Abstract
MicroRNAs play important roles in gene regulation, and their expression is frequently dysregulated in cancer cells. In a previous study, we reported that miR-193b represses cell proliferation and regulates cyclin D1 in melanoma cells, suggesting that miR-193b could act as a tumor suppressor. Herein, we demonstrate that miR-193b also down-regulates myeloid cell leukemia sequence 1 (Mcl-1) in melanoma cells. MicroRNA microarray profiling revealed that miR-193b is expressed at a significantly lower level in malignant melanoma than in benign nevi. Consistent with this, Mcl-1 is detected at a higher level in malignant melanoma than in benign nevi. In a survey of melanoma samples, the level of Mcl-1 is inversely correlated with the level of miR-193b. Overexpression of miR-193b in melanoma cells represses Mcl-1 expression. Previous studies showed that Mcl-1 knockdown cells are hypersensitive to ABT-737, a small-molecule inhibitor of Bcl-2, Bcl-XL, and Bcl-w. Similarly, overexpression of miR-193b restores ABT-737 sensitivity to ABT-737–resistant cells. Furthermore, the effect of miR-193b on the expression of Mcl-1 seems to be mediated by direct interaction between miR-193b and seed and seedless pairing sequences in the 3′ untranslated region of Mcl-1 mRNA. Thus, this study provides evidence that miR-193b directly regulates Mcl-1 and that down-regulation of miR-193b in vivo could be an early event in melanoma progression.
MicroRNAs (miRNAs) are ∼22-nucleotide noncoding RNAs that play important regulatory roles in animals and plants.1 miRNAs repress protein-coding genes posttranscriptionally by pairing to and destabilizing target mRNAs or decreasing the efficiency of target mRNA translation.2 Most miRNAs bind to conserved sequences in the 3′ untranslated region (3′ UTR) of target genes that form Watson-Crick base pairs with the miRNA “seed” (2 to 7 nucleotides at the 5′ end of the miRNA)3 or other highly complementary sequences outside the miRNA seed region.4 Typically, miRNAs regulate the expression of hundreds of genes,5, 6 and, to date, 1424 miRNA coding genes have been mapped in the human genome (http://www.mirbase.org/cgi-bin/mirna_summary.pl?org=hsa, last accessed September 1, 2011). It is estimated that up to 60% of human genes are regulated by miRNAs.7
Dysregulation of miRNAs is involved in human diseases, including cancer, and distinct miRNA expression profiles (signatures) have been identified in human tumors and tumor cell lines.8 Depending on tissue, cellular context, and specific target gene, miRNAs can act as oncogenes or as tumor suppressors.8, 9 Recent studies suggest that miRNA dysregulation may play a role in the progression of melanoma.10 For example, miR-137 targets microphthalmia-associated transcription factor, which is the master regulator of melanocyte development, survival, and function.11 The miR-221/222 oncogenes promote proliferation and dedifferentiation of melanoma cells by repressing c-Kit and p27Kip.12 miR-182 enhances melanoma metastasis by targeting FOXO3 and microphthalmia-associated transcription factor.13 On the other hand, tumor suppressors, such as let-7b and miR-193b, which are often down-regulated in melanoma, target cyclin D and other important cell-cycle regulators.14, 15 Three miRNAs, miR-34b, miR-34c, and miR-199a*, repress MET expression and weaken MET-mediated cell motility and invasion.16
Recent studies support the idea that miR-193b is a tumor suppressor. For example, overexpression of miR-193b represses estrogen receptor-α and urokinase-type plasminogen activator in breast cancer cell lines, subsequently inhibiting cell proliferation and invasion.17, 18 Our group first demonstrated that overexpression of miR-193b represses cell proliferation and arrests cells in G1 by targeting cyclin D in melanoma cells.15 More recently, Xu et al19 demonstrated that miR-193b targets cyclin D in hepatocellular carcinoma cells and regulates proliferation, migration, and invasion.
