Abstract
Chromosome segregation during mitosis depends on the action of the mitotic spindle, a self-organizing, bipolar protein machine which uses microtubules (MTs) and their associated motors1,2. Members of the BimC subfamily of kinesin-related MT–motor proteins are believed to be essential for the formation and functioning of a normal bipolar spindle3–14. Here we report that KRP130, a homotetrameric BimC-related kinesin purified from Drosophila melanogaster embryos13, has an unusual ultrastructure. It consists of four kinesin-related polypeptides assembled into a bipolar aggregate with motor domains at opposite ends, analogous to a miniature myosin filament15. Such a bipolar ‘minifilament’ could crosslink spindle MTs and slide them relative to one another. We do not know of any other MT motors that have a bipolar structure.
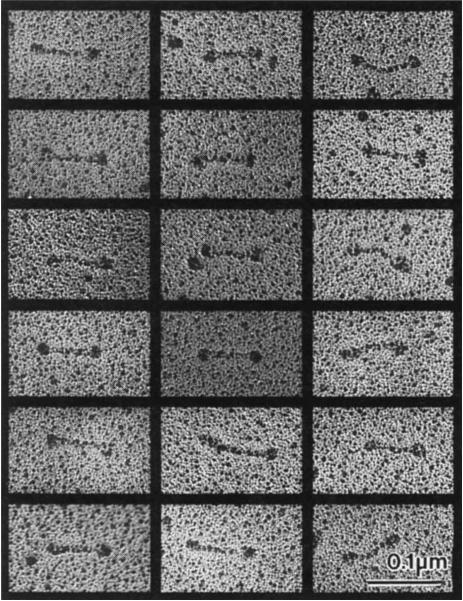
We investigated the structure of purified embryonic KRP130 holoenzymes from Drosophila melanogaster (Fig. 1a) using electron microscopy16–19 (Fig. 2). Single rotary-shadowed KRP130 molecules are visible as elongated bipolar structures with an overall length of 95.6 ± 9.8 nm (n = 131), consisting of two globular domains connected by a central rod with a length of 61.3 ± 8.3 nm (n = 130). The rod domain of KRP130 appeared less flexible than that of kinesin, although a small fraction of KRP130 molecules were slightly bent in the middle of the rod (average bend angle was 134°, compared with 180° for straight molecules; range 127°–154°). The dimensions of the globular domains at opposite ends of KRP130 molecules were indistinguishable, being 21.7 ± 3.7 nm in diameter (n = 154), approximately twice the diameter of a single rotary-shadowed kinesin motor domain17,18. This is consistent with there being two close-packed motor domains at each end of the tetrameric KRP130 molecule.
FIG. 1.
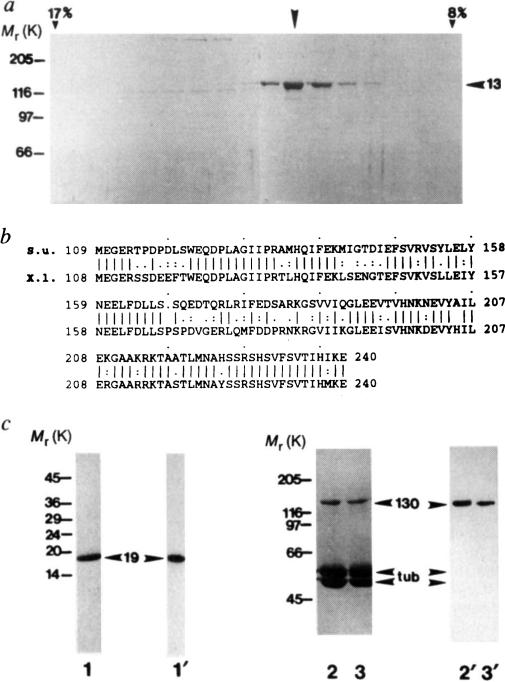
Purification of KRP130 and corresponding motor-domain antibodies for electron microscopy, a, SDS-polyacrylamide gel of fractions from sucrose density gradient centrifugation of KRP130 (ref. 13). Arrowhead indicates peak fraction. Percentage sucrose (w/v) is indicated for two fractions, b, Best-fit comparison of the sea urchin (S.u.) Eg5-related motor-domain subfragment used for raising Eg5 motor-domain antibodies, and the corresponding region of Xenopus laevis (X.I.) Eg5 (identity 69%, similarity 83%). c, Characterization of the Eg5 motor-domain antibody. SDS-polyacrylamide gels (1–3) and corresponding anti-Eg5 motor-domain immunoblots (1′–3′) of a purified fusion protein of Mr 19K, (His)6-SU Eg5 motor, comprising the sea-urchin Eg5-related motor-domain fragment fused to a polyhistidine tag (1,1′), and KRP130 pelleted with MTs in the presence of the nonhydrolysable ATP analogue, 5′adenylyl-β,γ-imidodiphosphate (AMP–PNP) (2,2′) or ATP (3,3′) in low salt. Numbers on the left refer to Mr of standards. Arrowheads indicate recombinant motor-domain fragment (19), tubulin (tub) and KRP130 (130).
