Investigation of extracts from the plant Athroisma proteiforme (Humbert) Mattf. (Asteraceae) for antimalarial activity led to the isolation of the five new sesquiterpene lactones 1-5 together with centaureidin (6). The structures of the new compounds were deduced from analyses of physical and spectroscopic data, and the absolute configuration of compound 1 was confirmed by an X-ray crystallographic study. Athrolides C (3) and D (4) both showed antiplasmodial activities with IC50 values of 6.6 μM (3) and 7.2 μM (4) against the HB3 strain and 5.5 μM (3) and 4.2 μM (4) against the Dd2 strain of the malarial parasite Plasmodium falciparum. The isolates 1-6 also showed antiproliferative activity against A2780 human ovarian cancer cells, with IC50 values ranging from 0.4 – 2.5 μM.
In our continuing search for biologically active natural products from tropical rainforests, we obtained an ethanol extract from the aerial parts of a plant identified as Athroisma proteiforme (Humbert) Mattf. (Asteraceae) from the Toliara dry forest in southwest Madagascar. The extract exhibited moderate antimalarial activities against HB3 (chloroquine sensitive; CQS) and Dd2 (chloroquine resistant; CQR) P. falciparum strains with IC50 values of less than 4 μg/mL to each strain. On the basis of these activities and the paucity of previous phytochemical studies on this genus, A. proteiforme was selected for bioassay-guided fractionation to isolate the antiplasmodial components.
Athroisma proteiforme was previously known as Polycline proteoformis, and the genus is represented by three endemic species in Madagascar. The only previous phytochemical work on it was the isolation of thymol and a menthene diol from A. gracile,2 and no phytochemical work has been reported on the genus Polycline. Previous phytochemical studies of plant species belonging to the family Asteraceae have revealed the presence of antimalarial sesquiterpene lactones3-7 and flavonoids.8,9 Among all natural products with antimalarial activity, including alkaloids, terpenoids, flavonoids, limonoids, chalcones, peptides, xanthones, quinones,2 and coumarins,10 the sesquiterpene artemisinin from the traditional Chinese medicinal plant Artemesia annua (Asteraceae) is one of the most important clinically used antimalarial agents,4 and it and its derivatives are currently used in artemisinin-based combination therapies (ACTs).11,12 The probability of isolating additional antimalarial sesquiterpenes thus provided a further incentive to investigate this plant.
Results and Discussion
The ethanol extract of the aerial parts of A. proteiforme was subjected to liquid-liquid partitioning to give hexanes, CH2Cl2, and MeOH fractions with IC50 values of 4.0 μg/mL, 1.5 μg/mL, and >10 μg/mL, respectively against the HB3 strain, and 2.0 μg/mL, 1.0 μg/mL, and 8.0 μg/mL, respectively against the Dd2 strain. Fractionation of the most active CH2Cl2 fraction by C-18 open column and HPLC yielded five new sesquiterpene lactones designated athrolides A – E (1-5), as well as the known flavonoid centureidin (6) (Figure 1). Herein we report the structural elucidation, the antimalarial properties, and the antiproliferative activities of the isolates.
Figure 1.
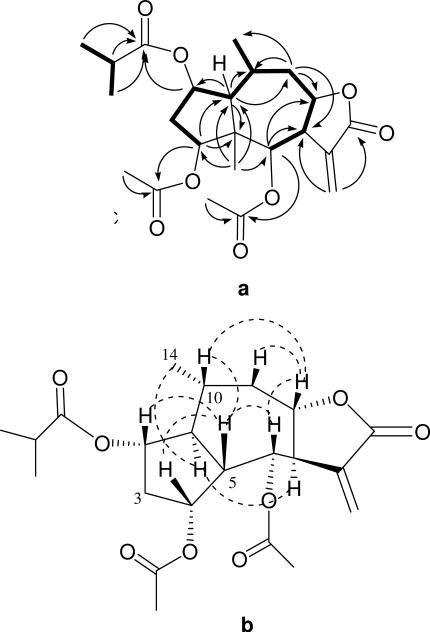
a) Key COSY (bold) and HMBC (arrows) correlations for 1. b) Key NOESY correlations for 1.
Athrolide A (1) was obtained initially as an off-white solid. Its positive ion HR-ESIMS revealed a pseudomolecular ion peak at m/z 437.2176 [M+H]+, corresponding to the molecular formula C23H32O8. The IR spectrum showed strong absorption in the range of 1740-1710 cm-1, consistent with the presence of ester and α,β-unsaturated lactone groups. Its 1H NMR spectrum in CDCl3 showed signals for four methyl singlets at δH 2.10, 2.09, 2.08, and 0.98, three methyl doublets at δH 1.16, 1.16, and 1.05, one vinyl methylene (two doublets at δH 6.27 and 5.65, J = 3.2 Hz, H-13a and H-13b), two pairs of methylenes at δH 2.64 (m, H-3a) and δH 1.32 (dd, J = 16.3, 2.7 Hz, H-3b), and at δH 2.33 (ddd, J = 12.8, 3.1, 3.1 Hz, H-9a) and δH 1.55 (m, H-9b), and eight methines [δH 1.88, m (H-10); δH 2.67, m (H-1); δH 2.52, septet, J = 7.0 Hz (H-2'); δH 3.48, m (H-7); δH 4.11, ddd, J = 12.2, 9.0, 3.5 Hz (H-8); δH 4.95, d, J = 4.9 Hz (H-4); δH 5.03, ddd, J = 8.7, 8.7, 2.7 Hz, (H-2) and δH 5.06, d, J = 11 Hz (H-6)]. The chemical shift data indicated that four of the methines were on oxygenated carbons (Table 1). The 13C NMR spectrum displayed signals for two acetoxy groups (δC 169.8 and 20.0 and δC 170.2 and 21.4) and one 2-methylpropanoyloxy group (δC 176.4, 33.9, 18.8, and 18.7) together with 15 other signals. These signals were assigned by an HMQC spectrum to two quaternary carbons at δC 51.5 (C-5) and 136.2 (C-11); a lactone carbonyl at δC 169.2 (C-12); one quaternary and one secondary methyl at δC 22.4 (C-15) and 20.0 (C-14); two methylenes at δC 37.9 (C-3) and 43.1 (C-9); four oxygen-bearing methines at δC 80.9 (C-8), 79.6 (C-4), 78.1 (C-6), and 75.5 (C-2); three methines at δC 51.3 (C-1), 46.3 (C-7), and 27.2 (C-10); and an vinylic methylene at δC 124.1 (C-13) (Table 6.1). These data are all interpretable by assignment of a sesquiterpene lactone structure to athrolide A.
