that Myc localizes in the cytoplasm in various cells including lymphoid and differentiated myeloid cells, fibroblasts, and HeLa cells (24–27). It has not been known why Myc is localized in the cytoplasm in some cells but not in the others. However, it has been suggested that hyperphosphorylation of Myc disrupts its interaction with α-tubulin during mitosis (26). This disruption may lead to the retention of Myc in the nucleus. As our results showed that differentiated human macrophages did not enter into the phases of DNA synthesis and mitosis, it would be intriguing to investigate whether Myc is unphosphorylated and resides in the cytoplasm of primary human macrophages via similar mechanisms.
Our results suggest that mycobacteria-induced Myc may play a positive feedback role in specifically augmenting the microbe-induced proinflammatory cytokines including TNF-α and IL-6, but not on the anti-inflammatory cytokine IL-10 in primary human macrophages. These findings support the hypothesis that the TLR signaling induces the expression of early factors including ATF3 and XBP1 for fine-tuning the downstream innate immune responses via negative or positive feedback controls (28, 29). Nevertheless, the cytokine expression was incompletely blocked by the two siRNA against Myc. This may be due to the limited efficacy of the siRNA for knocking down the Myc protein in primary human cells. As described, the siRNA concentration that we used in this study was already optimal for achieving the maximum knockdown of the protein with more than 90% transfection efficiency. This is consistent with similar efficacy of these specific siRNA in knocking down the Myc protein in human cancer cell lines (30, 31). In addition, another possibility is that mycobacteria-induced signaling can still be transduced by other alternative pathways when Myc was knocked down, leading to the low, albeit significant, expression of cytokines including TNF-α and IL-6.
Myc has been shown to induce apoptosis and cell death in mouse macrophages infected with H. pylori and S. typhimurium, respectively (20, 21). However, our results indicate that mycobacteria-induced Myc neither regulated the microbes-induced cell death nor the mycobacterial survival at the early phase of infection. Therefore, these processes are not involved in the Myc-enhanced cytokine expression. Furthermore, previous study has shown that TNF-α inhibits the intracellular growth of mycobacteria (32, 33). As our results indicate that Myc-induction of TNF-α preceded the Myc-suppression on the mycobacterial growth, it is possible that the oncogene-suppressed mycobacterial growth is TNF-α-dependent. Further study is required to examine this hypothesis. Nevertheless, Myc has been shown to repress the interaction of IRF8 with Miz1 in mouse macrophages, resulting in the inhibition of NRAMP1 expression, which is responsible for killing the intracellular S. enterica serovar typhimurium and BCG in IFN-γ/LPS-primed macrophages (17, 34). On the contrary, our results suggest that mycobacteria-induced Myc plays a role in the inhibition of intracellular mycobacterial growth at a later phase of infection in primary human macrophages without prior IFN-γ treatment. Additionally, this suppression is independent of NRAMP1 expression. These differences in the roles of Myc may be due to the different objectives examined between their and our studies. Indeed, our experiments revealed that in response to mycobacterial infection, Myc is induced in macrophages to mediate the induction of proinflammatory cytokine responses with consequent inhibition of the intracellular growth of the microbes. These innate immune responses may lead to the T cell induction of IFN-γ, which may in turn activate other uninfected macrophages (3). This finding is in agreement with a previous report showing that Myc plays a suppressive role against pathogens in IFN-γ-induced secondary responses (34), resulting in protecting the uninfected macrophages from infection by the microbes.
It has been suggested that, upon TLR/IL-1R activation, IRAK1 is phosphorylated and degraded, allowing the signaling complexes containing TRAF6, TAK1, TAB1, TAB2, and TAB3 to translocate from the cell membrane to the cytosol for subsequent activation of MAPK and NFκB pathways (5, 35). Hence, our findings suggest that Myc may play a novel role in enhancing the degradation of IRAK1, leading to the consecutive activation of ERK1/2, p38 kinases, IκBα and NFκB. As a consequence of these Myc-stimulated signaling cascades, induction of proinflammatory cytokines including TNF-α and IL-6 were augmented as illustrated by the experiments of either knocking down or overexpressing the Myc protein. However, further studies have to be done to delineate how Myc regulates the degradation or synthesis of IRAK1 and IκBα protein. Nevertheless, our results indicated that Myc protein knockdown only partially inhibited the TNF-α expression but much reduced the IL-6 expression. Indeed, previous studies from us and other groups have shown that the expression of TNF-α and IL-6 can be regulated by multiple pathways which involve different upstream signaling molecules including PKR, MKP-1, MEKK3, ASK1, TAK1, and TPL2 (6, 7, 36), and different downstream transcription factors such as C/EBPβ and ATF3 (29, 37). In addition, expressions of these cytokines can be regulated posttranscriptionally via controlling their mRNA stabilities (38). Therefore, it is plausible that Myc may differentially regulate these signaling molecules and the mRNA stability of these cytokines, leading to the specific greater reduction of expression of IL-6 than that of TNF-α by Myc siRNA knockdown.