METHODS KRP130 was purified from Drosophila melanogaster embryos13 and rotary-shadowed directly or following co-pelleting with MTs in AMP–PNP or ATP (KRP130 binding to MTs is ATP-sensitive at high but not low ionic strength; Fig. 1c and ref. 13). Antibody specific for KRP130 motor domains (Figs 1c and 4a) was raised against a recombinant motor-domain fragment of a sea-urchin egg protein, KRP170, that is a close relative of frog Eg5. Expression of a KRP170 motor-domain fragment complementary DNA in pRSETB vector yielded a (His)6-SU Eg5 motor fusion protein of Mr 19K, which was purified by Ni-nitrilotriacetic acid metal-chelate affinity chromatography followed by SDS-PAGE electroelution (Fig. 1c, lane 1) and used to raise rabbit polyclonal antibody. Specific antibody was affinity purified from serum using the 19K fusion protein coupled to agarose, analysed by immunoblotting (Fig. lc) and used for electron microscopy (Fig. 4a).
FIG. 2.
Electron micrographs of rotary-shadowed KPR130 molecules. These images, together with previous studies13, are consistent with the hypothesis that a KRP130 holoenzyme comprises four 130K subunits assembled into a structure 96 nm long, with a total Mr of roughly 500K, Stokes radius of 16.2 nm, and S-value of 7.6 S. For comparison, kinesin comprises two heavy chains of 110K–130K and two light chains of 55K–80K assembled into a structure 75–80 nm long with a total Mr of 350K–400K, Stokes radius of 9nm, and S-value of 9.0–9.6S. The dimensions of both rotary-shadowed KRP130 and kinesin appear slightly larger than their true dimensions owing to the coating of platinum, which in our experiments is routinely estimated to be approximately 2.5 nm thick.
METHODS. KRP130 alone or KRP130–MT complexes were mixed with an equal volume of 80% glycerol containing 0.6–1.0 M ammonium acetate and sprayed onto freshly cleaved mica plates. The plates were then vacuum dried, rotary shadowed with platinum at a 6° angle using a Balzers BAF 400T freeze-fracture device, and processed as described previously28. The replicas were visualized on a Philips 410LS electron microscope at 80 kV. No difference in the structure of KRP130 was observed with or without the MT binding step.
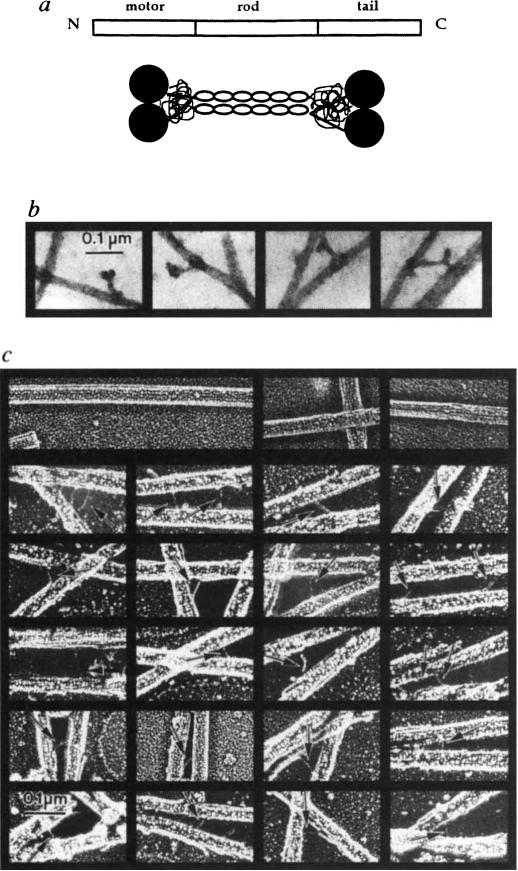
These rotary-shadowed images suggest that KRP130 holoenzymes have the structure shown in Fig. 3a. In this model, two KRP130 polypeptides associate by coiled-coil interactions to form a parallel dimer. Two dimers then associate by lateral interactions between their coiled-coil regions to form a bipolar tetramer, which consists of a central rod with two motor domains projecting from either end. To