Table 1.
1H and 13C NMR Data of Athrolides A (1), B (2), C (3), D (4) and E (5)a
| position | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1H (J, Hz) | 13C | 1H (J, Hz) | 13C | 1H (J, Hz) | 13C | 1H (J, Hz) | 13C | 1H (J, Hz) | 13C | |
| 1 | 2.67 m | 51.3 | 2.24 dd (10.8, 6.8) | 52.8 | 3.19 dm (12.6) | 50.5 | 3.19 dm (12.6) | 50.5 | 3.23 dm (12.8) | 50.2 |
| 2 | 5.03 td (8.7, 2.7) | 75.5 | 5.02 ddd (9.1, 6.9, 2.1) | 77.2 | 7.60 bd (6.0) | 161.0 | 7.60 bd (6.1) | 161.0 | 7.60 bd (6.0,) | 161.1 |
| 3 | 2.64 m, 1.32 dd (16.3, 2.7) | 37.9 | 2.66 ddd (16.4, 9.0, 4.8), 1.53 dd (16.5, 2.1) | 38.1 | 6.11 overlapped | 130.3 | 6.11 overlapped | 130.3 | 6.10 m | 130.3 |
| 4 | 4.95 d (4.9), | 79.6 | 4.90 d (4.8) | 82.4 | 207.7 | 207.7 | 207.8 | |||
| 5 | 51.5 | 51.3 | 55.3 | 55.3 | 55.2 | |||||
| 6 | 5.06 d (11.0) | 78.1 | 4.41 bs | 65.0 | 5.34 d (3.7) | 76.0 | 5.34 d(3.7) | 76.0 | 5.31 d (3.7) | 75.3 |
| 7 | 3.48 m | 46.3 | 3.04 m | 52.0 | 3.55 m | 45.5 | 3.56 m | 45.5 | 3.51 m | 45.7 |
| 8 | 4.11 ddd (12.2, 9.0, 3.5) | 80.9 | 4.64 ddd (12.1, 9.1, 3.2) | 75.7 | 5.48 s | 64.7 | 5.48 s | 64.7 | 5.49 s | 65.0 |
| 9 | 2.33 ddd (12.8, 3.1, 3.1), 1.55 m | 43.1 | 2.41 m, 1.41 m | 44.0 | 4.66 s | 88.5 | 4.66 s | 88.5 | 4.66 s | 88.6 |
| 10 | 1.88 m | 27.2 | 1.98 m | 29.0 | 2.43 m | 35.4 | 2.43 m | 35.4 | 2.43 m | 35.4 |
| 11 | 136.2 | 136.8 | 130.8 | 130.8 | 131.2 | |||||
| 12 | 169.2 | 169.3 | 162.4 | 162.4 | 162.5 | |||||
| 13 | 6.27 (d, 3.2), 5.65 (d, 3.2), | 124.1 | 6.40 (d, 3.6), 5.53(d, 3.6) | 121.0 | 6.80s, 6.11s | 134.4 | 6.80s, 6.11s | 134.3 | 6.78s, 6.10s | 134.1 |
| 14 | 1.05 d (6.8) | 20.0 | 1.02 d (6.6) | 20.9 | 1.41 d (7.2) | 19.2 | 1.41 d (7.2) | 19.2 | 1.41 (7.2) | 19.1 |
| 15 | 1.09 s | 22.4 | 0.98 s | 17.7 | 1.07 s | 18.8 | 1.07 s | 18.8 | 1.05 s | 18.9 |
| 1' | 176.4 | 176.4 | 166.1 | 166.1 | 165.5 | |||||
| 2' | 2.52 septet (7.0) | 33.9 | 2.50 septet (7.0) | 34.0 | 127.0 | 127.0 | 5.53 | 113.3 | ||
| 3' | 1.16 d (7.0) | 18.8 | 1.14 d (7.0) | 18.8 | 6.09 m | 139.7 | 6.09 m | 139.6 | 164.4 | |
| 4' | 1.16 d (7.0) | 18.7 | 1.13 d (7.0) | 18.7 | 1.94 dq (7.3, 1.5) | 15.7 | 1.93 dq (7.2, 1.5) | 15.7 | 2.16 q (7.3) | 33.9 |
| 5' | 1.78 m | 20.4 | 1.78 m | 20.4 | 1.05 t (7.3) | 11.8 | ||||
| 6' | 2.15 s | 19.0 | ||||||||
| 1” | 169.8 | 170.5 | 171.6 | 171.7 | 171.8 | |||||
| 2” | 2.08 s | 20.0 | 2.10 s | 21.3 | 2.49 s | 46.4 | 2.51 d (15.5) 2.42 d (15.5) |
44.5 | 2.51 d (15.2) 2.43d (15.2) |
44.6 |
| 3” | 69.2 | 71.4 | 71.5 | |||||||
| 4” | 1.27 s | 29.3 | 1.52 qd (7.5, 2.5) | 34.7 | 1.52 m | 34.7 | ||||
| 5” | 1.26 s | 29.2 | 0.90 t (7.5) | 8.2 | 0.90 t (7.5) | 8.3 | ||||
| 6” | 1.19 s | 26.2 | 1.19 s | 26.2 | ||||||
| 1’” | 2.09 s | 170.2 | ||||||||
| 2’” | 21.4 | |||||||||
In CDCl3; δ (ppm) 500 MHz for 1H and 125 MHz for 13C; multiplicities; J values (Hz) in parentheses.
