Abstract
The flavoprotein (R)-(+)-mandelonitrile lyase (MDL; EC 4.1.2.10), which plays a key role in cyanogenesis in rosaceous stone fruits, occurs in black cherry (Prunus serotina Ehrh.) homogenates as several closely related isoforms. Biochemical and molecular biological methods were used to investigate MDL microheterogeneity and function in this species. Three novel MDL cDNAs of high sequence identity (designated MDL2, MDL4, and MDL5) were isolated. Like MDL1 and MDL3 cDNAs (Z. Hu, J.E. Poulton [1997] Plant Physiol 115: 1359–1369), they had open reading frames that predicted a flavin adenine dinucleotide-binding site, multiple N-glycosylation sites, and an N-terminal signal sequence. The N terminus of an MDL isoform purified from seedlings matched the derived amino acid sequence of the MDL4 cDNA. Genomic sequences corresponding to the MDL1, MDL2, and MDL4 cDNAs were obtained by polymerase chain reaction amplification of genomic DNA. Like the previously reported mdl3 gene, these genes are interrupted at identical positions by three short, conserved introns. Given their overall similarity, we conclude that the genes mdl1, mdl2, mdl3, mdl4, and mdl5 are derived from a common ancestral gene and constitute members of a gene family. Genomic Southern-blot analysis showed that this family has approximately eight members. Northern-blot analysis using gene-specific probes revealed differential expression of the genes mdl1, mdl2, mdl3, mdl4, and mdl5.
Cyanogenesis, the release of the respiratory inhibitor HCN, is but one of many chemical defense systems used by higher plants against herbivory (Nahrstedt, 1985; Jones, 1988). Recognized in several thousand species, this phenomenon has caused numerous cases of acute and chronic cyanide poisoning in animals, including humans (Poulton, 1989). In most taxa, HCN arises during the catabolism of cyanogenic glycosides by specific β-glucosidases and HNLs. Among the most highly cyanogenic species known are the rosaceous stone fruits (e.g. apricots, peaches, and cherries), the kernels of which are a rich source of (R)-amygdalin (the β-gentiobioside of (R)-mandelonitrile) and its catabolic enzymes. In black cherry (Prunus serotina Ehrh.) seed macerates, this diglucoside is rapidly degraded to HCN, Glc, and benzaldehyde in three steps that are catalyzed by the enzymes amygdalin hydrolase, PH, and MDL (EC 4.1.2.10), respectively (Poulton, 1993). In common with MDLs from other members of the Prunoideae and Maloideae subfamilies (Poulton, 1988; Møller and Poulton, 1993), black cherry MDL exhibits several interesting features. Curiously, although it does not catalyze a redox reaction, this glycoprotein contains FAD bound noncovalently near its active site (Jorns, 1979). Constituting almost 10% of the soluble proteins of black cherry seeds, MDL is a highly expressed protein that is found in the protein bodies of the cotyledonary parenchyma cells (Swain et al., 1992b). In seed homogenates, it exists as at least five isoforms (molecular mass, 57–59 kD) whose chemical nature and physiological significance remain poorly understood (Yemm and Poulton, 1986). Finally, MDL also occurs in postembryonic tissues, but here it has a much higher molecular mass of 70 kD (Swain and Poulton, 1994b). It is unknown whether this significant difference in size reflects expression of one or more MDL genes that are unique to postembryonic tissues or whether differential posttranslational modifications (e.g. differential glycosylation or N-/C-terminal processing) of the same gene product are involved.
During the past several years, we have used molecular biological approaches to improve our understanding of MDL microheterogeneity and function in black cherry. This work has yielded two full-length MDL cDNA clones, designated MDL1 and MDL3, with ORFs that share 81% amino acid identity (Cheng and Poulton, 1993; Hu and Poulton, 1997). As expected, both cDNAs encode a signal sequence, a likely FAD-binding site, and several potential N-glycosylation sites. In this study, we describe the cloning and characterization of three additional cDNAs (MDL2, MDL4, and MDL5) from this species. Comparison of these sequences suggests that black cherry MDL is encoded by a gene family, the size of which was estimated by Southern-blot analysis. Genomic sequences corresponding to the MDL1, MDL2, and MDL4 cDNAs are also reported here, permitting the comparison of the organization of these genes with that of the previously described mdl3 gene (Hu and Poulton, 1997). Finally, northern-blot analyses were performed with gene-specific probes to investigate whether the genes mdl1, mdl2, mdl3, mdl4, and mdl5 show differential temporal and/or spatial expression.
MATERIALS AND METHODS
Plant Material
Inflorescences, leaves, and developing fruits of black cherry (Prunus serotina Ehrh.) were collected locally from a single tree, immediately frozen in liquid N2, and stored at −70°C until used for RNA and DNA extractions. Mature seeds were processed and stored for at least 3 months at 4°C, as described by Li et al. (1992). Seeds were germinated by soaking them overnight in aerated distilled water before planting them in Jiffy-mix Plus (Jiffy Products of America, Batavia, IL). After 4 to 6 weeks under fluorescent lights at room temperature, leaves, roots, and cotyledons were harvested and frozen in liquid N2 for later RNA isolation. MDL protein was isolated from aerial portions of seedlings harvested from local woods.
Isolation and Sequencing of MDL2 cDNA
An unamplified λZAP II cDNA library (2 × 105 plaque-forming units), constructed from poly(A+) RNA isolated from immature black cherry seeds (Zheng and Poulton, 1995), was screened by standard methods (Sambrook et al., 1989) using the MDL1 cDNA labeled with [α-32P]dCTP by random priming. After four rounds of plaque hybridization, phage DNA was isolated from positive clones using a liquid-culture method (Sambrook et al., 1989). Insert sizes were determined by PCR, and cDNA inserts greater than 1.7 kb in length were subcloned into pBluescript SK− by in vivo excision according to the manufacturer's instructions (Stratagene). Plasmids were isolated using the Wizard mini kit (Promega) and subjected to restriction analysis, followed in some cases by partial sequencing. One clone (pMDL2), whose insert differed from the MDL1 and MDL3 cDNAs in its restriction map and nucleotide sequence, was serially deleted by exonuclease III and S1 nuclease digestion (Sambrook et al., 1989) and sequenced in both directions by the dideoxy chain-termination method (Sanger et al., 1977) using Sequenase version 2.0 (United States Biochemical).
Isolation and Sequencing of MDL4 and MDL5 cDNAs
Isolation of Probe for Screening of the Leaf cDNA Library
Total RNA was isolated from seedling tops (uppermost 6 cm of 10-cm-tall seedlings) as described by Logemann et al. (1987). Reverse transcription was carried out at 37°C for 45 min in a 20-μL reaction volume containing 2 μg of heat-denatured (70°C for 10 min) RNA, 1 μg of oligo(dT)20 primer, 200 μm of each dNTP, 40 units of RNasin (Promega), and 200 units of SuperScript II RNase H− RT (GIBCO-BRL). After the reaction was stopped by heating at 70°C for 10 min, first-strand cDNA amplification was carried out using the primers 5′-TGGGTTGAAGACACTATTGTGT-3′ (sense) and 5′-AATGAAATTACGAGGATTGTCAT-3′ (antisense), which correspond to highly conserved regions of the mdl1, mdl2, and mdl3 genes (Hu and Poulton, 1997). Each reaction (100 μL) contained 10 μL of RT reaction product, 0.4 μm of each primer, 200 μm of each dNTP, 2 units of Taq DNA polymerase, and 1× PCR buffer. Reactions were cycled 34 times at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min (10 min for the final cycle). The resulting PCR product (0.5 kb) was purified by the GENECLEAN II kit (Bio 101, Inc., Vista, CA), blunt ended with Klenow enzyme (New England Biolabs), and ligated into the SmaI site of pBluescript SK− for transformation into Escherichia coli XL1-Blue competent cells. The insert was sequenced from both ends by primer walking using a dye-termination sequencing protocol run on the fluorescent automated sequencer at the University of Iowa DNA Facility (Iowa City).