On the contrary, Myc had only a slight inhibition on the mycobacteria activation of Akt and did not play a role in regulating the mycobacteria induction of IL-10. In line with our previous report (39), mycobacteria may use an alternative Akt signaling pathway for inducing anti-inflammatory cytokine, IL-10. This result suggests that Myc may play a role in specifically enhancing the proinflammatory, but not that of the anti-inflammatory pathways.
These Myc-augmented expressions of proinflammatory cytokines play crucial roles in combat against mycobacteria. For instance, in conjunction with TGF-β, IL-6 has been suggested to initiate the differentiation of TH17 cells, leading to the subsequent maintenance of granulomas and the expression of memory responses (40). Additionally, TNF-α produced from macrophages plays an important role in suppressing the intracellular growth of mycobacteria and involving in the formation of granulomas to prevent dissemination of mycobacterial infection (41, 42). Furthermore, these cytokines induced by TLR and its MyD88-IRAK1-dependent signaling pathway can lead to protection against not only mycobacteria (43) but also other intracellular pathogens such as Listeria monocytogenes and Toxoplasma gondii (44). Hence, our findings suggest the importance of Myc in regulating these proinflammatory cytokine responses by activating the IRAK1-dependent pathway, and in limiting the intracellular growth of the mycobacteria. It would be intriguing to investigate whether Myc has a similar role in other pathogens. Nevertheless, as Myc plays an essential role in regulating different cellular responses in different cell types (13), differentiated blood macrophages from mice with inducible conditional knockout of Myc need to be examined to identify the specific in vivo role of Myc in regulating innate immune responses. Before the macrophage and dendritic cells of these mice are examined, our report establishes a foundation for this future in vivo study. Understanding the mechanisms in inducing these responses may shed light on the development of Myc-enhancing drugs for tackling the epidemic problems of tuberculosis.
Materials and Methods
Cell Culture.
Isolation of primary human blood macrophages and culture of THP-1 cells were described in SI Materials and Methods.
Additional Materials and Methods.
Additional materials and procedures including siRNA transfection, electroporation of PBMac, Western blot, immunocytochemistry, ELISA, and statistical analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This project was supported in part by Hong Kong Research Grants Council Grant HKU 769810M (to A.S.Y.L.) and Hong Kong Research Fund for the Control of Infectious Diseases Grant 09080512 (to A.S.Y.L.). H.C.H.Y. is the recipient of a postgraduate studentship from the University of Hong Kong and is supported in part by the Chung Kue Suen Research Fund, Li Ka Shing Faculty of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104892108/-/DCSupplemental.
References
- 1.World Health Organization . Geneva: World Health Organization; 2009. Global tuberculosis control: Epidemiology, strategy, financing: WHO report 2009. P vii, 303. [Google Scholar]
- 2.McGrath EE, Blades Z, McCabe J, Jarry H, Anderson PB. Nontuberculous mycobacteria and the lung: From suspicion to treatment. Lung. 2010;188:269–282. doi: 10.1007/s00408-010-9240-9. [DOI] [PubMed] [Google Scholar]
- 3.Harding CV, Boom WH. Regulation of antigen presentation by Mycobacterium tuberculosis: A role for Toll-like receptors. Nat Rev Microbiol. 2010;8:296–307. doi: 10.1038/nrmicro2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 5.Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol. 2009;30:33–42. doi: 10.1016/j.it.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Cheung BK, Yim HC, Lee NC, Lau AS. A novel anti-mycobacterial function of mitogen-activated protein kinase phosphatase-1. BMC Immunol. 2009;10:64. doi: 10.1186/1471-2172-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung BK, Lee DC, Li JC, Lau YL, Lau AS. A role for double-stranded RNA-activated protein kinase PKR in Mycobacterium-induced cytokine expression. J Immunol. 2005;175:7218–7225. doi: 10.4049/jimmunol.175.11.7218. [DOI] [PubMed] [Google Scholar]
- 8.Vennstrom B, Sheiness D, Zabielski J, Bishop JM. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982;42:773–779. doi: 10.1128/jvi.42.3.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 10.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 11.Herold S, Herkert B, Eilers M. Facilitating replication under stress: An oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- 12.Garrison BS, Rossi DJ. Controlling stem cell fate one substrate at a time. Nat Immunol. 2010;11:193–194. doi: 10.1038/ni0310-193. [DOI] [PubMed] [Google Scholar]