Inspection of the 1H NMR data revealed that athrolide A is similar to the known pseudoguaianolides 6-angeloyloxypuchellin13 and 2-deacetyl-2-isobutyrylchamissonolide.14 The complete 1H and 13C NMR assignments and connectivities were established from a combination of HMQC, COSY, and HMBC data analyses. The COSY spectrum showed correlations that indicated the presence of the spin systems: H-4, H-3, H-2, H-1, H-10, H-9, H-8, H-7, H-6, and H-14, and H-2', H-3' and H-4' of the 2-methylpropanoyloxy moiety. In the HMBC spectrum, the correlations from H3-15 to C-4, C-5, C-1, and C-6 as well as the correlations from H-4 to C-1 and to C-5 corroborated the presence of a five-membered ring, fused at C-1 and C-5 with a seven-membered ring. The HMBC correlations from H2-9 to C-1, C-8, C-7, and C-14, and from H-6 to C-5, C-7, and C-8, and from H2-13 to C-7 and the lactone carbonyl at C-12 suggested the presence of a γ-lactone ring, fused at C-7 and C-8. These data indicated that 1 was a pseudoguaianolide.13, 14 The HMBC correlations from two methyl groups (H3-3' and H3-4'), one septet methine (H-2'), and H-2 to C-1', H-4 and H3-2” to C-1”, H-6 and H3-2’” to C-1’” indicated the presence of an 2-methylpropanoyl group at C-2, an acetate group at C-4, and another acetate group at C-6. In the NOESY spectrum of 1, the correlations from H-1 to H-7, H-9b, and H3-14, from H3-15 to H-2, H-4, H-6, H-8 and H-10, and from H-8 to H-6, H-9a, and H-10 suggested that H-1, H-7, and H3-14 were cofacial and that H-2, H-4, H-6, H-8, H-10, and H3-15 were on the opposite face.
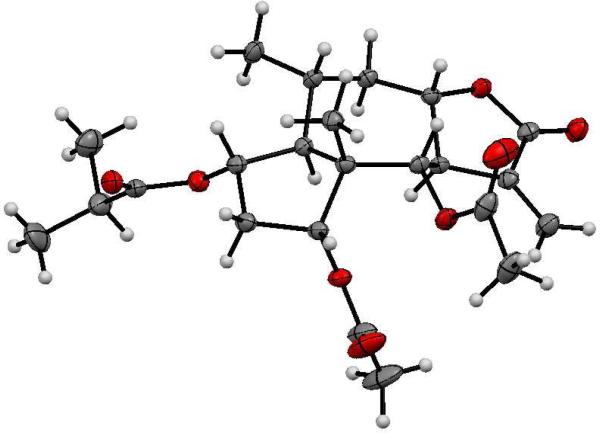
In order to determine the absolute configuration, compound 1 was crystallized from MeOH to afford good quality needle-shaped crystals, and its structure was confirmed by single crystal X-ray diffraction. An anisotropic displacement ellipsoid drawing is shown in Figure 2. Anomalous dispersion effects confirmed the absolute configuration of 1 to be (1S,2S,4R,5S,6S,7R,8S,10R)-2-(2-methylpropanoyloxy)-4-acetoxy-6-acetoxyguai-11(13)-en-8,12-olide (1, athrolide A).
Figure 2.
Anisotropic displacement ellipsoid drawing of 1
Athrolide B (2) was obtained as an off-white solid. Its positive ion HR-ESIMS revealed a pseudomolecular ion peak at m/z 417.1889 [M+Na]+, corresponding to the molecular formula C21H30O7. Its IR spectrum showed a hydroxyl stretch at 3474 cm-1 and strong absorption in the range 1740-1710 cm-1. Its 1H NMR spectrum in CDCl3 showed signals for two methyl singlets at δH 2.10 and 0.98, three methyl doublets at δH 1.14, 1.14 and 1.02, three methylene multiplets, two of which were olefinic, (δH 2.66, 1.53; 2.42, 1.41; and 5.53, 6.40), and eight methines (δH 1.98, 2.24, 2.50, 3.04, 4.41, 4.64, 4.90 and 5.02,), four of which were oxygenated (Table 1). Inspection of the 1H and 13C NMR data of 2 in CDCl3 showed a close similarity with the data of the previously isolated (lS,2S,4R,5R,6R,7S,8S,l0R)-2,4-diacetoxy-6-hydroxyguai-11(13)-en-8,12-olide (7).15 It differed from 7 by the presence of a 2-methylpropanoyl group at C-2 of 4 compared to an acetate group at C-2 of 7. HMBC correlations from H-2 to C-1', and of the methine septet at H-2' and of H3-3' and H3-4' to C-1', and from H-4 to C-1” indicated that the 2-methylpropanoyl group was located at C-2 and the acetate group at C-4. In the NOESY spectrum, the correlations from H-1 to H-6, H-7, H-9b, and H3-14, from H-6 to H-7 and H-1, and from H3-15 to H-2, H-4, H-8, and H-10, indicated that H-1, H-6, H-7 and H3-14 were cofacial and that H-2, H-4, H-8, H-10, and H3-15 were on the opposite face. The characteristic UV absorption of an α, β-unsaturated lactone chromophore was observed at 230 nm.16 The absolute configuration of 2 was deduced by the comparison of its CD spectrum with that of 1. The negative Cotton effect for 2 of [θ]230nm = -1.56 × 103 was similar to that of 1 ([θ]230 nm = -3.57 × 103), and enabled assignment of the S configuration to C-7 according to the back octant rule.17 Therefore, athrolide B (2) was determined to be (1S,2S,4R,5R,6R,7S,8S,10R)-2-(2-methylpropanoyloxy)-4-acetoxy-6-hydroxyguai-11(13)-en-8,12-olide.
