Abstract
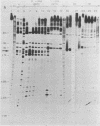
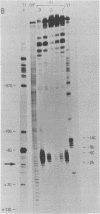
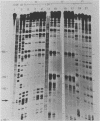
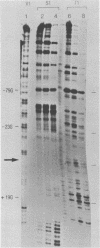
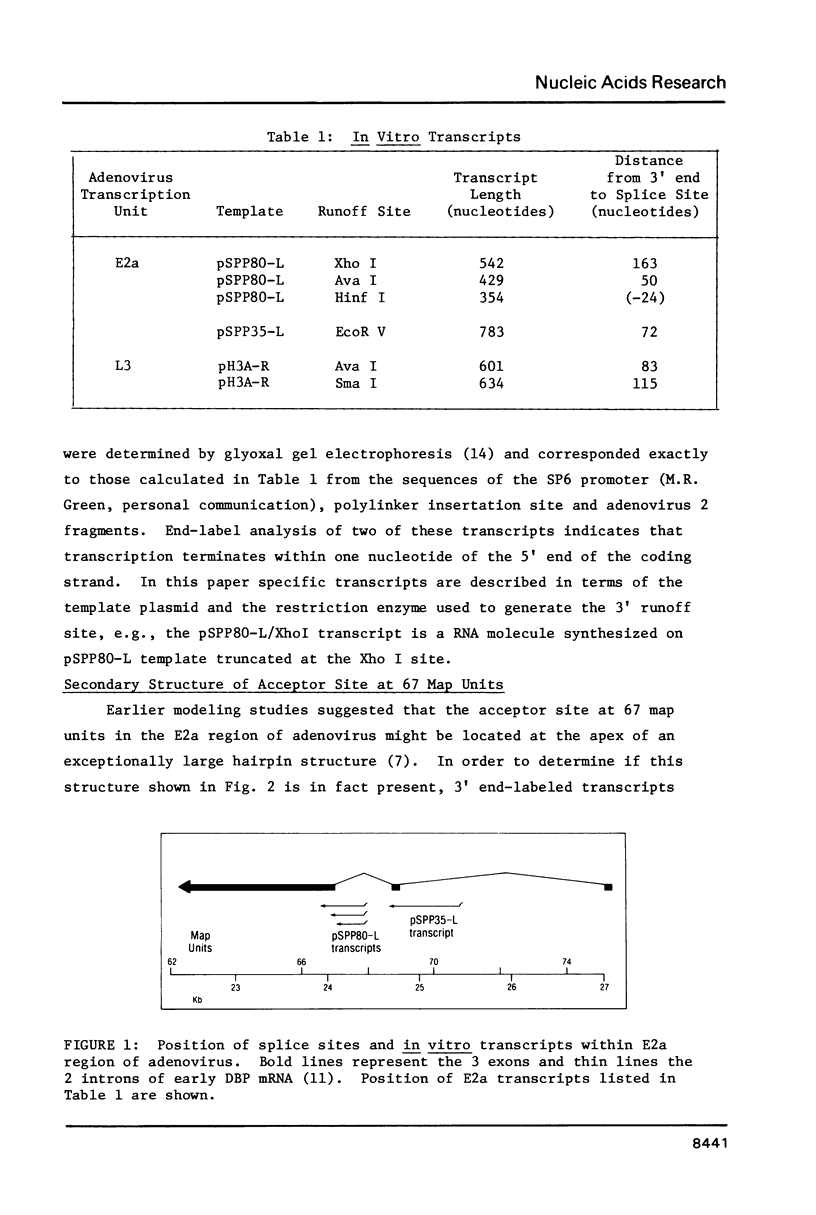
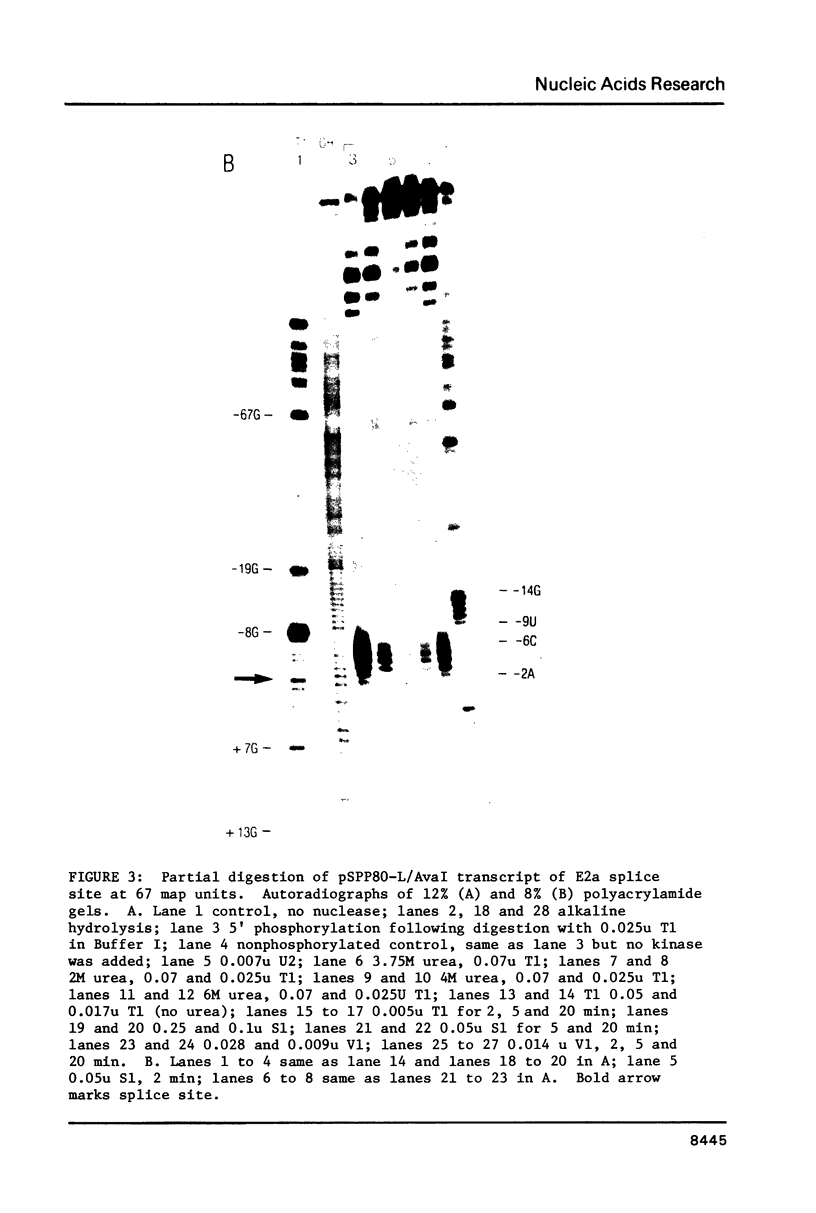
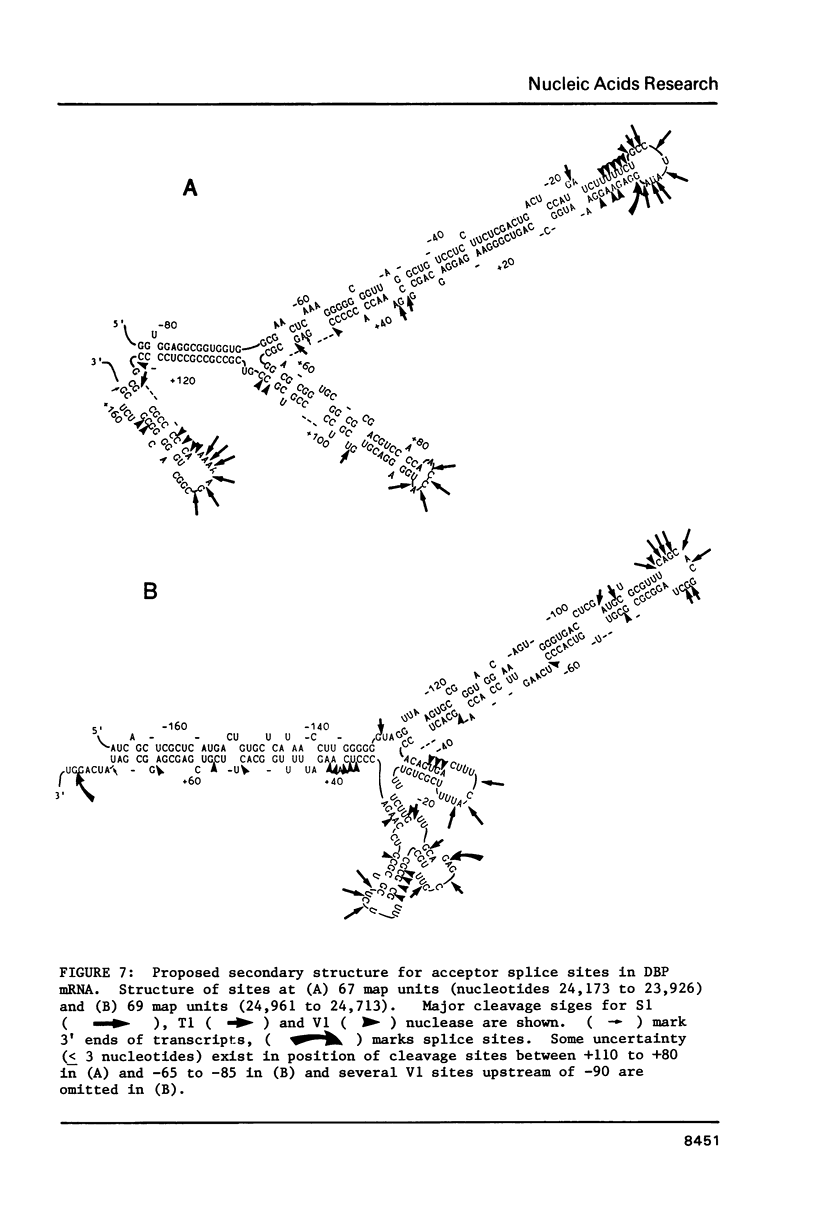
In order to investigate the possible role of RNA secondary structure in determining the efficiency and specificity of mRNA splicing, the structures of sequences at three acceptor splice sites in adenovirus were studied. Transcripts spanning intron-exon junctions were synthesized using SP6 RNA polymerase and analyzed using single and double-strand specific nucleases. Distinctive patterns of nuclease cleavage were observed for each of the 3 sites examined. At both sites in the E2a region sequences adjacent to the splice sites were particularly susceptible to digestion with T1 and S1 nucleases. In contrast, a splice site for hexon mRNA was largely resistant to these nucleases. The results obtained suggest that the conformation of the RNA at some, but not all, acceptor sites may enhance the accessibility of these sites to factors involved in splicing nuclear RNA and confirm the presence of a large, previously predicted hairpin structure centered on the acceptor site at 67 map units.