We previously identified distinct miRNA expression profiles in metastatic melanoma and demonstrated that miR-193b was down-regulated in metastatic melanoma relative to benign nevi.15 Having identified miR-193b as a potentially important miRNA in melanoma, the primary goal of this study was to explore its potential role in the regulation of a critical target gene, myeloid cell leukemia sequence 1 (Mcl-1), in melanoma. The results demonstrate that overexpression of miR-193b represses Mcl-1 expression and sensitizes melanoma cells to ABT-737–induced apoptosis. Luciferase reporter assay suggests that miR-193b directly regulates Mcl-1 by binding to seed and seedless pairing sequences in the Mcl-1 3′ UTR.
Materials and Methods
Clinical Specimens
In addition to the eight benign nevi and eight metastatic melanoma samples described in a previous study,15 15 primary melanoma samples were retrieved from the Department of Pathology and Molecular Medicine, Kingston General Hospital. All the samples were formalin-fixed, paraffin-embedded and reviewed by a dermatopathologist (V.A.T). The validity of using formalin-fixed, paraffin-embedded samples for Agilent microarray analysis (Agilent Technologies Inc., Santa Clara, CA) was confirmed by previous studies.20, 21 The Faculty of Health Sciences Ethics Board at Queen's University approved the study.
miRNA Microarray
Total RNA was isolated from formalin-fixed, paraffin-embedded samples using the RecoverAll total RNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. The procedure for miRNA microarray has been described in detail elsewhere.15 Raw microarray data from the present study were submitted to the National Center for Biotechnology Information Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo; accession number GSE24996).
Immunohistochemical Analysis
Immunohistochemical (IHC) analysis was performed on TMA slides with Mcl-1–specific antibody (1:800; Cell Signaling, Beverly, MA) according to a standard protocol. Mcl-1 staining was scored independently by one dermatopathologist (V.A.T.) and one other observer (M.A.-D.) using the following intensity scale: 0 = negative, 1 = weak, 2 = moderate, and 3 = strong. In cases of discordant scoring, both observers reviewed the slide, and a consensus score was determined.
Cell Culture, Reagents, and Expression Vectors
Melanoma cell lines Malme-3M, MeWo, SK-MEL-2, and SK-MEL-28 were grown in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (HyClone) at 37°C and 5% CO2. miRNA precusors (miR-193b or negative control) were obtained from Ambion. For miRNA overexpression, cells were seeded in 100-mm dishes at 6 × 105 cells per plate the day before transfection. Melanoma cells were transfected with 5 nmol/L miRNA precursors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
ABT-737 was supplied by Abbott Laboratories (Abbott Park, IL). Cells were transfected with 5 nmol/L miRNA precursors (either miR-193b or negative control) as described previously herein and were grown for 56 hours. ABT-737 was added to cell cultures at a final concentration of 10 μmol/L; cells were harvested after incubation for 16 hours. The level of apoptosis in ABT-737–treated cells was estimated by quantifying cleaved poly (ADP-ribose) polymerase (PARP) using Western blot analysis.
A sequence-validated Mcl-1 expression plasmid (SC315538) was purchased from OriGene Technologies (Rockville, MD). Malme-3M cells were seeded in 6-well plates at 3 × 105 cells per well the day before transfection. Expression vectors of 1-μg Mcl-1 were cotransfected with 5 nmol/L miRNA precursors using Lipofectamine 2000. ABT-737 was added to cell cultures as described previously herein.
Northern Blot Analysis
Total RNA from transfected cell lines was isolated using the miRNeasy mini kit (Qiagen, Valencia, CA). The miR-193b expression level was examined using an miRNA Northern blot assay kit (Signosis, Sunnyvale, CA) according to the manufacturer's protocol. U6 small nuclear RNA was used as an internal control.
Western Blot Analysis
Aliquots of cell lysates were separated on 10% SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA), and analyzed using a standard Western blot protocol. Blots were probed with Mcl-1–specific and cleaved PARP–specific antibodies (Cell Signaling); gamma tubulin was from Sigma-Aldrich (Oakville, ON, Canada). Densitometry was performed using Quantity One software (Bio-Rad, Mississauga, ON, Canada).