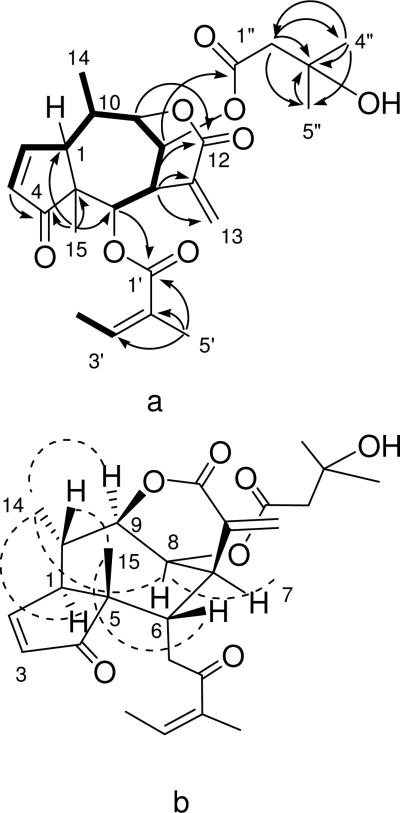
Athrolide C (3) was obtained as an off-white solid. Its positive ion HR-ESIMS revealed a pseudomolecular ion peak at m/z 483.2003 [M+Na]+, corresponding to the molecular formula C25H32O8. The IR spectrum showed a hydroxy absorption (3444 cm-1), and strong absorption in the range 1740-1710 cm-1, consistent with the presence of ester, ketocarbonyl, and lactone groups. Its 1H NMR spectrum in CDCl3 showed signals for three methyl singlets (δH 1.27, 1.26 and 1.07), one methyl doublet (δH 1.41 d, J = 7.2 Hz), two olefinic methyl groups (δH 1.78 m; 1.94 dq, J = 7.3, 1.5 Hz), one singlet methylene (δH 2.49), and three methines (δH 3.55 m, 3.19 m, and 2.43 m). Three oxygenated methines were also observed at δH 5.48 br s, 5.34 (d, J = 3.7 Hz), and δH 4.66 br s. Signals for five olefinic methines were observed at δH 7.60 dd (J = 6.0, 1.6 Hz), 6.80 s, 6.12 s, 6.12 m, and 6.10 m. The 13C NMR spectrum displayed a set of signals ascribable to a (Z)-2-methyl-2-butenoyl (angeloyl) group (δC 166.1, 127.0, 139.7, 15.7, 20.4)15 and a 3-hydroxy-3-methylbutanoyl group (δC 171.6, 46.4, 69.2, 29.3, 29.2)18 together with 15 signals of a sesquiterpene lactone [two quaternary carbons at δC 55.3 and 130.8; a lactone carbonyl at δC 162.4, and a conjugated carbonyl at δC 207.7; one quaternary and one secondary methyl (δC 18.8 and 19.2, respectively); two olefinic methines (δC 161.0 and 130.3); three oxygen-bearing methines (δC 88.5, 76.0, and 64.7); three methines (δC 50.5, 45.5, and 50.5); and an exocyclic methylene (δC 134.4), as indicated by the HMQC spectrum (Table 1)]. The complete 1H and 13C NMR assignments and connectivities were established from a combination of COSY, HMQC, and HMBC data. The COSY spectrum showed correlations that indicated the presence of the two spin systems: H-3, H-2, H-1, H-10, H-9, H-8, H-7, H-6, and H-14, and H-3' and H-4' of the angeloyl moiety. In the HMBC spectrum, the correlations from H3-15 to C-4, C-5, C-1, and C-6 as well as the correlations from H-3 to C-4 and C-5 corroborated the presence of an α, β-unsaturated cyclopentenone ring fused with a seven-membered ring. The HMBC correlations from H-9 to C-12 at δC 162.4 and H-7 to C-11 at δC 130.8, C-12 at δC 162.4 and C-13 at δC 134.4 suggested the presence of a δ-lactone ring, fused at C-7 and C-9 and substituted at C-8. These data indicated that 3 was a pseudoguaianolide analogue.19 Inspection of the 13C NMR data of 3 indicated a close similarity to the data of 1α,7α,10(H)β-4-oxo-6α-[(Z)-2-methyl-2-butenoyloxy]-8β-acetoxypseudoguaia-2(3),11(13)-dien-9β,12-olide (8), previously isolated from Hymenoxys ivesiana.19 These comparisons indicated that 3 differed from 8 only in the nature of the ester substituent at C-8. The locations of the (Z)-2-methyl-2-butenoate (angelate) group at C-6 and the 3-hydroxy-3-methyl butanoate group at C-8 were substantiated by the observation of HMBC cross peaks from H-6 to C-1', H-3' to C-1', H-4' to C-3' and C-2', and H-5' to C-1', C-2' and C-3' and from H-8 to C-1”, H2-2” to C-1”, C-3”, C-4” and C-5”, H3-4” to C-2”, C-3”, and C-5”, and H3-5” to C-2”, C-3”, and C-4”.
NOESY correlations of 3 from H3-15 to H-10 and H-6, and H3-14 to H-1, H-9 and H-8, as well as the cross peaks arising from H-7 to H-8 suggested that H-1, H-7, H-8, H-9 and H3-14 were cofacial and that H-6, H-10 and H3-15 were on the opposite face. Therefore, athrolide C (3) was determined to be 1S*,7R*,10(Η)R*-4-oxo-6S*-[(Z)-2-methyl-2-butenoyloxy]-8S*-(3-hydroxy-3-methylbutanoyloxy)pseudoguaia-2(3),11(13)-dien-9R*,12-olide.