Construction and Screening of the Leaf cDNA Library
Total RNA (1 μg) from seedling tops was used to construct a leaf cDNA library in λgt11 by the CapFinder PCR cDNA library construction method according to the manufacturer's suggestions (Clontech, Palo Alto, CA). Approximately 5 × 104 phage plaques were screened with the 0.5-kb PCR fragment described above, which had been labeled with [32P]dCTP by random priming (Boehringer Mannheim). After three successive rounds of plaque hybridization, phage DNA was isolated from positive clones by a plate-lysate method (Ausubel et al., 1995). After restriction analysis had identified two classes of recombinant phages corresponding to MDL4 and MDL5, cDNA inserts were recovered by NotI digestion, purified by the GENECLEAN II kit, and subcloned into the NotI site of pBluescript SK− for automated double-strand sequencing.
Isolation and Sequencing of mdl1, mdl2, and mdl4 Genomic Sequences
Genomic DNA was isolated from arborescent leaves, as described by Dellaporta et al. (1983). Genomic sequences corresponding to the MDL1, MDL2, and MDL4 cDNAs were obtained by PCR amplification using gene-specific primers. For mdl1 amplification, five parallel reactions were undertaken; each reaction contained up to 2 μg of genomic DNA, 0.1 μm each of the sense (5′- TTGAGTTATCAAAAAACATGGA-3′) and antisense (5′-GATAGTCTCGATATTACAAG-3′) primers, 200 μm of each deoxyribonucleotide triphosphate, 2 units of Pfu DNA polymerase, and 1× PCR buffer (Stratagene) in a total volume of 100 μL. Reactions were cycled 25 times at 94°C for 1 min, 53°C for 1 min, and 72°C for 3 min (10 min for the final cycle). All five reactions generated a major PCR product of approximately 2.3 kb, which was purified using the QIAquick gel- extraction kit (Qiagen, Dusseldorf, Germany) and individually ligated into the SmaI site of pBluescript SK−. After transformation into E. coli XL1-Blue competent cells, one clone resulting from each ligation was subjected to partial sequencing by the dideoxy chain-termination method. After confirmation that these PCR products had identical sequences in the regions examined, a single clone was selected for complete sequencing in both directions by primer walking. For comparative purposes, two other clones that originated from parallel amplification reactions were sequenced in specific regions as required. Genomic sequences corresponding to the MDL2 and MDL4 cDNAs were obtained by identical means using the following oligonucleotides as primers: MDL2, 5′-GCACGAGATTCAGAAACAT-3′ (sense) and 5′-AGAGAACTAGAACATCACAAAG-3′ (antisense); and MDL4, 5′-TGAAAGTGTGAAAGAAATTTTAAGAA-3′ (sense) and 5′-GAGTAAATAGAAAGCACAAG-G-3′ (antisense).
Isolation and N-Terminal Sequencing of a Major MDL Isoform from Young Seedlings
Enzyme Purification
All procedures were performed at 4°C. Seedling tops (180 g; uppermost 6 cm of 10-cm-tall seedlings) were homogenized using a blender with 500 mL of buffer A (0.1 m histidine-HCl, pH 6.0) containing 10 g of polyvinylpolypyrrolidone and 12 g of quartz sand. The homogenate was filtered through four layers of cheesecloth and centrifuged for 30 min at 20,000g. The resulting supernatant was dialyzed overnight against 4 L of buffer B (10 mm His-HCl, pH 6.0, containing 0.17 m NaCl) and applied to a Con A-Sepharose 4B column (1.6 × 10 cm) preequilibrated with buffer B. After the column was washed extensively with buffer B, bound proteins were eluted with 100 mL of 0.5 m α-methyl-d-glucoside in buffer B. Active MDL fractions derived from Con A-Sepharose chromatography were pooled and dialyzed overnight against 4 L of buffer C (20 mm sodium acetate, pH 5.0). The dialyzed pool was applied to a DEAE-cellulose column (1.6 × 12 cm) preequilibrated with buffer C. After unbound proteins were removed by washing with 500 mL of buffer C, MDL was eluted with a linear gradient (0–350 mm NaCl; total volume, 150 mL) in the same buffer. MDL fractions (7.5 mL each) from DEAE-cellulose chromatography were pooled, dialyzed overnight against 4 L of buffer C, and applied to a Reactive Red 120-agarose column (1.6 × 12 cm) preequilibrated with buffer C. The column was washed with 10 column volumes of buffer C to remove unbound proteins. Bound proteins were eluted with a linear gradient (0–0.5 m NaCl; total volume, 400 mL) in buffer C.
Enzyme Assay
MDL activity was measured as described previously (Yemm and Poulton, 1986).
SDS-PAGE and N-Terminal Sequence Analysis
To assess protein homogeneity, SDS-PAGE was performed on 10% resolving gels, as described by Sambrook et al. (1989), before staining with Coomassie brilliant blue. After electrophoresis, polypeptides were transferred to PVDF membranes (Bio-Rad) in 3-(cyclohexylamino)-1-propane-sulfonic acid transfer buffer and visualized by brief Coomassie brilliant blue staining (LeGendre et al., 1993). The MDL band was excised and subjected to sequence analysis by Edman degradation on a protein sequencer (model 475A, Applied Biosystems) at the University of Iowa Protein Structure Facility.
Sequence Analyses
Computer analyses of DNA and amino acid sequences were performed with the University of Wisconsin Genetics Computer Group sequence-analysis software package (Devereux et al., 1984). Comparison of coding regions was performed using the BestFit program, and intron sequences were compared by the GAP program (for both programs, gap creation penalty = 50 and gap extension penalty = 3). Deduced amino acid sequences were aligned by the Megalign Clustal program (DNASTAR, Inc., Madison, WI). For phylogenetic tree construction, the derived amino acid sequences of the black cherry and almond MDLs were aligned using the PileUp program (gap creation penalty = 12 and gap extension penalty = 4). Based on the corresponding amino acid sequence alignment, distance matrices were constructed using the Kimura (1980) protein-distance method, and phylogenetic reconstruction was performed using a neighbor-joining method (Saitou and Nei, 1987). Bootstrap analysis (100 replications; Felsenstein, 1985) was performed using the same tree reconstruction method to assess the stability of tree elements.
Northern-Blot Hybridization
Total RNA was extracted from immature seeds, inflorescences, and the leaves, roots, and cotyledons of young seedlings, as described by Logemann et al. (1987). After separation by electrophoresis (10 μg per lane) on denaturing 1.2% agarose gels containing 2.2 m formaldehyde, RNA was blotted onto membranes (Hybond N, Amersham) by standard methods (Sambrook et al., 1989). After cross-linking for 2 h at 80°C in a vacuum oven, blots were prehybridized for 1 h at 50°C in 20 mm Na2HPO4, pH 6.8, containing 6× SSC, 0.4% SDS, 5× Denhardt's solution, and 0.5 mg mL−1 denatured salmon sperm DNA. Overnight hybridization was carried out in this medium (without Denhardt's solution) with [γ-32P]ATP end-labeled, gene-specific oligonucleotide probes generated from the MDL1 (positions 489–466), MDL2 (positions 413–389), or MDL4 (positions 1839–1818) cDNAs. Blots were washed twice for 15 min in 6× SSC and 0.1% SDS at room temperature and twice for 15 min at 55°C before autoradiography. To study MDL3 and MDL5 expression, membranes were probed as described for Southern-blot analysis using either an MDL3 cDNA fragment (positions 1663–1878; Hu and Poulton, 1997) or an MDL5 cDNA fragment (positions 1962–2232) previously 32P labeled by random priming. Equal loading of RNA samples was assessed by hybridization with a 1-kb DNA fragment encoding soybean 28S rRNA labeled by random priming. Autoradiography was subsequently undertaken with intensifying screens at −70°C from overnight to up to 5 d.
Southern-Blot Hybridization
Genomic DNA was isolated from leaves of a single black cherry tree as described above, digested overnight with BamHI, HincII, or EcoRI, and separated by electrophoresis (10 μg per lane) on a 0.8% agarose gel. After blotting onto membranes (Hybond-N+, Amersham), the samples were UV cross-linked. Six different probes were generated for Southern-blot analysis. Highly conserved among all of the known MDL genes, nucleotides 453 to 534 of the mdl1 gene served as an 82-bp consensus probe that was labeled with [α-32P]dCTP by PCR (Mertz and Rashtchian, 1994). In addition, five gene-specific probes were generated by PCR amplification; these probes spanned the second introns of the mdl1, mdl2, and mdl4 genes, the first intron of the mdl3 gene, and the 3′-UTR of the MDL5 cDNA. These probes were separated by agarose-gel electrophoresis, purified using the GENECLEAN II kit, and labeled with [α-32P]dATP by random priming. Specificity of the gene-specific probes was confirmed by Southern-blot analysis against full-length fragments of the five mdl genes.
