Abstract
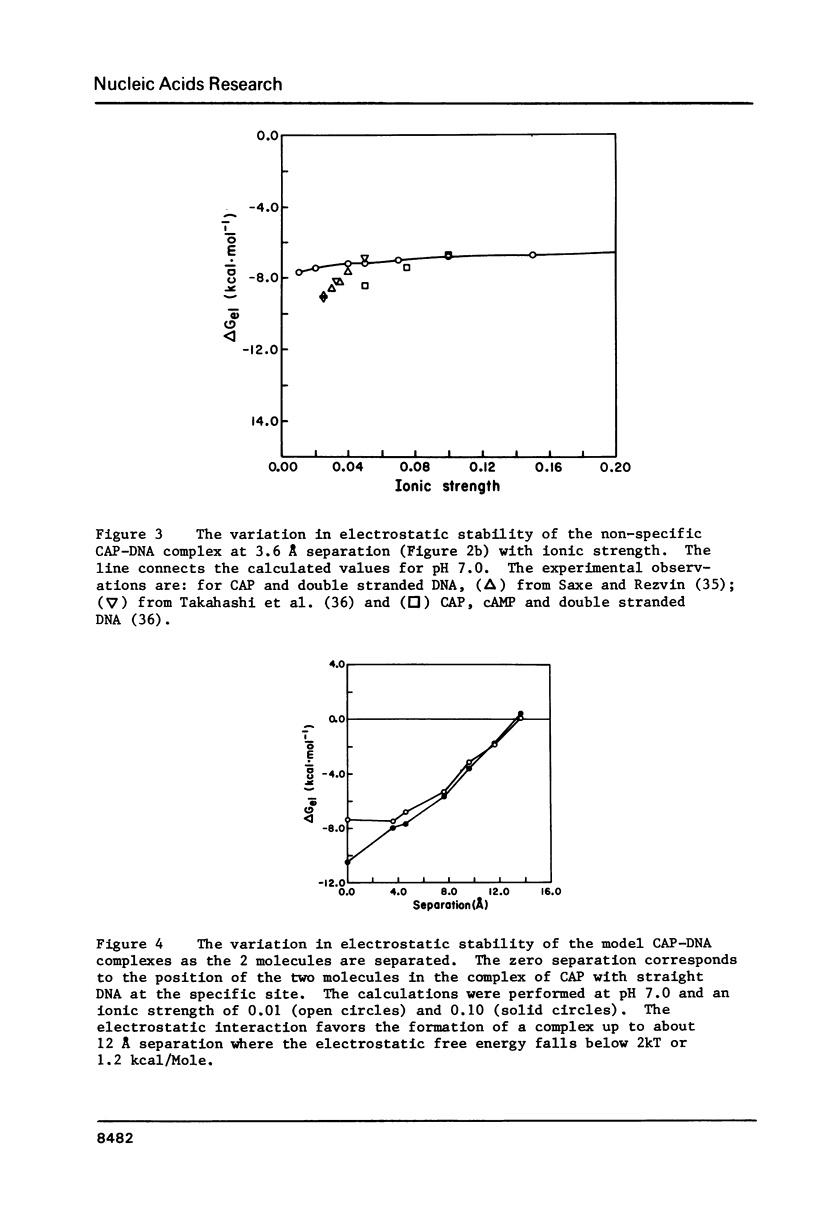
The binding of E. coli catabolite gene activator protein (CAP) to non-specific sequences of DNA has been modelled as an electrostatic interaction between four basic side chains of the CAP dimer and the charged phosphates of DNA. Calculation of the electrostatic contribution to the binding free energy at various separations of the two molecules shows that complex formation is favored when CAP and DNA are separated by as much as 12 A. Thus, the long range electrostatic interactions may provide the initial energy for complex formation and also the correct relative orientation of CAP and DNA. The non-specific complex does not involve the penetration of amino acid side chains into the major grooves of DNA and permits 'sliding' of the protein along DNA, which would enhance the rate of association of CAP with the specific site as has been proposed previously for lac repressor. We propose that, as it 'slides', CAP is moving in and out of the major grooves in order to sample the DNA sequence. Recognition of the specific DNA site is achieved by a complementarity in structure and hydrogen bonding between amino acids and the edges of base pairs exposed in the major grooves of DNA.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkley M. D., Lewis P. A., Sullivan G. E. Ion effects on the lac repressor--operator equilibrium. Biochemistry. 1981 Jun 23;20(13):3842–3851. doi: 10.1021/bi00516a027. [DOI] [PubMed] [Google Scholar]
- Barkley M. D. Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981 Jun 23;20(13):3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Epstein W., Rothman-Denes L. B., Hesse J. Adenosine 3':5'-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Spassky A., Chapon C., Blazy B., Buc H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1983 Nov 25;11(22):7833–7852. doi: 10.1093/nar/11.22.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Matthew J. B., Hanania G. I., Gurd F. R. Electrostatic effects in hemoglobin: hydrogen ion equilibria in human deoxy- and oxyhemoglobin A. Biochemistry. 1979 May 15;18(10):1919–1928. doi: 10.1021/bi00577a011. [DOI] [PubMed] [Google Scholar]
- Matthew J. B., Richards F. M. Anion binding and pH-dependent electrostatic effects in ribonuclease. Biochemistry. 1982 Sep 28;21(20):4989–4999. doi: 10.1021/bi00263a024. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Weber I. T., Steitz T. A. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982 Aug 25;257(16):9518–9524. [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Lewis M., Pabo C. O., Matthews B. W. Comparison of the structures of cro and lambda repressor proteins from bacteriophage lambda. J Mol Biol. 1983 Sep 25;169(3):757–769. doi: 10.1016/s0022-2836(83)80169-7. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Takeda Y., Matthews B. W. High resolution structural studies of Cro repressor protein and implications for DNA recognition. J Biomol Struct Dyn. 1983 Oct;1(2):553–563. doi: 10.1080/07391102.1983.10507461. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Revzin A., von Hippel P. H. Direct measurement of association constants for the binding of Escherichia coli lac repressor to non-operator DNA. Biochemistry. 1977 Nov 1;16(22):4769–4776. doi: 10.1021/bi00641a002. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Steitz T. A. Protein-DNA interaction investigated by binding Escherichia coli lac repressor protein to poly(d(A-U-HgX)). J Mol Biol. 1976 May 5;103(1):25–38. doi: 10.1016/0022-2836(76)90050-4. [DOI] [PubMed] [Google Scholar]