Athrolide D (4) was obtained as an off-white solid. Its positive ion HR-ESIMS revealed a pseudomolecular ion peak at m/z 497.2143 [M+Na]+, corresponding to the molecular formula C26H34O8. Its IR spectrum and 1H NMR data were similar to those of athrolide C (3). The only significant difference was the presence of a set of signals at δH 2.51 (1H, d J = 15.5 Hz), 2.42 (1H, d J = 15.5 Hz), 1.52 (2H, qd J = 7.5, 2.6 Hz), 1.19 (3H, s), and 0.90 (3H, t J = 7.5 Hz) instead of those for the 3-hydroxy-3-methylbutanoate group of 3. The 13C NMR spectrum displayed signals ascribable to a 3-hydroxy-3-methylpentanoate group20 (δC 171.7, 71.4, 44.5, 34.7, 26.2, and 8.2). The HMBC correlations from H-8 to C-1”; H-2” to C-1” and C-3”; H-4” to C-2”, C-3”, C-5” and C-6”, H-5” to C-3”, and H-6” to C-2” and C-3” as well as the COSY cross peak from H-5” and H-6” confirmed the presence of a 3-hydroxy-3-methylpentanoate group and its location at C-8. The NOESY spectrum of 4 showed similar correlations to those observed for 3. The correlations from H3-15 to H-10 and H-6, and H3-14 to H-1, H-9 and H-8, as well as the correlation from H-7 to H-8 were observed. Thus the structure of 4 was assigned as 1S*,7R*,10(Η)R*-4-oxo-6S*-[(Z)-2-methyl-2-butenoyloxy]-8S*-(3-hydroxy-3-methylpentanoyloxy)pseudoguaia-2(3),11(13)-dien-9R*,12-olide.
Athrolide E (5) was obtained as an off-white solid. Its positive ion HR-ESIMS revealed a pseudomolecular ion peak at m/z 489.2496 [M+H]+, corresponding to the molecular formula C27H36O8. Its IR spectrum was similar to those of athrolides C (3) and D (4). The similarity of its 1H and 13C NMR spectroscopic data to those of 4 (Table 1) suggested that 5 was a closely related pseudoguaianolide analogue. The presence of an (E)-3-methyl-2-pentenoate group at C-6 was indicated by the observation of a set of signals at δH 5.53 (1H, s), 2.16 (2H, q J = 7.3 Hz), 2.15 (3H, s), and 1.05 (3H, t J = 7.3 Hz) in the 1H NMR spectrum, and the corresponding 13C NMR data (δC 165.5, 164.4, 113.3, 33.9, 19.0, and 11.8) supported this conclusion.21 The HMBC correlations from H-6 to C-1', H-2' to C-1' and C-3', H-4' to C-2', C-3', C-5', and C-6', H-5' to C-3' and C-4', and H-6' to C-2', C-3', and C-4' as well as the COSY cross peak between H-4' and H-5' confirmed the location of the (E)-3-methyl-2-pentenoate group at C-6. A NOESY correlation from H-2' to H-4' assigned the configuration of the double bond as E. Finally, the NOESY correlations of the ring protons observed in 5 were similar to those observed in 3 and 4. Clear correlations from H3-15 to H-10 and H-6, from H3-14 to H-1, H-9 and H-8, and from H-7 to H-8 of 5 confirmed its configuration to be the same as that of 4. Thus the structure of 5 was concluded to be 1S*,7R*,10(Η)R*-4-oxo-6S*-(E)-3-methyl-2-pentenoyloxy-8S*-(3-hydroxy-3-methylpentanoyloxy)pseudoguaia-2(3),11(13)-dien-9R*,12-olide.
The structure of the known compound (6) was determined to be centaureidin by comparison of its MS and 13C NMR data with literature data.22
Athrolides A (1), C (3), D (4) and E (5) were tested for their antimalarial activities against the drug sensitive HB3 and drug resistant Dd2 strains of P. falciparum (Table 2); a lack of material prevented the assay of athrolide B (2). Athrolide D (4) showed the strongest activities against the drug-resistant Dd2 strain with an IC50 value of 4.2 μM, while athrolide C (3) had similar activity (IC50 5.5 μM) against the same strain. Interestingly both compounds were less potent against the drug-sensitive HB3 strain, with IC50 values of 6.6 and 7.2 μM, respectively. These potencies are less than those obtained for the partially purified fractions described earlier, but no other active materials could be obtained. It is possible that the original extract contained a highly active but unstable compound, but it is also possible that the preliminary assays, which were obtained over a limited dose range, indicated erroneously higher potencies than the true values.
Table 2.
Antiplasmodial and Antiproliferative Data for Athrolides A - E (1 - 5)
| Compound | Antiplasmodial activity (IC50, μM)a | Antiproliferative activity (IC50, μM)b A2780 | ||
|---|---|---|---|---|
| HB3 | Dd2 | Rfc | ||
| 1 | 34.3 ± 0.3 | 32.7 ± 0.3 | 1.0 | 2.1 ± 0.2 |
| 2 | NDd | NDd | 2.5 ± 0.12 | |
| 3 | 6.6 ± 0.1 | 5.5 ± 0.03 | 0.8 | 0.57 ± 0.05 |
| 4 | 7.2 ± 0.2 | 4.2 ± 0.7 | 1.7 | 0.38 ± 0.02 |
| 5 | 16.0 ± 0.3 | 11.7 ± 0.3 | 0.7 | 1.9 ± 0.2 |
| Chloroquine | 0.0147 ± 0.0007 | 0.224 ± 0.009 | NDd | |
| Paclitaxel | NDd | NDd | 0.017 ± 0.006 | |
Data ± standard error of the mean
Data ± standard deviation
Resistance factor
Not determined
The antiproliferative activities of compounds 1-6 were also evaluated against the A2780 human ovarian cancer cell line. Athrolides A-E had IC50 values of 2.1, 2.5, 0.57, 0.38, and 1.9 μM, respectively, against this cell line, while centaureidin was also weakly active with an IC50 value of 3.9 μM. The fact that athrolides C and D were more potent towards the drug-resistant Dd2 line than towards the drug-sensitive HB3 line is an interesting observation, but the fact that their antiproliferative IC50 values are lower than their antimalarial IC50 values suggests that these compounds are not likely to be useful lead compounds because of potential toxicity concerns. The fact that the most potent antiproliferative compounds 3 and 4 were also the most potent antimalarial compounds also suggests that it will prove difficult to separate these two activities in this class of compounds.