Prehybridization (30 min) and hybridization were performed at 65°C in 0.25 m Na2HPO4, pH 7.4, containing 1 mm EDTA, 1% BSA, and 7% SDS. After overnight hybridization to the consensus probe, membranes were washed twice for 20 min with 0.5× SSC and 0.1% SDS at 50°C and exposed to x-ray film at −70°C with intensifying screens. Subsequently, they were washed twice for 15 min with 0.1× SSC and 0.1% SDS at 65°C (high-stringency conditions) and reexposed overnight. Finally, blots were stripped by incubation in 0.1% SDS for 30 min at 95°C, hybridized with the 32P-labeled gene-specific probes under identical conditions, and subjected to autoradiography after high-stringency washing.
RESULTS AND DISCUSSION
In cyanophoric plants, α-hydroxynitriles arising from the catabolism of cyanoglycosides and cyanolipids are further degraded to HCN and a carbonyl compound by HNLs. First described in almonds by Rosenthaler (1908), these enzymes are currently the focus of increased attention not only because of their role in herbivore deterrence (Nahrstedt, 1992) but also because of their potential use as biocatalysts in the synthesis of chiral cyanohydrins (Effenberger, 1994; Griengl et al., 1997). HNLs have been purified extensively and characterized from many taxa that accumulate aliphatic and aromatic cyanoglycosides (Møller and Poulton, 1993; Wajant et al., 1994). They fall into two broad groups based on their FAD content (Wajant and Effenberger, 1996). The first group consists of the FAD-containing MDLs from seeds of the Prunoideae and Maloideae subfamilies (Poulton, 1988; Møller and Poulton, 1993). Requiring only 5- to 35-fold purification to reach homogeneity, these monomeric glycoproteins are major seed constituents and may additionally serve as storage proteins (Swain and Poulton, 1994a). The second group of HNLs, which includes those isolated from sorghum, cassava, and linen flax, are less prevalent proteins that lack FAD and are more diverse in their physicochemical properties (Hickel et al., 1996). In recent cloning studies, no sequence similarities were found between the flavoprotein and FAD-independent HNLs (Trummler and Wajant, 1997).
Microheterogeneity of Rosaceous Stone-Fruit MDLs
One puzzling aspect of the biochemistry of rosaceous stone-fruit MDLs is their microheterogeneity. With only one known exception (Xu et al., 1986), all MDLs isolated from seeds of members of the Prunoideae and Maloideae subfamilies exist as multiple forms (Hickel et al., 1996). Within a given species, these isoforms display slight differences in molecular mass, pI, electrophoretic mobility, and specific activity, but they have identical antigenic properties. Further microheterogeneity is observed when these seed MDLs are compared with their counterparts in postembryonic tissues. For example, in black cherry the hypocotyl and epicotyl MDLs (70 kD) are much larger than the seed isoforms (57–59 kD). To date, few studies have focused on the structural differences between the MDL isoforms within a species or addressed the possibility that individual isoforms may have unique functions (Gerstner et al., 1971; Yemm and Poulton, 1986). Likely sources of MDL microheterogeneity include allelic differences at the structural locus for the polypeptide (i.e. allozymes), alteration of the translated polypeptide (e.g. N- and C-terminal processing, glycosylation, and phosphorylation), and gene duplication leading to multigene families (Weeden, 1983). In higher plants, polyploidy is perhaps the most conspicuous mechanism for gene duplication. Several rosaceous stone fruits are polyploid (Darlington, 1928), including black cherry, which is a tetraploid species (2n = 4x = 32) of ancient origin whose chromosomes pair as bivalents during meiosis (Maynard et al., 1991). Thus, it is possible that allozymic forms could contribute significantly to the observed MDL multiplicity of some Prunus species.
In our laboratory, five MDL isoforms (two major and three minor) were purified to near homogeneity from mature black cherry seeds by Con A-Sepharose 4B chromatography and chromatofocusing (Yemm and Poulton, 1986). Differing only slightly in molecular mass (57–59 kD) and pI (4.58–4.63), these forms are monomers whose multiplicity is not attributable to partial proteolysis during enzyme isolation. Although differential glycosylation may underlie some of the observed microheterogeneity (Yemm and Poulton, 1986), chemical deglycosylation of MDL by trifluoromethanesulfonic acid did not significantly reduce this multiplicity, as shown by two-dimensional SDS-PAGE of deglycosylated MDL (Wu and Poulton, 1991). The existence of so many structurally similar isoforms raises the question of whether they might nevertheless differ in enzymatic properties. However, detailed comparative kinetic analysis of the two major seed forms revealed no significant differences in their Km values, pH optima, stability, metal-ion requirements, and sensitivity to inhibitors (Yemm and Poulton, 1986).
Wishing to take molecular biological approaches to elucidate the chemical nature and possible physiological significance of MDL microheterogeneity in a Prunus species, our laboratory first constructed a λgt11 expression library using poly(A+) RNA from mid-maturation black cherry seeds, a developmental stage at which MDL is highly expressed (Swain et al., 1992a). Screening of this library with anti-MDL antiserum yielded a full-length MDL cDNA designated MDL1 (Cheng and Poulton, 1993). We later obtained a second MDL cDNA (MDL3) by RT-PCR using mRNA from immature seeds as a template (Hu and Poulton, 1997).
Isolation and Characterization of MDL2 cDNA
In the present study, a novel MDL cDNA, designated MDL2, was obtained by probing a new seed cDNA library (Zheng and Poulton, 1995) with the radiolabeled MDL1 cDNA under low-stringency conditions. The MDL2 cDNA was sequenced in both directions, and its nucleotide sequence was assigned accession number AF040078. As shown in Table I, the cDNA is 1932 nucleotides in length and consists of 17 nucleotides of 5′-UTR, a 1731-nucleotide ORF, and a 3′-UTR of 184 nucleotides terminated by a 9-nucleotide poly(A+) tail. The ORF encodes a polypeptide of 576 amino acids with a predicted molecular mass of 62.7 kD and a pI of 4.39. Five putative polyadenylation (AATAAA) signals are located 3, 108, 144, 148, and 152 nucleotides downstream of the TAA stop codon.
Table I.
Major features of the black cherry (MDL1–MDL5) and almond (MDLa) cDNAs and their deduced polypeptide sequences
| Clone Designation | Accession No. | Sequence Length | ORF | Polypeptide Length | Molecular Mass | pI | Predicted Features |
|---|---|---|---|---|---|---|---|
| bp | amino acids | kD | |||||
| MDL1 | X72617 | 1856 | 1692 | 563 | 61.2 | 5.38 | FAD-binding site, 5 N-glycosylation sites |
| MDL2 | AF040078 | 1932 | 1731 | 576 | 62.7 | 4.39 | FAD-binding site, 15 N-glycosylation sites |
| MDL3 | AF013161 | 1948 | 1722 | 573 | 62.2 | 4.40 | FAD-binding site, 13 N-glycosylation sites |
| MDL4 | AF053384 | 1940 | 1725 | 574 | 61.7 | 4.72 | FAD-binding site, 10 N-glycosylation sites |
| MDL5 | AF053386 | 2243 | 1680 | 559 | 61.1 | 4.80 | FAD-binding site, 15 N-glycosylation sites |
| MDLaa | Y08211 | 1965 | 1680 | 559 | 61.1 | 4.79 | FAD-binding site, 12 N-glycosylation sites |
Isolation and Characterization of MDL4 and MDL5 cDNAs
Isolation of the MDL4 cDNA Fragment by RT-PCR
To assess rapidly which MDL genes are expressed in black cherry leaves, RT-PCR was used with total RNA from this source acting as the template. Using two primers based on highly conserved regions of the MDL1, MDL2, and MDL3 cDNAs, a 0.5-kb fragment was obtained that was subcloned into the pBluescript vector for double-strand sequencing. This partial-length cDNA (495 bp) shared 84% to 86% similarity with the MDL1, MDL2, and MDL3 cDNAs. In view of the uniqueness of this MDL sequence, a seedling-leaf cDNA library was constructed with the goal of obtaining full-length clones.