test the hypothesis that the bipolar appearance of KRP130 molecules reflects the presence of globular MT-binding motor domains at both ends, we first examined MT–KRP130 complexes by negative stain and rotary-shadow electron microscopy (Fig. 3b, c). Bipolar, negatively stained KRP130 molecules were observed to bind to microtubules by one or both ends (Fig. 3b). Rotary shadowing of MT–KRP130 complexes revealed aperiodic MT–MT crossbridges of length 52.7 ± 8.9 nm (n = 47), which were absent in preparations of MTs alone (Fig. 3c). However, kinesin itself, which is thought to crosslink vesicles to MTs in vivo, forms similar MT–MT crossbridges under some experimental conditions20. It is therefore difficult to interpret our observation that KRP130 can form crossbridges to adjacent MTs.
FIG. 3.
KRP130 molecules are bipolar structures capable of crosslinking microtubules, a, Drosophila KRP130 is probably a homologue of Xenopus Eg5, a member of the BimC subfamily, consisting of 1,060 amino acid residues arranged in a tripartite motor–rod–tail manner, according to the map (top). The model (bottom) shows how four KRP130 polypeptides could be arranged to produce bipolar homotetramers. Two parallel KRP130 dimers assemble in an antiparallel fashion to form bipolar tetramers 96 nm in length with two 10-nm motor-domain ‘heads’ protruding on each end. The ‘tails’ of the KRP130 subunits would be juxtaposed to the ‘heads’; close packing of two heads and two tails would produce a structure visible in rotary-shadowed specimens as a single, globular domain approximately 20 nm × 20 nm on each end of the 60-nm rod. This is consistent with the dimensions of the rotary-shadowed KRP130 molecules, b, Negatively stained images of complexes of KRP130 with MTs. Sometimes the ‘dumbbell-shaped’ KRP130 molecules bind by opposite ends to adjacent MTs (right), producing MT–MT crossbridges, whereas other molecules bind to MTs with one end only, producing projections from the MT wall, c, Individual fine, aperiodic cross-bridges are observed in rotary-shadowed KRP130–MT pellets (arrows). The reduced length of the crossbridges (53 nm) compared to the ‘rods’ of individual molecules (60 nm) may be due to occlusion of part of the ‘rod’ domain by MTs. Crossbridges were not observed in preparations of MTs alone (top row).
METHODS. Rotary shadowing was performed as described (Fig. 2 legend). The aperiodic crossbridges are difficult to shadow because they are obscured by MTs during platinum spraying. For negative staining, KRP130–MT complexes were resuspended in buffer, loaded onto formvar-coated grids, then stained with 2% aqueous uranyl acetate for 3 min.