Experimental Section
General Experimental Procedures
Optical rotations were recorded on a JASCO P-2000 polarimeter. UV and IR spectra were measured on a Shimadzu UV-1201 spectrophotometer and a MIDAC M-series FTIR spectrophotometer, respectively. CD analyses were performed on a JASCO J-810 spectropolarimeter with a 1.0 cm cell in MeOH. NMR spectra were recorded in CDCl3 on either JEOL Eclipse 500 or Bruker 600 spectrometers. The chemical shifts are given in δ (ppm) and coupling constants (J) are reported in Hz. Mass spectra were obtained on an Agilent 6220 TOF Mass Spectrometer. HPLC was performed on a Shimadzu LC-10AT instrument with a semi-preparative C18 Varian Dynamax column (5 m, 250 × 10 mm).
Plant Material
The aerial parts of Athroisma proteiforme (Humbert) Mattf. (formerly Polycline proteiformis Humbert) (Asteraceae) were collected on April 24th, 1998 near Toliara, Madagascar, at coordinates 23°24'30”S 043°46'40”E and an elevation of 47 m. This aromatic herbaceous plant can grow up to 60 cm height and bears white flowers. Voucher specimens have been deposited at the Smithsonian Institution, Washington D.C., and at the Missouri Botanical Garden under voucher number Richard Randrianaivo 197.
Extraction and Isolation
Dried aerial parts of A. proteiforme (approximately 500 g) were ground in a hammer mill, then extracted with EtOH by percolation for 24 h at room temperature to give the crude extract N110635 (5 g), of which 3 g was shipped to Virginia Polytechnic Institute and State University for bioassay guided isolation. The extract N110635 [IC50: 1.9 μg/mL (HB3), 1.6 μg/mL (Dd2)], (2 g) was suspended in aqueous MeOH (MeOH-H2O, 9:1, 100 mL) and extracted with hexanes (3 × 100 mL portions). The aqueous layer was then diluted to 60% MeOH (v/v) with H2O and extracted with CH2Cl2 (3 × 150 mL portions). The hexanes extract was evaporated in vacuo to leave 186.5 mg with IC50 values of 4.0 (HB3) and 2.0 (Dd2) μg/mL. The residue from the CH2Cl2 extract (470.1 mg) had IC50 values of 1.5 (HB3) and <1.0 (Dd2) μg/mL. The aqueous MeOH extract (1.392 g) was less active with IC50 values of >10.0 (HB3) and 8.0 (Dd2). The CH2Cl2 extract was selected for fractionation, and a separation on a C18 open column gave the four fractions I - IV (155.7, 149.7, 54.2 and 19.3 mg). IC50 values were as follows: fractions I, >5.0 (HB3) and >5.0 (Dd2); fraction II, 0.45 (HB3), 0.42 (Dd2); fraction III, 5.0 (HB3), 3.5 (Dd2); fraction IV, > 5.0 (HB3), > 5.0 (Dd2) μg/mL. Fraction II was selected for further separation by C-18 preparative HPLC (60% CH3CN-H2O). Compounds 1 (2.2 mg, tR 22.1 min), 2 (2.2 mg, tR 14.6 min), 3 (2.3 mg, tR 13.2 min), 4 (2.9 mg, tR 16.5 min), 5 (1.1 mg, tR 20.2 min), and 6 (2.3 mg, tR 10.0 min) were isolated as the major components of this fraction.
Bioassays
Antiplasmodial assays were performed against the chloroquine-sensitive HB3 strain and the chloroquine-resistant Dd2 strain of P. falciparum at Georgetown University. The assay utilized the previously reported protocol23 with minor modifications. Typically, the original dried extract was dissolved in DMSO to give stock solutions. Further dilutions of these stock solutions were performed using complete media, finally resulting working stocks. Samples of the working stock solutions (100 μL) were transferred into 96 well plates which were pre-warmed to 37°C prior to the addition of the cultures. Sorbitol synchronized cultures were utilized for the assays with >95% of the parasites in the ring stage. Usually, cultures were diluted to give a working stock of 2% parasitemia and 4% hematocrit and 100 μL was transferred into each drug pre-loaded well (final 1% hematocrit and 0.5% parasitemia). The plates were transferred to an airtight chamber which was gassed (90% N, 5% O2, 5% CO2 gas mixture) and incubated at 37 °C. After 72 h, 50 μL of 10× SYBR Green I dye (diluted using complete media from a 10,000X DMSO stock) was added, the plates incubated for an additional 1 h at 37 °C to allow DNA intercalation, and fluorescence measured at 530 nm (490 nm excitation) using a Spectra GeminiEM plate reader (Molecular Devices). IC50 values from assays done in triplicate were averaged and are shown +/- S.E.M. In these assays, chloroquine (CQ) was included as a positive control. For IC50 calculations, data analysis was performed using Sigma Plot 10.0 software after downloading data in Excel format.23
The A2780 ovarian cancer cell line assay was performed at Virginia Polytechnic Institute and State University as previously reported.24 The A2780 cell line is a drug-sensitive ovarian cancer cell line.25
Athrolide A, (1S,2S,4R,5S,6S,7R,8S,10R)-2-(2-methylpropanoyloxy)-4-acetoxy-6-acetoxyguai-11(13)-en-8,12-olide (1)
White solid; [α]D23 -53 (c 0.1, CHCl3); CD [θ]230 -3570 (MeOH); UV (MeOH) λmax nm (log ε) 211 (4.0); IR νmax cm-1: 3463, 2965, 1733, 1464, 1375, 1258, 1156, 1052, 1018 cm-1. 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; HR-ESIMS m/z 437.2176 [M+H]+, (calcd for C23H33O8, 437.2175).