Isolation of MDL4 and MDL5 cDNAs by Screening of a Leaf cDNA Library
Using the CapFinder PCR cDNA library construction system, a leaf cDNA library was constructed in the λgt11 vector using mRNA isolated from leaves of 3- to 5-week-old seedlings. Despite its small size (5 × 104 plaque-forming units), this library yielded 26 putative MDL clones when screened with the 495-bp MDL fragment described above. Restriction analysis using SalI, EcoRI, and HindIII revealed that these clones fell into two groups, one with 25 members and one with 1 member. DNA inserts of two clones from the first group and of the sole member of the second group were obtained by NotI digestion and subcloned into the pBluescript vector. Partial sequencing revealed that the two clones from the first group were identical and matched the 495-bp fragment obtained by RT-PCR; they were designated MDL4. The single clone from the second group was clearly dissimilar from clones MDL1 to MDL4 and was designated MDL5. The cDNA inserts of one MDL4 clone and the single MDL5 clone were fully sequenced and subsequently assigned accession nos. AF053384 and AF053386, respectively. The major features of these cDNAs and of the predicted proteins that they encode are summarized in Table I.
Comparison of the Six Known MDL cDNA Sequences
The three novel black cherry MDL cDNAs reported here were compared with the previously reported black cherry MDL1 and MDL3 cDNAs (Cheng and Poulton, 1993; Hu and Poulton, 1997) and the almond MDL cDNA (Suelves and Puigdomènech, 1998) by the BestFit algorithm of the Genetics Computer Group sequence-analysis software package. Table II shows the high similarity between these sequences. Among black cherry MDLs, the amino acid identity ranged from 75.3% to 87.6%. This degree of similarity suggests that they represent members of a multigene family rather than multiple alleles. This notion is also strongly supported by the intron data and differential expression patterns presented below. Even greater sequence identity was observed between MDL5 and almond MDL (93.7% identity and 97% similarity). Because both were expressed in postembryonic tissues, this pair probably represents orthologs. The lowest similarity was observed with MDL2 and almond MDL, yet they still shared 75.1% identity and 84.6% similarity. When amino acid alignment was performed using the Megalign Clustal program, the most obvious differences between the six deduced sequences were (a) the presence in MDL2 and MDL4 of an additional amino acid at positions 11 and 34, respectively, (b) a single amino acid deletion in MDL1 at position 251, and (c) an additional six to nine amino acids at or near the C termini of MDL2, MDL3, and MDL4 (Fig. 1).
Table II.
Identity and similarity (%) of the deduced amino acid sequences of the unprocessed MDLs from black cherry (MDL1–MDL5) and almond (MDLa)
| Enzyme | MDL2 | MDL3 | MDL4 | MDL5 | MDLa |
|---|---|---|---|---|---|
| MDL1 | 78.7 (86.3) | 81.0 (88.3) | 78.2 (88.6) | 76.1 (85.8) | 75.8 (85.8) |
| MDL2 | 87.6 (91.8) | 78.9 (88.5) | 75.3 (84.4) | 75.1 (84.6) | |
| MDL3 | 78.9 (88.7) | 78.6 (87.6) | 78.0 (87.6) | ||
| MDL4 | 77.6 (85.7) | 76.9 (85.7) | |||
| MDL5 | 93.7 (97.0) |
Figure 1.
Comparison of the deduced amino acid sequences of MDLs encoded by the black cherry MDL1 to MDL5 and almond MDL cDNAs. Amino acid alignment was performed using the Megalign Clustal program (DNASTAR, PAM 250 residue weight table). MDLa denotes the almond MDL cDNA sequence. Gaps introduced to optimize alignments are designated by dashes. Shaded boxes enclose amino acid residues that are identical in at least two sequences. The N termini of the mature MDL1 and MDL4 proteins are marked by the arrow, and conserved Cys residues are indicated by asterisks. The βαβ-fold comprising the putative FAD-binding site is underlined. Accession numbers for the cDNA sequences are: MDL1, X72617 (Cheng and Poulton, 1993); MDL2, AF040078; MDL3, AF013161; MDL4, AF053384; MDL5, AF053386; and almond MDL (MDLa), Y08211 (Suelves and Puigdomènech, 1998).
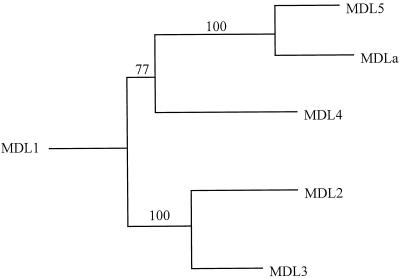
An unrooted phylogenetic tree was constructed with the neighbor-joining method using as input a matrix of protein distances calculated according to the method of Kimura (1980). The amino acid alignment used for this analysis is similar to that shown in Figure 1. In this tree, MDL5 and the almond MDL appear as closely related sequences within a clade that includes MDL4 (Fig. 2). MDL2 and MDL3 form another clade that is distinct from MDL5. The relationship between these proteins suggests that the black cherry MDLs arose through a series of gene duplications followed by extensive sequence divergence. There is strong bootstrap support for the grouping of MDL2 and MDL3 and that of MDL5 and the almond MDL. The latter two share a high amino acid similarity (97%) and appear to form an orthologous lineage that existed before the divergence of black cherry and almond. This result suggests that almond may also contain an MDL-encoding multigene family, members of which are orthologs of the black cherry MDL1 to MDL4 sequences. This prediction needs to be studied in detail to resolve the origin of the complex MDL gene family in black cherry. It is also possible, for example, that some genes in black cherry arose through polyploidization and have since undergone functional diversification, leading to the distinct gene sequences found in our study.
Figure 2.
Unrooted phylogenetic tree showing the evolutionary relationship between the mdl genes. This tree was constructed using a neighbor-joining method from the predicted amino acid sequences of the five black cherry MDLs (MDL1–MDL5) and the almond MDL (MDLa). Numbers at the internal nodes represent percentages from 100 bootstrap replicates.
The availability of six MDL cDNA sequences from black cherry and almond provides important insights into several major features of rosaceous stone-fruit HNLs that were first noted in studies conducted at the protein level. These are discussed in turn below.
N-Terminal Signal Sequence
Analysis of the six derived MDL polypeptide sequences according to the rules of von Heijne (1986) leads to the prediction that each of them will possess an N-terminal signal sequence of 27 or 28 amino acids. In support of this view, N-terminal sequencing of the major seed (MDL1) and leaf (MDL4) isoforms has now confirmed the cleavage of signal sequences at the predicted sites (Fig. 1); this is consistent with the protein body and vacuolar locations, respectively, of these two proteins (Swain et al., 1992b; Swain and Poulton, 1994b).
FAD-Binding Site
As mentioned above, a hallmark of rosaceous HNLs is their FAD content, although the enzymatic reaction that they catalyze does not involve a net oxidation-reduction (Jorns, 1979). Available evidence suggests that this dinucleotide binds to a hydrophobic region near the enzyme's catalytic site (Baerwald and Jaenicke, 1978; Jorns, 1979). Sequence analysis now shows that a highly conserved (>90%) βαβ-fold, which probably serves as the FAD-binding site, lies close to the predicted N termini of all six mature MDL proteins (Fig. 1). This fold exhibits most of the consensus residues recommended by Wierenga et al. (1986) as a diagnostic “fingerprint” for binding of the ADP moieties of FAD and NAD+, including the sequence G-X-G-X-X-G.
N-Glycosylation Sites
As noted above, all black cherry MDL isoforms bind to Con A-Sepharose, indicative of their glycosylation by α-d-Man and/or α-d-Glc moieties (Yemm and Poulton, 1986). Consistent with this observation, the five black cherry MDL cDNA sequences predicted several potential N-glycosylation sites (N-X-S/T), although the number varied widely (Fig. 1). Although the MDL2, MDL3, MDL4, and MDL5 sequences include 15, 13, 10, and 15 potential sites, respectively, the MDL1 sequence has only 5. This striking difference may underlie the differential glycosylation detected by Con A-Sepharose chromatography (Yemm and Poulton, 1986) and will be pursued further. It is noteworthy that the five sites shown by MDL1 are conserved in at least four of the six known MDL sequences.
Catalytically Active Cys Residue
In 1984, Jaenicke and Preun reported that bitter almond HNL was inhibited in 1:1 stoichiometry by the active-site-directed, irreversible inhibitor 3-OPP. This pseudo-first-order inactivation process was inhibited by substrate and competitive inhibitors. Complete hydrolysis of the boronate-reduced, 3-OPP-modified apoprotein yielded l-2-amino-4-thia-dl-7-hydroxy-7-phenylheptanoic acid, the reduced linear addition product of 3-OPP to a Cys-SH group. This suggests that a catalytically important Cys is involved in the active site of this enzyme. In support of this idea, the almond and black cherry MDLs were inhibited by iodoacetamide and iodoacetic acid, respectively (Yemm and Poulton, 1986; Wajant et al., 1995).