Vector Construction and Luciferase Assay
PCR was performed using KOD hot start DNA polymerase (Novagen, Madison, WI). DNA fragments were PCR amplified from human genomic DNA and were cloned into the multiple cloning site (XhoI and NotI) distal to the Renilla luciferase coding region of the psiCHECK-2 vector (Promega, Madison, WI). The primer sequences used to construct the four vectors containing the wild-type Mcl-1 3′ UTR fragments were as follows: i) psiCHECK-2-Mcl-1_3′UTR, XhoI-Mcl-1-FW 5′-ACGCCTCGAGGCAGTTGGACTCCAAGCTGTAAC-3′, NotI-Mcl-1-RV 5′-ATAAGAATGCGGCCGCGGTCCTAACCCTTCCTGGCACAGC-3′; ii) psiCHECK-2-Mcl-1_UTR1, XhoI-UTR1-FW 5′-ACGCCTCGAGGCAGTTGGACTCCAAGCTGTAAC-3′, NotI-Mcl-1-UTR1-RV 5′-ATAAGAATGCGGCCGCAGTAAGAATCATGGAAACCAAGCC-3′; iii) psiCHECK-2-Mcl-1_Mcl-1 UTR2, XhoI-UTR2-FW 5′-ACGCCTCGAGGGCTTGGTTTCCATGATTCTTACT-3′, NotI-Mcl-1-UTR2-RV 5′-ATAAGAATGCGGCCGCGGAAACTTTAGAGAAAGCCTC-3′; and iv) psiCHECK-2-Mcl-1_Mcl-1 UTR3, XhoI-UTR3-FW 5′-ACGCCTCGAGGAGGCTTTCTCTAAAGTTTCC-3′, NotI-Mcl-1-UTR2-RV 5′-ATAAGAATGCGGCCGCGGTCCTAACCCTTCCTGGCACAGC-3′.
Overlapping PCR was used to introduce mutations at the seed matching site of psiCHECK-2-Mcl-1_3′UTR and psiCHECK-2-Mcl-1_UTR1,22 generating psiCHECK-2-Mcl-1_3′UTR M, psiCHECK-2-Mcl-1_UTR1 M1, and psiCHECK-2-Mcl-1_UTR1 M2. To generate deletions, two additional primers were used: Mcl-1-ΔmiR-193b-FW 5′-CCTTGTTGAGAACAGGAAAGTGCCAGGCAAGTCATAGAATTG-3′, Mcl-1-ΔmiR-193b-RV 5′-ACTTTCCTGTTCTCAACAAGG-3′. To generate double point mutations, two additional primers were used: Mcl-1-M2-FW 5′-GAGAACAGGAAAGTGGACAGAAGCCCAGGCAAGTCATAG-3′, Mcl-1-M2-RV 5′-CTATGACTTGCCTGGCTTCTGTCCACTTTCCTGTTCTC-3′ (mismatched nucleotides are shown in italics).
Malme-3M cells were seeded at 75,000 cells per well in a 12-well plate the day before transfection. The cells were cotransfected with 5 nmol/L miRNA precursor (either miR-193b or negative control) and 100 ng of psiCHECK-2 using Lipofectamine 2000 (Invitrogen). Renilla luciferase activity was measured 24 hours after transfection using the Dual-Glo luciferase assay system (Promega). Data were normalized to firefly luciferase activity.
miRNA Real-Time PCR
Total RNA from transfected cell lines was isolated using the miRNeasy mini kit (Qiagen) according to the manufacturer's protocol. miR-193b level was determined using a TaqMan miRNA assay (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. miRNA expression was assayed in triplicate, and data were normalized to endogenous RNU6B. The relative level was calculated using the ΔΔCT method.
Statistical Analysis
Data were analyzed using student's t-test and Pearson correlation in SPSS 17.0 (SPSS Inc., Chicago, IL). P ≤ 0.05 was considered statistically significant.