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Persson H. Gene and mRNA for precursor polypeptide VI from adenovirus type 2. J Virol. 1981 May;38(2):469–482. doi: 10.1128/jvi.38.2.469-482.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Role of RNA structure in splicing: excision of the intervening sequence in yeast tRNA3leu is dependent on the formation of a D stem. Cell. 1983 Nov;35(1):109–115. doi: 10.1016/0092-8674(83)90213-1. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burnett L. A model for the mechanism and control of eukaryote gene splicing. J Theor Biol. 1982 Aug 7;97(3):351–366. doi: 10.1016/0022-5193(82)90370-8. [DOI] [PubMed] [Google Scholar]
- Butler E. T., Chamberlin M. J. Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. J Biol Chem. 1982 May 25;257(10):5772–5778. [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Garrett R. A., Olesen S. O. Structure of eukaryotic 5S ribonucleic acid: a study of Saccharomyces cerevisiae 5S ribonucleic acid with ribonucleases. Biochemistry. 1982 Sep 14;21(19):4823–4830. doi: 10.1021/bi00262a047. [DOI] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Van Schaik F. M., Sussenbach J. S. Nucleotide sequence of the gene encoding adenovirus type 2 DNA binding protein. Nucleic Acids Res. 1982 Aug 11;10(15):4493–4500. doi: 10.1093/nar/10.15.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H. A large inverted repeat sequence overlaps two acceptor splice sites in adenovirus. Nucleic Acids Res. 1983 Dec 20;11(24):8891–8900. doi: 10.1093/nar/11.24.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H. Ribonucleoprotein structure of adenovirus nuclear RNA probed by nuclease digestion. J Mol Biol. 1982 Dec 15;162(3):585–606. doi: 10.1016/0022-2836(82)90390-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Qu H. L., Michot B., Bachellerie J. P. Improved methods for structure probing in large RNAs: a rapid 'heterologous' sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5' terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Schmidt C., May K., Schweyen R. J. Determination of functional domains in intron bI1 of yeast mitochondrial RNA by studies of mitochondrial mutations and a nuclear suppressor. EMBO J. 1983;2(11):2047–2052. doi: 10.1002/j.1460-2075.1983.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Thomas J. O., Raziuddin, Sobota A., Boublik M., Szer W. A RNA helix-destabilizing protein is a major component of Artemia salina nuclear ribonucleoproteins. Proc Natl Acad Sci U S A. 1981 May;78(5):2888–2892. doi: 10.1073/pnas.78.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]