Although the catalytically active Cys residue has not yet been identified unequivocally, a small number of likely candidates may be pinpointed by directly examining the available MDL cDNA sequences shown in Figure 1 for absolutely conserved Cys residues. If one assumes that the same Cys is functional in both black cherry and almond MDLs, only three candidates present themselves. The first residue is located within the N-terminal βαβ-fold previously identified as the putative FAD-binding site. The remaining absolutely conserved Cys residues are located in the C-terminal region of the polypeptides. Distinguishing between these candidates might be achieved in several ways. In our laboratory, we are attempting to label black cherry seed MDL with 3-OPP and subsequently identify the modified Cys residue by sequencing of tryptic peptides, but this strategy has proven unsuccessful. An alternative approach is being taken by Lauble et al. (1994), who have obtained x-ray diffraction data at 2.6 Å resolution after crystallizing the almond MDL.
Isolation and Characterization of the mdl1, mdl2, and mdl4 Genomic Sequences and Comparison with the mdl3 Gene
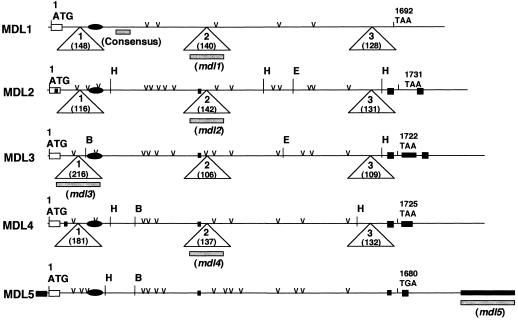
In an earlier publication (Hu and Poulton, 1997), we reported the sequences of the mdl3 gene and its respective cDNA (MDL3). Representing the first information about the gene organization of any plant HNL, we showed that the 2156-bp coding region of this gene is interrupted by three short introns (217, 107, and 110 bp in length). To determine whether other black cherry MDL genes are similarly organized, genomic sequences corresponding to the MDL1, MDL2, and MDL4 cDNAs were obtained by PCR amplification using black cherry genomic DNA as the template. Designated mdl1, mdl2, and mdl4, respectively, these nucleotide sequences were assigned accession nos. U78814, AF040079, and AF053385. The major features of the mdl1, mdl2, mdl3, and mdl4 genes are summarized in Table III. Comparison of these four genomic sequences, both among themselves and with their respective cDNAs, revealed that the ORF of each gene is interrupted at identical positions by three AT-rich introns (Fig. 3). This constitutes strong, additional evidence that these genes were derived from a common ancestral gene. In contrast to the conservation of intron locations, the lengths of the introns varied considerably among the four genes, presumably as a result of insertions and/or deletions (Table III). For example, intron I varied in length from 116 to 216 bp. Nevertheless, within matched regions individual introns showed high homology (45%–85% identity) across the four genes. With the exception of intron II of mdl3, which is flanked by the dinucleotides GC and AG, all 12 exon-intron junction sequences conform to the GT-AG rule for RNA splicing (Mount, 1982). A GC-AG junction sequence has been reported for several other plant genes, including seven myrosinase genes (Xue and Rask, 1995).
Table III.
Major features of the genomic sequences of mdl1 to mdl4
| Clone Designation | Accession No. | Sequence Length | Intron I
|
Intron II
|
Intron
III
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Intron length | AT | Exon-intron boundaries | Intron length | AT | Exon-intron boundaries | Intron length | AT | Exon-intron boundaries | |||
| bp | bp | % | bp | % | bp | % | |||||
| mdl1 | U78814 | 2247 | 148 | 66 | catgGT..AGattt | 140 | 70 | ccagGT..AGgttt | 128 | 64 | ggagGT..AGgtat |
| mdl2 | AF040079 | 2278 | 116 | 66 | catgGT..AGattt | 142 | 72 | tcagGT..AGgtgt | 131 | 69 | ggagGT..AGgtac |
| mdl3 | U51562 | 4748 | 216 | 70 | catgGT..AGattt | 106 | 77 | tcagGC..AGgtgt | 109 | 66 | ggagGT..AGgtat |
| mdl4 | AF053385 | 2380 | 181 | 67 | catgGT..AGattt | 137 | 69 | tcagGT..AGgttt | 132 | 69 | gcagGT..AGgtat |
Figure 3.
Comparison of the major features of the MDL1 to MDL5 cDNAs, including their initiation codons (ATG), termination codons (TAA/TGA), signal sequences (open rectangles), putative FAD-binding sites (closed ovals), insertions (closed rectangles), and potential N-glycosylation sites (v). Triangles indicate where the ORFs of the mdl1 to mdl4 genes are interrupted in the genome by three introns. The intron sizes are shown in parentheses within each triangle. The positions of fragments used as probes in Southern-blot analysis are indicated by gray bars. B, E, and H denote BamHI, EcoRI, and HincII restriction sites, respectively. Not indicated are additional EcoRI (−594) and HincII (−1941) sites in the mdl3 promoter region.
N-Terminal Sequencing of Seedling MDL
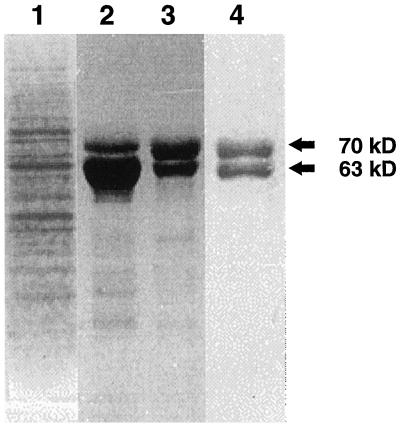
To ascertain whether either of the MDL4 and MDL5 cDNAs encodes the 70-kD MDL polypeptide detected in young seedlings by immunoblotting studies (Swain and Poulton, 1994b), MDL protein was extensively purified from this source to obtain N-terminal sequence information. The purification protocol used here mirrored that of Wu and Poulton (1991) for the purification of MDL from black cherry seeds. When the dialyzed crude extract from seedlings was applied to a Con A-Sepharose 4B column, MDL bound to this matrix, thereby demonstrating its glycoprotein character. This step provided an effective means of MDL purification, because the majority of seedling proteins failed to bind to this affinity resin (Fig. 4). After its elution by α-methyl-d-glucoside, the leaf MDL was subjected to DEAE-cellulose and Reactive Red 120-agarose chromatography. Unlike seed MDL (Li et al., 1992), the seedling MDL did not bind to the Reactive Red 120-agarose column. Nevertheless, this column was retained in the purification protocol because it removed many contaminating proteins. Subsequent analysis of the highly purified seedling MDL by SDS-PAGE revealed two polypeptides with molecular masses of 70 and 63 kD (Fig. 4). These are believed to represent the seedling MDL and PH, respectively. After these polypeptides were electroblotted onto PVDF membranes, the 70-kD band was excised and subjected to 15 cycles of N-terminal sequencing. Its N terminus was LAXTSSEHDFGYLKF. Excluding the third residue, for which an assignment could not be unequivocally made, this sequence matches exactly the derived amino acid sequence of the MDL4 cDNA beginning at residue 28 and differs significantly from those of the other known black cherry MDL isoforms. Failure to assign the third residue may be the result of glycosylation, because in the MDL4 cDNA this residue is an Asp that lies within the context of a putative N-glycosylation site (N-X-S/T).
Figure 4.
SDS-PAGE analysis of purification of MDL4 isoform from black cherry seedlings. Seedling MDL at various stages of isolation was subjected to SDS-PAGE with Coomassie brilliant blue staining. Lane 1, Crude extract; lane 2, Con A-Sepharose 4B chromatography; lane 3, DEAE-cellulose chromatography; and lane 4, Reactive Red 120-agarose chromatography.