Results
miR-193b Expression Is Down-Regulated in Primary Melanoma and Is Inversely Correlated with Expression of Mcl-1 in Melanoma Tissue
miR-193b expression is significantly reduced in primary (159.85 ± 50.36, mean ± SEM) and metastatic (160.58 ± 36.09) melanomas relative to its expression in benign nevi (583.09 ± 50.71) (Figure 1A). miR-193b is expressed at a similar level between primary and metastatic melanoma, suggesting that down-regulation of miR-193b is an early event during melanoma progression.
Figure 1.
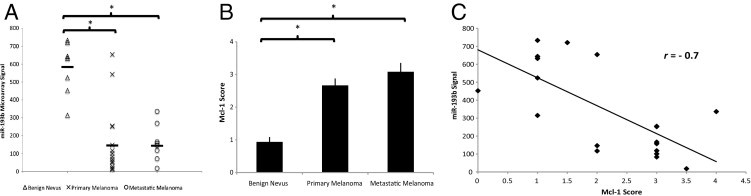
Expression of miR-193b and Mcl-1 in melanoma tissue samples. A: miR-193b expression was measured by Agilent miRNA microarray. The black line indicates the mean value for each group. B: Mcl-1 staining was scored in metastatic melanomas, primary melanomas, and benign nevi. Data shown are mean ± SEM. C: Pearson correlation analysis was performed to determine the relationship between miR-193b expression and Mcl-1 expression. The results show an inverse correlation (r = −0.7 and P < 0.001). *P < 0.0001 was calculated using the independent samples t-test.
Mcl-1, which is highly expressed in malignant melanoma,23, 24 is a predicted target of miR-193b (TargetScanHuman release 5.1, http://www.targetscan.org, last accessed September 1, 2011). Overexpression of miR-193b also correlated with a lower level of Mcl-1 mRNA in previous gene array studies.15 Thus, we speculated that dysregulation of miR-193b might play a role in malignant melanoma by altering the level of Mcl-1. Herein, we examined Mcl-1 expression in a subgroup of melanocytic tissues used for previous and current microarray studies, including eight benign nevi, six primary melanomas, and six metastatic melanomas. The results show that the mean ± SEM Mcl-1 staining score was significantly higher in metastatic (3.08 ± 0.27) and primary (2.67 ± 0.21) melanomas than in benign nevi (0.94 ± 0.15; Figure 1B). Furthermore, expression of miR-193b correlated inversely with expression of Mcl-1 in these 20 melanocytic samples with a Pearson correlation coefficient r = −0.7 and P < 0.001 (Figure 1C), despite a relatively small sample size.
miR-193b Represses Mcl-1 and Sensitizes Melanoma Cells to ABT-737
As mentioned earlier, a previous study showed that overexpression of miR-193b in Malme-3M cells correlated with a reduced Mcl-1 mRNA level.15 To confirm this result, we extended the study to four melanoma cell lines: Malme-3M, MeWo, SK-MEL-2, and SK-MEL-28 were transfected with miR-193b or negative control miRNA. Ectopic expression of miR-193b in melanoma cells was confirmed by Northern blot analysis (see Supplemental Figure S1 at http://ajp.amjpathol.org). Figure 2A shows that overexpression of miR-193b reduced the level of Mcl-1 30% to 60% in all four melanoma cell lines tested. Consistent with the low level of Mcl-1, cleaved PARP increased 4.55-fold in SK-MEL-2 cells and 1.26-fold in SK-MEL-28 cells. However, cleaved PARP did not increase in Malme-3M or MeWo cells expressing a high level of miR-193b (see Supplemental Figure S1 at http://ajp.amjpathol.org) and a low level of Mcl-1.
Figure 2.