- Richter P. H., Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor--operator association and membrane bound enzymes. Biophys Chem. 1974 Oct;2(3):255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Newby R. F., Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. Interaction of the cAMP receptor protein with the lac promoter. Nucleic Acids Res. 1980 Feb 25;8(4):759–766. [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Harrison R., Weber I. T., Leahy M. Ligand-induced conformational changes in proteins. Ciba Found Symp. 1983;93:25–46. doi: 10.1002/9780470720752.ch3. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Weber I. T., Matthew J. B. Catabolite gene activator protein: structure, homology with other proteins, and cyclic AMP and DNA binding. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):419–426. doi: 10.1101/sqb.1983.047.01.049. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A., Hillen W. On the origin of selectivity in recognition by cyclic adenosine 3',5'-monophosphate receptor protein of its specific binding site of the lactose promoter region. J Mol Biol. 1983 Jul 15;167(4):895–899. doi: 10.1016/s0022-2836(83)80118-1. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A. Non-specific interactions of CRP from E. coli with native and denatured DNAs: control of binding by cAMP and cGMP and by cation concentration. Nucleic Acids Res. 1979 Nov 24;7(6):1699–1712. doi: 10.1093/nar/7.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., McKay D. B., Steitz T. A. Two helix DNA binding motif of CAP found in lac repressor and gal repressor. Nucleic Acids Res. 1982 Aug 25;10(16):5085–5102. doi: 10.1093/nar/10.16.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R. B., Berg O. G., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981 Nov 24;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Le Bret M. Counter-ion condensation and system dimensionality. J Biomol Struct Dyn. 1983 Oct;1(2):461–471. doi: 10.1080/07391102.1983.10507455. [DOI] [PubMed] [Google Scholar]
- Zubay G., Schwartz D., Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci U S A. 1970 May;66(1):104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiderweg E. R., Kaptein R., Wüthrich K. Secondary structure of the lac repressor DNA-binding domain by two-dimensional 1H nuclear magnetic resonance in solution. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5837–5841. doi: 10.1073/pnas.80.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHaseth P. L., Lohman T. M., Record M. T., Jr Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. Biochemistry. 1977 Nov 1;16(22):4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Revzin A., Gross C. A., Wang A. C. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]