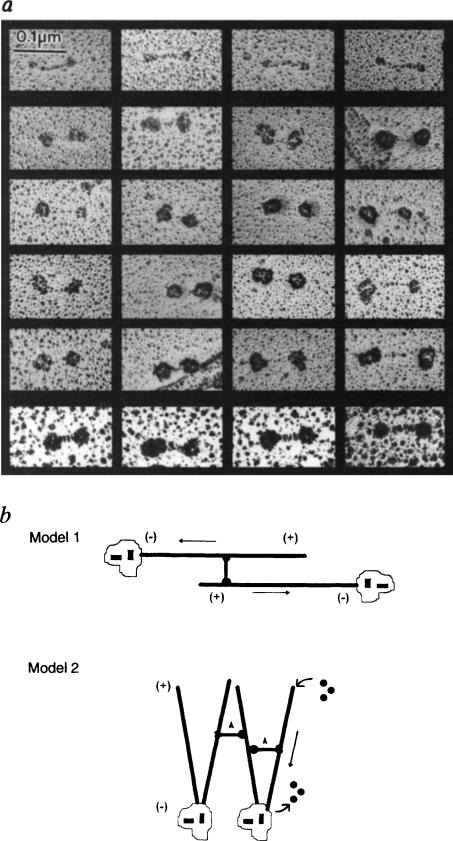
Therefore, to test whether KRP130 is a bipolar tetramer, we used electron microscopy to study rotary-shadowed KRP130 molecules decorated with an antibody that reacts specifically with the motor domains of Eg5 and its close relatives, all members of the BimC subfamily of kinesins (Fig. 1b, c). We had previously observed that the motor-domain monoclonal IgG, SUK4, decorates only one end of the conventional kinesin molecule17. In contrast, the Eg5 motor-domain antibodies used here usually bound to both ends of KRP130 molecules (60–70% of those molecules examined), decorating the globular domains and resulting in a marked increase in their dimensions (Fig. 4a). For example, we examined 70 randomly selected KRP130 molecules that had been incubated with motor-domain antibody before rotary shadowing, of which 18 were undecorated (overall length, 102 ± 9.9nm; rod length, 62.6 ± 11.3 nm; globular domain diameter, 22.6 ± 3.7 nm), 6 were decorated on one end only, and 46 were decorated on both ends (overall length, 127 ± 18.7nm; rod length, 43.1 ± 13.3 nm; decorated globular domain diameter, 44.7 ± 7.2 nm). By comparison, under similar conditions, Hirokawa and co-workers18 found that approximately 50% of conventional kinesin molecules were labelled with antibody, and the decorated ends of these molecules displayed an overall diameter of approximately 30 nm, two-thirds the size of the corresponding dimension observed here.
FIG. 4.
Antibody decoration of bipolar KRP130 tetramers and models of KRP130 function, a, Rotary-shadowed KRP130 decorated with anti-Eg5 motor-domain antibody (Fig. 1c). About one-quarter of the molecules on the grid were undecorated (top row). Decorated molecules show the characteristic ‘dumbbell’ shape but the heads are enlarged. Because of the large size of the heads and low angle of shadowing, the contrast of the ‘rod’ of decorated molecules (middle four rows) is hard to visualize, but was clearer when rotary shadowing was prolonged to produce larger platinum grain (bottom row). b, Models for KRP130 function. In model 1, KRP130 crosslinks two antiparallel MTs emanating from opposite spindle poles with MT plus ends distal to the poles. The motor domains at opposite ends of KRP130 molecules move towards the plus ends of the crosslinked MTs, causing the MTs themselves to slide apart, minus ends leading (arrows), resulting in spindle-pole separation during the formation, maintenance or elongation of a bipolar mitotic spindle. In model 2, KRP130 could crosslink parallel MTs emanating from one or both poles and drive movements necessary for spindle formation and maintenance. For example, by moving towards the plus ends of crosslinked MTs (arrowheads) it may ‘zip’ together MTs into kinetochore fibres or, as proposed for Xenopus Eg5 (ref. 5), it could exert pushing forces (straight arrow) that drive MT flux towards the poles and take part in organizing spindle poles. We favour model one, which is more consistent with the mechanism of action of the bipolar actin-based motor, myosin II.
METHODS. Purified KRP130 from the sucrose gradients was mixed with MTs and anti-Eg5 motor-domain antibody (Fig. 1) in amounts approximately equimolar to the amount of KRP130. The MT–KRP130–antibody complexes were pelleted through a glycerol cushion and processed for rotary shadowing as described (Fig. 2 legend).
Our results are consistent with the hypothesis that at least two Eg5 motor-domain antibody molecules have decorated each end of a KRP130 molecule, and that each KRP130 holoenzyme has two motor domains at either end. Thus, based on electron microscopy and previous hydrodynamic studies13, we conclude that KRP130 holoenzymes are extended, elongated molecules consisting of four motor polypeptides assembled into bipolar ‘minifilaments’ (Fig. 3a). These ‘minifilaments’ could crosslink adjacent MTs and slide them relative to one another (Fig. 4b). This model is consistent with recent evidence that an intact carboxy-terminal tail is a prerequisite for Eg5 localization and function in the spindle21, presumably because the tail region participates in the self-assembly of functional bipolar holoenzymes.