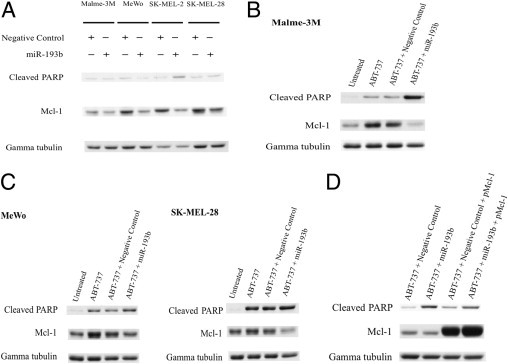
miR-193b down-regulates Mcl-1 and sensitizes melanoma cells to ABT-737. A: Western blot analysis of Mcl-1 and cleaved PARP expression in Malme-3M, MeWo, SK-MEL-2, and SK-MEL-28 cells transfected with miR-193b or negative control. Cells were transfected with 5 nmol/L miRNA precursor (miR-193b or negative control) and were harvested 72 hours after transfection. B: Western blot analysis of cleaved PARP and Mcl-1 in Malme-3M cells transfected with miR-193b or negative control, as indicated, incubated for 56 hours, and treated with 10 mmol/L ABT-737, as indicated. Cells were harvested 72 hours after transfection. C: Western blot analysis of MeWo and SK-MEL-28 cells, as indicated. The procedure was performed as described in B. D: One microgram of Mcl-1 plasmid (pMcl-1) was cotransfected with 5 nmol/L miRNA precursors, as indicated, and treated with 10 nmol/L ABT-737 as described in B. Gamma tubulin was used as the loading control. Representative data from one of three independent experiments are shown.
Previous studies established that cells that express a high level of Mcl-1 are resistant to ABT-737–induced apoptosis25 and that small-interfering RNA–mediated knockdown of Mcl-1 restores the sensitivity of ABT-737–resistant melanoma cells to ABT-737.24 Therefore, we speculated that miR-193b, which down-regulates Mcl-1, would also sensitize ABT-737–resistant melanoma cells, such as Malme-3M, to ABT-737. Consistent with this idea, exposure of Malme-3M cells to ABT-737 caused a small increase in cleaved PARP (Figure 2B), but in miR-193b–transfected Malme-3M cells, Mcl-1 expression was reduced and PARP cleavage was increased, indicating that sensitivity to ABT-737–induced apoptosis was restored (Figure 2B). Although, ABT-737 induced a significant amount of PARP cleavage and apoptosis in MeWo and SK-MEL-28 cells, overexpression of miR-193b enhanced the effect of ABT-737 in these cell lines (Figure 2C). Exposure to ABT-737 increased the level of Mcl-1 in all three cell lines (Figure 2, B and C). Furthermore, the addition of ABT-737 alone had no effects on miR-193b expression (see Supplemental Figure S2 at http://ajp.amjpathol.org).
To address the question of whether miR-193b functions through Mcl-1, a rescue experiment was undertaken using an Mcl-1–expressing plasmid. Ectopic expression of Mcl-1 reduced the extent of apoptosis in Malme-3M cells treated with both miR-193b and ABT-737, as PARP cleavage was notably decreased compared with control (Figure 2D). As a control, overexpression of Mcl-1 did not reduce apoptosis in cells treated with ABT-737 alone (Figure 2D). This is predicted because ABT-737 functions through inhibition of Bcl-2 and Bcl-xl.
miR-193b Interacts with Seeded and Seedless Sequences in the Mcl-1 3′ UTR
TargetScan analysis identified an evolutionarily conserved seed binding site for miR-193b in the 3′ UTR of Mcl-1 mRNA (Figure 3A). To test whether this predicted seed binding site is functional and to determine whether miR-193b directly regulates Mcl-1, wild-type and a seed binding site mutant form of the full-length Mcl-1 3′ UTR were cloned downstream of the Renilla luciferase gene in psiCHECK-2. As expected, miR-193b repressed ∼60% of the activity of the reporter construct containing the full-length wild-type Mcl-1 3′ UTR. However, the mutant Mcl-1 3′ UTR, carrying a deletion in the seed region binding site, still mediated a somewhat attenuated repressive effect of miR-193b, resulting in an ∼30% reduction in luciferase reporter gene activity. This result suggests that miR-193b has multiple binding sites in the Mcl-1 3′ UTR, one of which seems to be a seedless target.
Figure 3.