X-ray Crystallography of 1
Compound 1 crystallized from MeOH as colorless needles. One needle (0.03 × 0.03 × 0.33 mm3) was centered on the goniometer of an Oxford Diffraction SuperNova diffractometer operating with CuKα radiation. The data collection routine, unit cell refinement, and data processing were carried out with the program CrysAlisPro.26 The Laue symmetry and systematic absences were consistent with the monoclinic space groups C2, Cm, and C2/m. As the molecule was known to be enantiomerically pure, the chiral space group, C2, was chosen. The structure was solved using SHELXS-9727 and refined using SHELXL-9727 via OLEX2.28 The final refinement model involved anisotropic displacement parameters for non-hydrogen atoms and a riding model for all hydrogen atoms. The absolute configuration was established from anomalous dispersion effects [Flack x = 0.02(15);29 Hooft P2(true) = 1.000, P3(true) = 1.000, P3(rac-twin) = 0.3×10-5; P3(false) = 0.7×10-25, y = 0.06(9)].30, 31
Crystal data
C23H32O8, Mr =436.49, monoclinic C2, a = 32.7331(12) Å, b = 7.1315(3) Å, c = 9.7799(3) Å, α = 90.00, β = 92.961, γ = 90.00, V = 2279.94(13) Å3, 13543 reflections, 287 parameters. The atomic coordinates and equivalent isotropic displacement parameters, as well as a full list of bond distances and angles, are deposited as supplementary material at the Cambridge Crystallographic Data Centre (Deposition No. CCDC 802814).
Athrolide B, (1S,2S,4R,5R,6R,7S,8S,10R)-2-(2-methylpropanoyloxy)-4-acetoxy-6-hydroxyguai-11(13)-en-8,12-olide (2)
White solid; [α]D23 -4 (c 0.2, CHCl3); CD [θ]230 -1560 (MeOH); UV (MeOH) λmax nm (log ε) 211 (3.9); IR νmax cm-1: 3474, 2972, 1729, 1464, 1376, 1250, 1160, 1044, 1017 cm-1. 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; HR-ESIMS m/z 417.1889 [M+Na]+, (calcd for C21H30NaO7, 417.1889).
Athrolide C, 1S*,7R,10(H)R*-4-oxo-6S*-[(Z)-2-methyl-2-butenoyloxy]-8S*-(3-hydroxy-3-methylbutanoyloxy)pseudoguaia-2(3),11(13)-dien-9R*,12-olide. (3)
White solid; [α]D23 -69 (c 0.1, CHCl3); UV (MeOH) λmax nm (log ε) 220 (4.2); IR νmax cm-1: 3444, 2924, 1721, 1458, 1382, 1229, 1154, 1035, 999.8 cm-1. 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; HR-ESIMS m/z 483.2003 [M+Na]+ (calcd for C25H32NaO8, 483.1995).
Athrolide D, 1S*,7R*,10(H)R*-4-oxo-6S*-[(Z)-2-methyl-2-butenolyloxy]-8S*-(3-hydroxy-3-methylpentanoyloxy)pseudoguaia-2(3),11(13)-dien-8R*,12-olide (4)
White solid; [α]D23 -66 (c 0.2, CHCl3); UV (MeOH) λmax nm (log ε) 220 (4.3); IR νmax cm-1: 3445, 2927, 1721, 1458, 1382, 1216, 1154, 1035, 999.5 cm-1. 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; HR-ESIMS m/z 497.2143 [M+Na]+ (calcd for C26H34NaO8, 497.2151) .
Athrolide E, 1S*,7R*,10(H)R*-4-oxo-6S*-(E)-3-methyl-2-pentenoyloxy-8S*-(3-hydroxy-3-methyl-pentanoyloxy)pseudoguaia-2(3),11(13)-dien-9R*,12-olide (5)
White solid; [α]D23 -33 (c 0.1, CHCl3); UV (MeOH) λmax nm (log ε) 221 (4.3); IR νmax cm-1: 3441, 2923, 1721, 1458, 1380, 1216, 1142, 1034, 1005.5 cm-1. 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3), see Table 1; HRESIMS m/z 489.2496 [M+H]+ (calcd for C27H37O8, 489.2488).
Supplementary Material
Figure 3.
a) Key COSY (bold) and HMBC (arrows) correlations for 3. b) Key NOESY correlations for 3.
Acknowledgments
This project was supported by the Fogarty International Center, the National Cancer Institute, the National Science Foundation, the National Heart, Lung and Blood Institute, the National Institute of Mental Health, the Office of Dietary Supplements, and the Office of the Director of NIH, under Cooperative Agreement U01 TW000313 with the International Cooperative Biodiversity Groups. This project was also supported by the Agricultural Food Research Initiative of the National Institute of Food and Agriculture, USDA, Grant #2008-35621-04732 and by the National Science Foundation under Grant No CHE-0619382 for purchase of the Bruker Avance 600 NMR spectrometer and Grant No. CHE-0722638 for the purchase of the Agilent 6220 mass spectrometer. These supports are gratefully acknowledged. We thank Mr. B. Bebout and Dr. Mehdi Ashraf-Khorassani for obtaining the mass spectra, and Dr. Hugo Azurmendi for assistance with the NMR spectra. We thank Dr. David Newman (National Cancer Institute) for the supply of the crude extract N110635.