How could such an assembly function in the spindle? The only other motor protein known to assemble into bipolar aggregates is myosin II (ref. 15), which crosslinks actin filaments and slides them relative to one another during muscle contraction and cytokinesis22–24. Similarly, we propose that KRP130 crosslinks adjacent MTs and slides them relative to one another. We propose that the bipolar KRP130 tetramer binds to two antiparallel MTs emanating from opposite spindle poles and, by walking towards their plus ends, pushes the poles apart (Fig. 4b, model 1). This model is supported by the observation that several antibodies to members of the BimC subfamily react with KRP130 and stain mitotic spindles, and by genetic studies which suggest that members of the BimC subfamily participate in separation of the spindle poles3–6,11. However, our data do not exclude the possibility that KRP130 drives other forms of MT motility. For example, by cross-linking parallel MTs emanating from one or both poles it could drive MT flux and organize spindle poles5 or kinetochore fibres (Fig. 4b, model 2). It will now be interesting to test these models and determine whether other motors assemble into bipolar structures capable of driving MT–MT sliding in the mitotic spindle3–12,14,25,26 or interphase cytoplasm27.
ACKNOWLEDGEMENTS
We thank R. S. Hawley and F. McNally for discussion, and R. Harris and K. Sheehan for technical assistance. This work was supported by grants from the March of Dimes Birth Defects Foundation (J.M.S.), the NIH (J.M.S. and W.M.S.), the Whitaker foundation (R.J.B.), and the American Cancer Society (J.M.S.). W.M.S. is an established investigator of the American Heart Association.
References
- 1.Mitchison TJ. Phil. Trans. R. Soc. Lond. 1992;B336:99–106. doi: 10.1098/rstb.1992.0049. [DOI] [PubMed] [Google Scholar]
- 2.Vernos I, Karsenti E. Trends Cell Biol. 1995;5:297–301. doi: 10.1016/s0962-8924(00)89045-5. [DOI] [PubMed] [Google Scholar]
- 3.Enos AP, Morris NR. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- 4.LeGuellec R, Paris J, Couturier A, Roghi C, Philippe M. Molec. cell. Biol. 1991;11:3395–3398. doi: 10.1128/mcb.11.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 6.Roof DM, Meluh PB, Rose MD. J. Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyt MA, He L, Loo KK, Saunders WS. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan I, Yanagida M. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- 9.Hagan I, Yanagida M. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- 10.Sanders WS, Hoyt MA. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 11.Heck MMS, et al. J. Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houliston E, LeGuellec R, Kress M, Philippe M, LeGuellec K. Devl Biol. 1994;164:147–159. doi: 10.1006/dbio.1994.1187. [DOI] [PubMed] [Google Scholar]
- 13.Cole DG, Saxton WM, Sheehan KB, Scholey JM. J. biol. Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 14.Barton NR, Pereira AJ, Goldstein LSB. Molec. Biol. Cell. 1995;6:1563–1574. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huxley HE. J. molec. Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 16.Amos LA. J. Cell Sci. 1987;87:105–111. doi: 10.1242/jcs.87.1.105. [DOI] [PubMed] [Google Scholar]
- 17.Scholey JM, Heuser J, Yang JT, Goldstein LSB. Nature. 1989;338:355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa N, et al. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 19.Tsuprun VL, Kashina AS, Gelfand VI. Dokl. Akad. Nauk SSSR. 1991;320:1270–1272. [PubMed] [Google Scholar]
- 20.Andrews SB, Gallant PE, Leapman RD, Schnapp BJ, Reese TS. Proc. natn. Acad. Sci. U.S.A. 1993;90:6503–6507. doi: 10.1073/pnas.90.14.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawin KE, Mitchison TJ. Proc. natn. Acad. Sci. U.S.A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huxley AF, Nieredgerke RM. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 23.Huxley HE, Henson J. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 24.Yumura S, Fukui Y. Nature. 1985;314:194–196. doi: 10.1038/314194a0. [DOI] [PubMed] [Google Scholar]
- 25.Hogan CJ, et al. Proc. natn. Acad. Sci. U.S.A. 1993;90:6611–6615. doi: 10.1073/pnas.90.14.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- 27.Koonce MP, Tong J, Euteneuer U, Schliwa M. Nature. 1987;328:737–739. doi: 10.1038/328737a0. [DOI] [PubMed] [Google Scholar]
- 28.Tyler JM, Branton D. J. Ultrastruct. Res. 1980;71:95–102. doi: 10.1016/s0022-5320(80)90098-2. [DOI] [PubMed] [Google Scholar]