These data allow several significant conclusions to be drawn. First, the identification of the N terminus of the seedling MDL protein within the ORF of the MDL4 cDNA confirms the identity of the MDL4 clone. Second, our ability to isolate easily large amounts of MDL4 protein from seedling leaves points to its high expression in this organ, a feature confirmed by northern-blot analysis. Third, the fact that the N-terminal Leu of the MDL4 protein corresponds to residue 28, as predicted by the cDNA, strongly suggests that the first 27 amino acids act as a signal peptide to facilitate intracellular movement of the polypeptide to the vacuole (Swain and Poulton, 1994b) via the ER. Fourth, cloning of the MDL4 gene has provided an important insight into the reported size difference between seed (57–59 kD) and seedling (70 kD) MDLs in black cherry (Swain and Poulton, 1994a). With the knowledge that after N-terminal processing the molecular masses of the mature MDL1 and MDL4 polypeptides, as deduced from their cDNAs, would be 58.1 and 58.6 kD, respectively, it is clear that this modest difference cannot readily explain the significant discrepancy in the observed sizes of seed- and seedling-expressed MDLs. Rather, it points to the likelihood of differential posttranslational modifications within these two plant parts. Two likely (and not mutually exclusive) possibilities are being considered at this time. The first is differential glycosylation. This is particularly attractive because the MDL4 cDNA sequence predicts 10 N-glycosylation sites, whereas only five sites are predicted by the MDL1 cDNA. Alternatively, the discrepancy in MDL protein size may result from differential C-terminal processing of polypeptides, given their dissimilar intracellular destinations (i.e. protein bodies in seeds, vacuoles in leaves). Precedence for the C-terminal processing of storage proteins after their arrival in protein bodies of seeds is provided by Chrispeels et al. (1982). To assess this possibility, we intend to sequence the C terminus of both isoforms.
Black Cherry MDL Is Encoded by a Small Gene Family
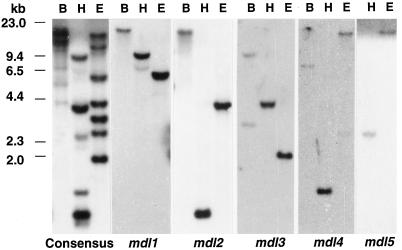
The size of the putative MDL gene family was assessed by Southern-blot analysis. Genomic DNA from a single tree was digested with BamHI, HincII, or EcoRI and probed with an 82-bp fragment corresponding to nucleotides 453 to 534 of the mdl1 gene (hereafter designated the consensus probe). This region was selected because comparison of the MDL1 to MDL5 cDNAs showed that it is highly conserved (approximately 91% identity) and lacks restriction sites for these endonucleases. Moreover, the short length of this probe would minimize the possibility of other target sequences in the genome containing these sites. As a result, the number of bands hybridizing to the consensus probe would more likely reflect the actual size of the MDL gene family. After the blot was washed at low stringency (0.5× SSC and 0.1% SDS at 50°C), approximately eight bands were detected on the membrane in BamHI-, HincII-, or EcoRI-digested DNA (data not shown). These patterns remained unchanged after the blot was washed twice at high stringency (0.1× SSC and 0.1% SDS at 65°C) (Fig. 5). These data suggest that black cherry MDL is encoded by a multigene family of approximately eight members.
Figure 5.
Southern-blot analysis of the black cherry mdl genes. Genomic DNA (10 μg) from an individual tree was digested with BamHI (B), HincII (H), or EcoRI (E), electrophoresed on 0.8% agarose gels, and blotted onto nylon membranes. Subsequently, the blots were hybridized at high stringency with the consensus or gene-specific probes. DNA molecular-mass markers are shown on the left.
To better interpret these banding patterns and to assign identities to bands hybridizing to the consensus probe, gene-specific probes were generated from intron II of the mdl1, mdl2, and mdl4 genes, from intron I of the mdl3 gene, and from the 3′-UTR of mdl5. These probes were 137 to 271 bp in length and were sufficiently distinct from one another (only 51%–65% identity) that cross-hybridization was eliminated when hybridization and washing were performed at high stringency (data not shown). The probes were hybridized to DNA blots that were replicas of the ones used for the consensus-probe hybridizations. With HincII- and EcoRI-restricted genomic DNA, each of the gene-specific probes gave a single band, which was also visible in the consensus-probe blot (Fig. 5). In complete agreement with the restriction map of these genes (Fig. 3), the mdl2-specific probe detected a 1-kb fragment and the mdl4-specific probe detected a 1.4-kb fragment after HincII digestion. Furthermore, after HincII and EcoRI digestion, the mdl3-specific probe detected fragments of the predicted sizes (4.1 and 2.1 kb, respectively). When the BamHI-digested genomic DNA was hybridized with either the mdl1- or the mdl2-specific probe, a single 21.3-kb band was seen in each case, raising the interesting possibility that the mdl1 and mdl2 genes may be tandemly arranged. A similar situation may exist for the mdl4 and mdl5 genes, because a single 19-kb band was detected when EcoRI-digested genomic DNA was hybridized with either the mdl4- or the mdl5-specific probe. Further restriction of these large bands and Southern-blot analysis will be required to confirm or refute the possibility of tandem gene arrangement. The mdl3-specific probe detected two bands of approximately equal intensity at 9.8 and 3.0 kb when cut with BamHI (Fig. 5); these bands are not visible in the consensus-probe pattern because a BamHI site occurs between the regions corresponding to the consensus and the mdl3-specific probes. The origin of this multiplicity is unknown, but it might be caused by polymorphism outside of the MDL gene.
Northern-Blot Analysis Reveals Differential Expression of Known MDL Genes
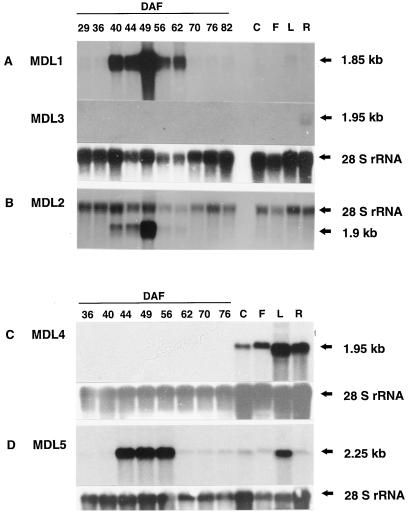
In previous work, the temporal expression of MDL in developing black cherry seeds was analyzed by northern-blot analysis using the full-length MDL1 cDNA as a probe (Zheng and Poulton, 1995). MDL transcripts were first detectable approximately 40 DAF, when embryos became visible macroscopically, and peaked around 49 DAF before declining to negligible levels by fruit maturity (82 DAF). This time course is consistent with the levels of MDL proteins measured during fruit ripening by direct enzyme assay of seed homogenates (Zheng and Poulton, 1995). In agreement with previously published data (Swain et al., 1992a), MDL activities were undetectable until 44 DAF. They then increased rapidly during mid-phase II, essentially reaching a plateau at full fruit maturity. Despite this excellent correlation, the high degree of sequence identity now demonstrated for mdl1 to mdl5 makes it highly unlikely that the MDL1 cDNA probe was specific for any one MDL gene. Thus, it remains unclear how many MDL genes are being expressed during fruit maturation.
To overcome this shortcoming, gene-specific probes with specificities verified by Southern-blot analysis (data not shown) were used here to measure the steady-state transcript levels corresponding to mdl1 to mdl5 in various black cherry organs. As seen in Figure 6, both mdl1 and mdl2 were highly expressed in immature seeds from 40 to 70 DAF and exhibited essentially similar time courses and transcript sizes (approximately 1.9 kb). With its significantly larger transcript size of 2.25 kb, mdl5 was also highly expressed in developing seeds, with maximum expression occurring between 44 and 56 DAF. In contrast to the strong expression of mdl1, mdl2, and mdl5 in immature seeds, mdl3 and mdl4 transcripts were undetectable by northern-blot analysis (even when blots were exposed with intensifying screens to x-ray film for 7 d). Although at first glance this appears to contradict the fact that the MDL3 cDNA clone was isolated from mid-maturation seeds by RT-PCR, it should be stressed that its cloning required high levels of template mRNA. We believe, therefore, that our failure to detect mdl3 expression in total RNA samples from developing seeds was the result of the transcript levels of this gene lying below the detection limits of northern-blot analysis. In support of this belief, when mRNA from immature seeds was used as a template, RT-PCR using mdl3-specific primers generated a fragment of the expected size that hybridized to the mdl3-specific probe (data not shown).
Figure 6.