Footnotes
Supporting Information Available: 1H, 13C, COSY, HMBC, HMQC, and NOESY spectra of Athrolides A-E (1-5). This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Biodiversity Conservation and Drug Discovery in Madagascar, Part 49. For Part 48, see Harinantenaina L, Brodie PJ, Callmander MW, Randrianaivo R, Rakotonandrasana S, Rasamison VE, Rakotobe E, Kingston DGI. Nat. Prod. Commun. 2011;6:1259–1262.
- 2.Zdero C, Bohlmann F, Mungai GM. Phytochemistry. 1991;30:3297–3303. [Google Scholar]
- 3.Goffin E, Ziemons E, De Mol P, de Madureira M. d. C., Martins AP, da Cunha AP, Philippe G, Tits M, Angenot L, Frederich M. Planta Med. 2002;68:543–545. doi: 10.1055/s-2002-32552. [DOI] [PubMed] [Google Scholar]
- 4.Haynes RK, Vonwiller SC. Acc. Chem. Res. 1997;30:73–79. [Google Scholar]
- 5.Nour AMM, Khalid SA, Kaiser M, Brun R, Abdallah WE, Schmidt TJ. Planta Med. 2009;75:1363–1368. doi: 10.1055/s-0029-1185676. [DOI] [PubMed] [Google Scholar]
- 6.Pillay P, Vleggaar R, Maharaj VJ, Smith PJ, Lategan CA. J. Ethnopharmacol. 2007;112:71–76. doi: 10.1016/j.jep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Pillay P, Vleggaar R, Maharaj VJ, Smith PJ, Lategan CA, Chouteau F, Chibale K. Phytochemistry. 2007;68:1200–1205. doi: 10.1016/j.phytochem.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Nour AMM, Khalid SA, Kaiser M, Brun R, Abdalla W. l. E., Schmidt TJ. J. Ethnopharmacol. 2010;129:127–130. doi: 10.1016/j.jep.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Andrade-Neto VF, Brandão MGL, Oliveira FQ, Casali VWD, Njaine B, Zalis MG, Oliveira LA, Krettli AU. Phytother. Res. 2004;18:634–639. doi: 10.1002/ptr.1510. [DOI] [PubMed] [Google Scholar]
- 10.Kaur K, Jain M, Kaur T, Jain R. Bioorg. Med. Chem. 2009;17:3229–3256. doi: 10.1016/j.bmc.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 11.Haynes RK, Fugmann B, Stetter J, Rieckmann K, Heilmann HD, Chan HW, Cheung MK, Lam WL, Wong HN, Croft SL, Vivas L, Rattray L, Stewart L, Peters W, Robinson BL, Edstein MD, Kotecka B, Kyle DE, Beckermann B, Gerisch M, Radtke M, Schmuck G, Steinke W, Wollborn U, Schmeer K, Römer A. Angew. Chem. Int. Ed. 2006;45:2082–2088. doi: 10.1002/anie.200503071. [DOI] [PubMed] [Google Scholar]
- 12.Mutabingwa TK. Acta Trop. 2005;95:305–315. doi: 10.1016/j.actatropica.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Bohlmann F, Zdero C, King RM, Robinson H. Phytochemistry. 1984;23:1979–1988. [Google Scholar]
- 14.Gao F, Wang H, Mabry TJ, Bierner MW. Phytochemistry. 1990;29:895–899. [Google Scholar]
- 15.Silva GL, Pacciaroni A. d. V., Oberti JC, Espinar LA, Diáz JG, Herz W. Phytochemistry. 1992;31:1621–1630. [Google Scholar]
- 16.Atta-ur-Rahman, Choudhary MI. Pure Appl. Chem. 1998;70:385–389. [Google Scholar]
- 17.Moffitt W, Woodward RB, Moscowitz A, Klyne W, Djerassi C. J. Am. Chem. Soc. 1961;83:4013–4018. [Google Scholar]
- 18.An S, Park Y-D, Paik Y-K, Jeong T-S, Lee WS. Bioorg. Med. Chem. Lett. 2007;17:1112–1116. doi: 10.1016/j.bmcl.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Gao F, Wang H, Mabry TJ, Jakupovic J. Phytochemistry. 1991;30:553–562. [Google Scholar]
- 20.Harvala E, Aligiannis N, Skaltsounis A-L, Pratsinis H, Lambrinidis G, Harvala C, Chinou I. J. Nat. Prod. 2002;65:1045–1048. doi: 10.1021/np010569t. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, Su Z, Xin X, Aisa HA. Helv. Chim. Acta. 2010;93:414–421. [Google Scholar]
- 22.Flamini G, Antognoli E, Morelli I. Phytochemistry. 2001;57:559–564. doi: 10.1016/s0031-9422(01)00066-8. [DOI] [PubMed] [Google Scholar]
- 23.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Antimicrob. Agents Chemother. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao S, Brodie PJ, Miller JS, Randrianaivo R, Ratovoson F, Birkinshaw C, Andriantsiferana R, Rasamison VE, Kingston DGI. J. Nat. Prod. 2007;70:679–681. doi: 10.1021/np060627g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Research. 1985;45:2110–2115. [PubMed] [Google Scholar]
- 26.CrysAlisPro 171.33.31 Oxford Diffraction. Wroclaw; Poland: 2009. [Google Scholar]
- 27.Sheldrick G. Acta Crystallogr., Sect. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H. J. Appl. Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889811041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flack H. Acta Crystallogr., Sect. A. 1983;39:876–881. [Google Scholar]
- 30.Hooft RWW, Straver LH, Spek AL. J. Appl. Crystallogr. 2008;41:96–103. doi: 10.1107/S0021889807059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spek AL. J. Appl. Crystallogr. 2003;36:7–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.