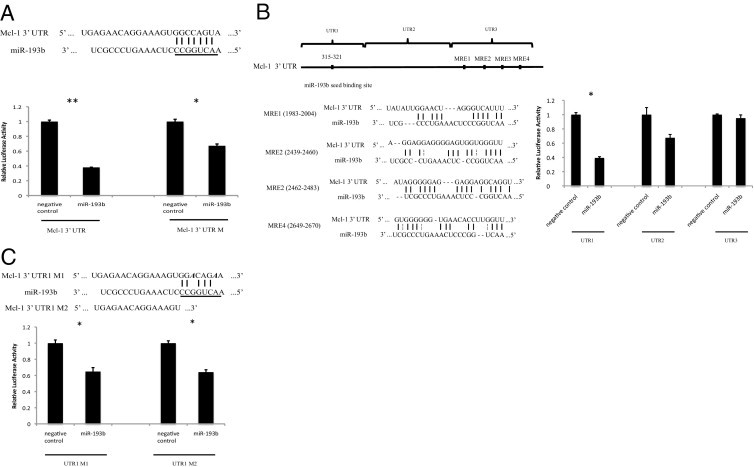
miR-193b directly targets the 3′ UTR of Mcl-1. A: miR-193b and its predicted seed binding site in the 3′ UTR of Mcl-1 (top). The underlined 8-nucleotide sequence indicates the miR-193b seed region. Relative luciferase activity is shown for reporter constructs containing 3′ UTR or 3′ UTR M (with deleted seed pairing region) (bottom) in cells transfected with miR-193b or negative control. B: As in A, except reporter constructs carried the indicated fragment of the Mcl-1 3′ UTR (top). Four MREs predicted by RNA22 in the 3′ UTR of Mcl-1 (middle). C: As in A, except reporter constructs carried UTR1 M1 or UTR1 M2 (top). UTR1 M1 contains two point mutations, as indicated in bold italic. UTR1 M2 contains no seed binding region. Renilla luciferase activity was measured 24 hours after transfection using the Dual-Glo luciferase assay system (Promega). Data were normalized to firefly luciferase. Data shown are the mean ± SEM of three replicates and are representative of three independent experiments. *P < 0 0.05 and **P < 0.001 were calculated using the paired samples t-test.
To validate the existence of a putative seedless miR-193b binding site in the Mcl-1 3′ UTR, the 3′ UTR sequence was reanalyzed using another target-predicting algorithm, RNA22, which allows seed mismatches and does not rely on cross-species conservation.4 RNA22 identified four putative miR-193b recognition elements (MREs) between coordinates 1983 and 2670 of the Mcl-1 3′ UTR (Figure 3B). To test the function of these putative MREs, reporter constructs were generated carrying the following three segments of the Mcl-1 3′ UTR: UTR1 (start to ∼600 bp, containing the seed binding site), UTR2 (∼600 to ∼1800 bp, between the seed binding site and the MREs), and UTR3 (∼1800 to ∼2700 bp, containing the putative MREs). Each segment was cloned into the luciferase reporter vector, and its susceptibility to miR-193b–mediated repression was tested. As expected, overexpression of miR-193b resulted in ∼60% repression of the construct containing UTR1 (Figure 3B). In contrast, no statistically significant repression was observed in constructs containing UTR2 or UTR3 (Figure 3B). Because miR-193b repressed ∼35% of the reporter activity of constructs carrying UTR1 with a seed binding site mutation or deletion, we conclude that the UTR1 region may contain both seeded and seedless binding sites for miR-193b (Figure 3C). However, none of the available target site prediction algorithms are capable of identifying target binding sites that lack stringent or highly compensatory seed pairing. Therefore, additional experiments are needed to confirm the existence and the exact location of the seedless binding site(s) in the Mcl-1 3′ UTR.