Northern-blot analysis of mdl1 to mdl5 gene expression in different black cherry organs. Total RNA was isolated from leaves (L), cotyledons (C), and roots (R) of young seedlings; inflorescences (F); and developing seeds at the times indicated (DAF). Samples (10 μg) were electrophoresed on denaturing agarose gels, blotted onto nylon membranes, and hybridized with gene-specific probes under conditions described in Methods. Approximate transcript sizes are indicated by black arrows. A, Hybridization with mdl1- and mdl3-specific probes. Loading equivalents for RNA samples were assessed by hybridization with a 1-kb fragment of the gene encoding soybean 28S rRNA. B, Cohybridization with the mdl2-specific probe and soybean 28S rDNA fragment. C, Hybridization with the mdl4-specific probe. D, Hybridization with the mdl5-specific probe.
Postembryonic tissues of black cherry are also cyanogenic but, in contrast to cotyledons, which accumulate the diglucoside (R)-amygdalin, they contain (R)-prunasin. Upon tissue disruption, this monoglucoside is degraded to HCN by PH and MDL. In earlier studies, MDL enzyme activity was detected in the epicotyls, hypocotyls, and green cotyledons of young seedlings and was immunolocalized to the phloem parenchyma cells of arborescent leaves (Swain and Poulton, 1994a, 1994b). Northern-blot analysis revealed that the five mdl genes also exhibit differential expression in postembryonic tissues (Fig. 6). In sharp contrast to its high transcript levels in immature seeds, the mdl1 gene exhibits barely perceptible expression in seedling leaves and roots. mdl2, which was also highly expressed in mid-maturation seeds, was undetectable in all postembryonic tissues tested. mdl3 differs from the other four MDL genes by having detectable expression only in roots, and this only at low levels (Fig. 6). Although mdl4 transcripts were undetectable in developing seeds, the mdl4 gene was strongly expressed in all postembryonic tissues examined, especially leaves and roots. This finding agrees with the ease with which MDL4 protein was purified from extracts of seedling tops. mdl5 transcripts were detectable in all postembryonic tissues but showed the highest level in leaves. Taken together, to our knowledge, these data provide the first evidence at the molecular level for organ-specific expression of individual mdl genes in a Prunus species.
Physiological Significance of the MDL Gene Family
The data described here provide unequivocal evidence that the reported MDL isoforms do not simply reflect differential posttranslational modifications of the product of a single gene. Instead, screening of both leaf and seed cDNA libraries, augmented by RT-PCR-based cloning, has led to the recognition of five distinct MDL genes. Sharing 75% to 88% amino acid identity, they are believed to be members of a previously unrecognized multigene family of approximately eight members.
Whether through duplication of specific chromosomal regions or genome polyploidy, gene duplication with subsequent differentiation by nucleotide substitution provides the major path leading to formation of multigene families (Li, 1997). Several advantages may be conferred upon plants having such families. First, if the duplicate copies retain their original function, they may enable the organism to produce increased quantities of certain RNA species or proteins that are required for the normal functioning of that individual. Representative examples are the rRNA and histone gene families (Elgin and Weintraub, 1975). A second advantage gained by having multigene families is the increase in functional diversity of the organism. Often, only one of the duplicate loci is necessary to fulfill the cell's need for the gene product, permitting the other locus (loci) to collect “forbidden” mutations (Ohno, 1970). Such isozyme systems might exhibit differences in specificity, in regulatory controls, or in developmental expression without reducing the fitness of the individual. Indeed, the fitness may often increase with such divergence if it bestows greater metabolic flexibility. In some cases, a completely new enzymatic activity may result, as exemplified by the evolution of the stilbene synthase gene (Schröder et al., 1993). Catalyzing synthesis of the stilbene phytoalexins, stilbene synthase is believed to have evolved several times from chalcone synthase, which catalyzes the first committed step to the flavonoid and anthocyanin pigments in plants (Tropf et al., 1994). In other cases, the duplicated copies in the gene family may maintain their original biochemical function but allow for fine tuning of the organism's metabolism through changes in their regulatory regions. A classic example in animal systems is lactate dehydrogenase, the two genes of which have become specialized to different tissues and to different developmental stages (Markert, 1984).
What advantage(s) might black cherry gain from possession of an MDL gene family as opposed to having MDL encoded at a single locus? Perhaps this question might be rephrased as: Why is MDL so highly prevalent, especially when one considers that its substrate, mandelonitrile, decomposes nonenzymatically? It should be noted, however, that the rate of nonenzymatic dissociation of mandelonitrile is low at the slightly acidic pH values typical of plant macerates (Gross et al., 1982; Selmar et al., 1989). Because in some cyanogenic systems the rate of HCN release, rather than the concentration of cyanogenic compounds, is more important for herbivore deterrence (Hsieh, 1989), the high levels of MDL seen in tissues of black cherry may be an adaptation that ensures maximum rates of HCN liberation upon tissue disruption. The observed high-level expression of the mdl1, mdl2, and mdl5 genes in immature seeds and of the mdl4 and mdl5 genes in leaves may represent the mechanism that guarantees optimum concentrations of MDL protein for posttrauma cyanogenesis and herbivore repellence. Aside from the considerations of herbivore deterrence, MDL also fulfills many of the stated criteria of typical plant storage proteins, i.e. accumulation in mid-maturation seeds at high levels (>5% of seed soluble proteins) and sequestration in protein bodies. This led Swain and Poulton (1994a) to suggest that MDL might serve an additional role as a storage protein. In support of this notion, the mdl1, mdl2, and mdl5 genes show high expression in immature seeds at the same time that the traditional black cherry storage proteins (amandins) are being sequestered in these organelles.
ACKNOWLEDGMENTS
We thank Drs. Debashish Bhattacharya, Chi-Lien Cheng, and Ming-Che Shih for their helpful suggestions and for carefully reviewing the manuscript. We are also grateful to Dr. Bhattacharya for assistance with sequence analysis.
Abbreviations:
- 3-OPP
3-oxo-3-phenylpropene
- Con A
concanavalin A
- DAF
days after flowering
- HNL
α-hydroxynitrile lyase
- MDL
(R)-(+)-mandelonitrile lyase
- ORF
open reading frame
- PH
prunasin hydrolase
- RT
reverse transcriptase
- UTR
untranslated region
Footnotes
This research was supported by the National Science Foundation (grant nos. IBN 9630935 and MCB 9723302).