- 13.Laurenti E, Wilson A, Trumpp A. Myc's other life: Stem cells and beyond. Curr Opin Cell Biol. 2009;21:844–854. doi: 10.1016/j.ceb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Vriz S, Lemaitre JM, Leibovici M, Thierry N, Méchali M. Comparative analysis of the intracellular localization of c-Myc, c-Fos, and replicative proteins during cell cycle progression. Mol Cell Biol. 1992;12:3548–3555. doi: 10.1128/mcb.12.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papoulas O, Williams NG, Kingston RE. DNA binding activities of c-Myc purified from eukaryotic cells. J Biol Chem. 1992;267:10470–10480. [PubMed] [Google Scholar]
- 16.Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen H, et al. c-Myc represses and Miz-1 activates the murine natural resistance-associated protein 1 promoter. J Biol Chem. 2002;277:34997–35006. doi: 10.1074/jbc.M204232200. [DOI] [PubMed] [Google Scholar]
- 18.Greenwell-Wild T, et al. Mycobacterium avium infection and modulation of human macrophage gene expression. J Immunol. 2002;169:6286–6297. doi: 10.4049/jimmunol.169.11.6286. [DOI] [PubMed] [Google Scholar]
- 19.Blumenthal A, et al. Common and unique gene expression signatures of human macrophages in response to four strains of Mycobacterium avium that differ in their growth and persistence characteristics. Infect Immun. 2005;73:3330–3341. doi: 10.1128/IAI.73.6.3330-3341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seong J, et al. Expression of c-Myc is related to host cell death following Salmonella typhimurium infection in macrophage. J Microbiol. 2009;47:214–219. doi: 10.1007/s12275-008-0308-7. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, et al. Helicobacter pylori-induced macrophage apoptosis requires activation of ornithine decarboxylase by c-Myc. J Biol Chem. 2005;280:22492–22496. doi: 10.1074/jbc.C500122200. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, et al. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig RW, Buchan HL, Civin CI, Kastan MB. Altered cytoplasmic/nuclear distribution of the c-myc protein in differentiating ML-1 human myeloid leukemia cells. Cell Growth Differ. 1993;4:349–357. [PubMed] [Google Scholar]
- 26.Niklinski J, et al. Disruption of Myc-tubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt's lymphoma. Mol Cell Biol. 2000;20:5276–5284. doi: 10.1128/mcb.20.14.5276-5284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohan JN, Weigel NL. 1Alpha,25-dihydroxyvitamin D3 reduces c-Myc expression, inhibiting proliferation and causing G1 accumulation in C4-2 prostate cancer cells. Endocrinology. 2009;150:2046–2054. doi: 10.1210/en.2008-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bermudez LE, Young LS. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-gamma, is associated with macrophage killing of Mycobacterium avium complex. J Immunol. 1988;140:3006–3013. [PubMed] [Google Scholar]
- 34.Alter-Koltunoff M, et al. Innate immunity to intraphagosomal pathogens is mediated by interferon regulatory factor 8 (IRF-8) that stimulates the expression of macrophage-specific Nramp1 through antagonizing repression by c-Myc. J Biol Chem. 2008;283:2724–2733. doi: 10.1074/jbc.M707704200. [DOI] [PubMed] [Google Scholar]
- 35.Xiao H, et al. Pellino 3b negatively regulates interleukin-1-induced TAK1-dependent NF kappaB activation. J Biol Chem. 2008;283:14654–14664. doi: 10.1074/jbc.M706931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su B. Linking stress to immunity? Nat Immunol. 2005;6:541–542. doi: 10.1038/ni0605-541. [DOI] [PubMed] [Google Scholar]
- 37.Hu HM, Baer M, Williams SC, Johnson PF, Schwartz RC. Redundancy of C/EBP alpha, -beta, and -delta in supporting the lipopolysaccharide-induced transcription of IL-6 and monocyte chemoattractant protein-1. J Immunol. 1998;160:2334–2342. [PubMed] [Google Scholar]
- 38.Neininger A, et al. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J Biol Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 39.Chan MM, Cheung BK, Li JC, Chan LL, Lau AS. A role for glycogen synthase kinase-3 in antagonizing mycobacterial immune evasion by negatively regulating IL-10 induction. J Leukoc Biol. 2009;86:283–291. doi: 10.1189/jlb.0708442. [DOI] [PubMed] [Google Scholar]
- 40.Cooper AM, Khader SA. The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol Rev. 2008;226:191–204. doi: 10.1111/j.1600-065X.2008.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flynn JL, et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 42.Jacobs M, et al. Correction of defective host response to Mycobacterium bovis BCG infection in TNF-deficient mice by bone marrow transplantation. Lab Invest. 2000;80:901–914. doi: 10.1038/labinvest.3780094. [DOI] [PubMed] [Google Scholar]
- 43.Bafica A, et al. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.