Discussion
This study examined the expression of miR-193b in 15 primary melanoma tissue samples using Agilent miRNA microarray. The data show that miR-193b is expressed at a significantly lower level in primary melanoma than in benign nevi, suggesting that down-regulation of miR-193b could occur early in melanoma progression (Figure 1). This is consistent with the observation that miRNA genes, including miR-193b, exhibit frequent (85.9%) copy number variations in primary cultured melanoma cell lines.26 Five of 45 melanoma samples (11%) were reported to have copy number reduction at the miR-193b locus. Epigenetic factors could also lead to dysregulated miRNA expression.27 For example, a recent study suggested that miR-193b is epigenetically silenced in prostate cancer cells.28
Several technical issues arise in studies of this nature. One is the choice of control for the comparison of expression levels. Nevus tissue was used as a benign comparator in these studies for several reasons. First, cultured melanocytes growing in artificial conditions proliferate at rates that far exceed in vivo rates. Second, obtaining single melanocytes for study from tissue sections would be practically impossible using current technology, although we would aspire to doing this when feasible. Third, evidence suggests that melanoma arises from a precursor lesion, such as a nevus. In particular, the high frequency of BRAF mutations in benign nevi suggests that these may represent precursor lesions that need to undergo additional mutations before becoming malignant. Overall, it was believed that benign nevi represented the best currently feasible control for this study.
In this study, as with our previous work,15 we profiled miRNA expression in formalin-fixed melanoma samples. Unlike mRNAs, miRNAs are well preserved in formalin-fixed, paraffin-embedded tissues, and formalin fixation does not seem to significantly alter the expression of miRNAs.20, 21, 29 The ability to use archival samples is particularly critical in studies of primary melanoma, where the samples are typically small and often come from community clinics, where tissue is typically formalin fixed. In addition, the small lesion size means that the entire primary lesion is usually required for accurate diagnosis, leaving no additional tissue for freezing. Therefore, the use of frozen material, although optimal, is likely not possible in the present clinical setting.
Mcl-1, an antiapoptotic protein belonging to the Bcl-2 family, plays a important role in regulating apoptosis.30, 31, 32 We observed that Mcl-1 was up-regulated in malignant melanoma, which agreed with previous studies.23, 24 Furthermore, this study showed that expression of miR-193b correlates inversely with expression of Mcl-1 in melanoma tissue samples (Figure 1) and that overexpression of miR-193b represses Mcl-1 levels in multiple melanoma cell lines.
ABT-737, a small-molecule inhibitor of the antiapoptotic proteins Bcl-2, Bcl-XL, and Bcl-w, has had limited success as a single agent in tumors with low Mcl-1 expression.33 However, overexpression of Mcl-1 confers resistance to ABT-737,25, 34 and knockdown of Mcl-1 restores ABT-737 sensitivity to ABT-737–resistant cells.24, 25 We, therefore, predicted and confirmed that overexpression of miR-193b could sensitize ABT-737–resistant melanoma cells to ABT-737 by repressing Mcl-1 (Figure 2). Furthermore, overexpression of Mcl-1 notably attenuated miR-193b–induced apoptosis in ABT-737–treated Malme-3M cells (Figure 2D). In addition, ABT-737 stimulates expression of Mcl-1 in Malme-3M, MeWo, and SK-MEL-28 cells (Figure 2C), as observed also in hepatoma cells.35 Although ABT-737 could potentially stabilize Mcl-1 protein, the exact mechanism by which ABT-737 interacts directly or indirectly with Mcl-1 is not yet known.