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. [Google Scholar]
- Baerwald KR, Jaenicke L. d-Hydroxynitrile lyase: involvement of the prosthetic flavin adenine dinucleotide in enzyme activity. FEBS Lett. 1978;90:255–260. doi: 10.1016/0014-5793(78)80380-9. [DOI] [PubMed] [Google Scholar]
- Cheng I-P, Poulton JE. Cloning of cDNA of Prunus serotina (R)-(+)-mandelonitrile lyase and identification of a putative FAD-binding site. Plant Cell Physiol. 1993;34:1139–1143. [Google Scholar]
- Chrispeels MJ, Higgins TJV, Spencer D. Assembly of storage protein oligomers in the endoplasmic reticulum and processing of the polypeptides in the protein bodies of developing pea cotyledons. J Cell Biol. 1982;93:306–313. doi: 10.1083/jcb.93.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington CD. Studies in Prunus I, II. J Genet. 1928;19:213–256. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger F. Synthesis and reactions of optically active cyanohydrins. Angew Chem Int Ed Engl. 1994;33:1555–1564. [Google Scholar]
- Elgin SCR, Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gerstner E, Steffen A, Pfeil E. Zur Entwicklung der multiplen Formen des Flavin-Enzyms D-Oxynitrilase bei Prunus spinosa. Naturwissenschaften. 1971;58:269–270. [Google Scholar]
- Griengl H, Hickel A, Johnson DV, Kratky C, Schmidt M, Schwab H. Enzymatic cleavage and formation of cyanohydrins: a reaction of biological and synthetic relevance. Chem Comm. 1997;20:1933–1951. [Google Scholar]
- Gross M, Jacobs GH, Poulton JE. A rapid and sensitive spectrophotometric assay for prunasin hydrolase activity employing purified mandelonitrile lyase. Anal Biochem. 1982;119:25–30. doi: 10.1016/0003-2697(82)90660-1. [DOI] [PubMed] [Google Scholar]
- Hickel A, Hasslacher M, Griengl H. Hydroxynitrile lyases: functions and properties. Physiol Plant. 1996;98:891–898. [Google Scholar]
- Hsieh JS. Cyanogenesis and aphid resistance in sorghum (abstract no. 74934) Chemical Abstracts. 1989;111:462. [Google Scholar]
- Hu Z, Poulton JE. Sequencing, genomic organization, and preliminary promoter analysis of a black cherry (R)-(+)-mandelonitrile lyase gene. Plant Physiol. 1997;115:1359–1369. doi: 10.1104/pp.115.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke L, Preun J. Chemical modification of hydroxynitrile lyase by selective reaction of an essential cysteine-SH group with α,β-unsaturated propiophenones as pseudosubstrates. Eur J Biochem. 1984;138:319–325. doi: 10.1111/j.1432-1033.1984.tb07917.x. [DOI] [PubMed] [Google Scholar]
- Jones DA. Cyanogenesis in animal-plant interactions. In: Evered D, Harnett S, editors. Cyanide Compounds in Biology: Ciba Foundation Symposium No. 140. Chichester, UK: John Wiley & Sons; 1988. pp. 151–176. [DOI] [PubMed] [Google Scholar]
- Jorns M. Mechanism of catalysis by the flavoenzyme oxynitrilase. J Biol Chem. 1979;254:12145–12152. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of sequence evolution. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lauble H, Müller K, Schindelin H, Förster S, Effenberger F. Crystallization and preliminary X-ray diffraction studies of mandelonitrile lyase from almonds. Proteins. 1994;19:343–347. doi: 10.1002/prot.340190410. [DOI] [PubMed] [Google Scholar]
- LeGendre N, Mansfield M, Weiss A, Matsudaira P (1993) Purification of proteins and peptides by SDS-PAGE. In P Matsudaira, ed, A Practical Guide to Protein and Peptide Purification for Microsequencing. Academic Press, New York, pp 71–101
- Li CP, Swain E, Poulton JE. Prunus serotina amygdalin hydrolase and prunasin hydrolase: purification, N-terminal sequencing and antibody production. Plant Physiol. 1992;100:282–290. doi: 10.1104/pp.100.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Molecular Evolution. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Markert CL. Lactate dehydrogenase: biochemistry and function of lactate dehydrogenase. Cell Biochem Funct. 1984;2:131–134. doi: 10.1002/cbf.290020302. [DOI] [PubMed] [Google Scholar]
- Maynard CA, Kavanagh K, Fuerukranz H, Drew AP (1991) I.1 Black cherry (Prunus serotina Ehrh.) In YPS Bajaj, ed, Biotechnology in Agricultural Forestry, Vol 16. Springer-Verlag, Berlin, pp 3–22
- Mertz LM, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- Møller BL, Poulton JE. Cyanogenic glycosides. In: Lea PJ, editor. Methods in Plant Biochemistry, Vol 9. San Diego, CA: Academic Press; 1993. pp. 183–207. [Google Scholar]
- Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrstedt A. Cyanogenic compounds as protecting agents for organisms. Plant Syst Evol. 1985;150:35–47. [Google Scholar]
- Nahrstedt A. The biology of the cyanogenic glycosides: new developments. In: Mengel K, Pilbeam DJ, editors. Nitrogen Metabolism of Plants: Proceedings of the Phytochemical Society of Europe, Vol 33. Oxford, UK: Clarendon Press; 1992. pp. 249–269. [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- Poulton JE. Localization and catabolism of cyanogenic glycosides. In: Evereds D, Harnett S, editors. Cyanide Compounds in Biology: Ciba Foundation Symposium No. 140. Chichester, UK: John Wiley & Sons; 1988. pp. 67–91. [DOI] [PubMed] [Google Scholar]
- Poulton JE (1989) Toxic compounds in plant foodstuffs: cyanogens. In JE Kinsella, WG Soucie, eds, Food Proteins. American Oil Chemists' Society, Champaign, IL, pp 381–401
- Poulton JE. Enzymology of cyanogenesis in rosaceous stone fruits. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology. ACS Symposium Series No. 533. Washington, DC: American Chemical Society; 1993. pp. 170–190. [Google Scholar]
- Rosenthaler L. Asymmetric syntheses produced by enzymes. Biochem Z. 1908;14:238–253. [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J, Schamz S, Tropf S, Kircher B, Schröder G. Phytoalexin biosynthesis: stilbene synthase and co-action of a reductase with chalcone synthase. In: Fritig B, Legrand M, editors. Mechanisms of Plant Defense Response. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 257–267. [Google Scholar]
- Selmar D, Lieberei R, Biehl B, Conn EE. α-Hydroxynitrile lyase in Hevea brasiliensis and its significance for rapid cyanogenesis. Physiol Plant. 1989;75:97–101. [Google Scholar]
- Suelves M, Puigdomènech Molecular cloning of the cDNA coding for the (R)-(+)-mandelonitrile lyase of Prunus amygdalus: temporal and spatial expression patterns in flowers and mature seeds. Planta. 1998;206:388–393. doi: 10.1007/s004250050414. [DOI] [PubMed] [Google Scholar]
- Swain E, Li CP, Poulton JE. Development of the potential for cyanogenesis in maturing black cherry (Prunus serotina Ehrh.) fruits. Plant Physiol. 1992a;98:1423–1428. doi: 10.1104/pp.98.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Li CP, Poulton JE. Tissue and subcellular localization of enzymes catabolizing (R)-amygdalin in mature Prunus serotina seeds. Plant Physiol. 1992b;100:291–300. doi: 10.1104/pp.100.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Poulton JE. Utilization of amygdalin during seedling development of Prunus serotina. Plant Physiol. 1994a;106:437–445. doi: 10.1104/pp.106.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Poulton JE. Immunocytochemical localization of prunasin hydrolase and mandelonitrile lyase in stems and leaves of Prunus serotina. Plant Physiol. 1994b;106:1285–1291. doi: 10.1104/pp.106.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropf S, Lanz T, Rensing SA, Schröder J, Schröder F. Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol. 1994;38:610–618. doi: 10.1007/BF00175881. [DOI] [PubMed] [Google Scholar]
- Trummler K, Wajant H. Molecular cloning of acetone cyanohydrin lyase from flax (Linum usitatissimum) J Biol Chem. 1997;272:4770–4774. doi: 10.1074/jbc.272.8.4770. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Effenberger F. Hydroxynitrile lyases of higher plants. Biol Chem. 1996;377:611–617. [PubMed] [Google Scholar]
- Wajant H, Förster S, Selmar D, Effenburger F, Pfizenmaier K. Purification and characterization of a novel (R)-mandelonitrile lyase from the fern Phlebodium aureum. Plant Physiol. 1995;109:1231–1238. doi: 10.1104/pp.109.4.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Mundry K-W, Pfizenmaier K. Molecular cloning of hydroxynitrile lyase from Sorghum bicolor (L.): homologies to serine carboxypeptidases. Plant Mol Biol. 1994;26:735–746. doi: 10.1007/BF00013758. [DOI] [PubMed] [Google Scholar]
- Weeden NF (1983) Evolution of plant isozymes. In SD Tanksley, TJ Orton, eds, Developments in Plant Genetics and Breeding: Isozymes in Plant Genetics and Breeding, Part A. Elsevier, New York, pp 175–205
- Wierenga RK, Terpstra P, Hol WGJ. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wu H-C, Poulton JE. Immunocytochemical localization of mandelonitrile lyase in mature black cherry (Prunus serotina Ehrh.) seeds. Plant Physiol. 1991;96:1329–1337. doi: 10.1104/pp.96.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L-L, Singh BK, Conn EE. Purification and characterization of mandelonitrile lyase from Prunus lyonii. Arch Biochem Biophys. 1986;250:322–328. doi: 10.1016/0003-9861(86)90733-2. [DOI] [PubMed] [Google Scholar]
- Xue J, Rask L. The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol Biol. 1995;29:167–171. doi: 10.1007/BF00019128. [DOI] [PubMed] [Google Scholar]
- Yemm RS, Poulton JE. Isolation and characterization of multiple forms of mandelonitrile lyase from mature black cherry (Prunus serotina Ehrh.) seeds. Arch Biochem Biophys. 1986;247:440–445. doi: 10.1016/0003-9861(86)90604-1. [DOI] [PubMed] [Google Scholar]
- Zheng L, Poulton JE. Temporal and spatial expression of amygdalin hydrolase and (R)-(+)-mandelonitrile lyase in black cherry (Prunus serotina Ehrh.) seeds. Plant Physiol. 1995;109:31–39. doi: 10.1104/pp.109.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]