The data presented herein suggest that miR-193b represses Mcl-1 by binding to both seed and seedless matching sites in the Mcl-1 3′ UTR (Figure 3). Although most miRNA targets have sites that are perfectly complementary to the seed region, experiments have shown that miRNAs may directly interact with seedless (imperfect homology to the seed) binding sequences on mRNA 3′ UTRs.36 Lal et al37 showed that miR-24 regulates a series of genes via base pairing to highly complementary but seedless sequences. Although RNA22 identified four putative MREs for miR-193b in the Mcl-1 3′ UTR, luciferase reporter gene assays could not confirm that these MREs mediate miR-193b–dependent posttranscriptional repression. In contrast, deletion and point mutagenesis analysis suggested that the seedless binding site(s) for miR-193b maps to the first 600 nucleotides of the Mcl-1 3′ UTR. Thus, the present study indicates that the available algorithms are inadequate for accurately predicting functional seedless miRNA targets that lack highly complementary seed pairing.38 Note that an earlier study showed that miR-193b targets Mcl-1 in malignant hepatocyte cells via the matched seed sequence that was essential for posttranscriptional repression.39 Three scenarios could explain this discrepancy. First, the present study analyzes the interaction between miR-193b and the full length Mcl-1 3′ UTR, whereas Braconi et al39 examined a short fragment of the Mcl-1 3′ UTR that included the seed pairing site. Thus, the seedless miR-193b binding site in the Mcl-1 3′ UTR could have been missing from the fragment studied by the Braconi group. Second, the Braconi study was performed in hepatocytes, whereas the present study was performed in melanoma cells. Thus, the discrepancy could be explained by cell type–specific miRNA-mRNA interactions in melanoma cells and hepatocytes. Third, the analysis applied in Braconi's study would mask the effect that we observed because they did not compare the effects of miR-193 with negative control on the same construct, ie, containing either the wild-type or the mutant Mcl-1 3′ UTR.
Recent publications on the role of miR-193b have consistently suggested that it plays an important role, although the data have been somewhat contradictory. Whereas Caramuta and colleagues40 reported that high expression levels of miR-193b were associated with shorter survival after diagnosis, a more recent study showed that miR-193b was overexpressed in metastatic melanoma tissues from patients with longer survival times.41 Further studies with large patient cohorts, using primary melanoma tissues, are needed to clarify the association between miR-193b level and melanoma patient survival.
In summary, this study demonstrates that miR-193b is down-regulated in primary melanoma and suggests that Mcl-1 is directly regulated by miR-193b in melanoma cells. Reduced miR-193b expression could contribute directly to elevated expression of Mcl-1 in melanoma cells. The mechanism by which miR-193b is repressed in melanoma cells is not yet known. The present and previous data suggest that miR-193b represses cyclin D and Mcl-1, which, in turn, inhibits growth of melanoma cells and sensitizes them to ABT-737–induced apoptosis. Therefore, miR-193b has potential application as a therapeutic agent for melanoma.
Acknowledgment
We thank Evan Rees for assistance with Northern blot analysis.
Footnotes
Supported in part by a Canadian Institutes for Health Research grant (H.E.F. and V.A.T.). J.C. is funded in part by the Queen's University Terry Fox Foundation Training Program in Transdisciplinary Cancer Research in partnership with the Canadian Institutes for Health Research.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.07.010.
Supplementary Data
miR-193b levels in melanoma cell lines. Northern blot analysis of miR-193b expression in Malme-3M, MeWo, SK-MEL-2, and SK-MEL-28 cells transfected with either miR-193b or negative control or untreated. Cells were transfected with 5 nmol/L miRNA precursor (miR-193b or negative control) and were harvested 72 hours after transfection. U6 small nuclear RNA was used as an internal control.
The miR-193b expression level was quantified using the TaqMan miRNA assay. RNU6B was used as an internal control. Data are shown as the mean ± SEM of three independent experiments.
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 4.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 5.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 7.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A., Slack F.J. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Mueller D.W., Bosserhoff A.K. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551–556. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bemis L.T., Chen R., Amato C.M., Classen E.H., Robinson S.E., Coffey D.G., Erickson P.F., Shellman Y.G., Robinson W.A. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 12.Felicetti F., Errico M.C., Bottero L., Segnalini P., Stoppacciaro A., Biffoni M., Felli N., Mattia G., Petrini M., Colombo M.P., Peschle C., Care A. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 13.Segura M.F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., Polsky D., Wei J., Lee P., Belitskaya-Levy I., Bhardwaj N., Osman I., Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz J., Lorenz P., Gross G., Ibrahim S., Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 15.Chen J., Feilotter H.E., Pare G.C., Zhang X., Pemberton J.G., Garady C., Lai D., Yang X., Tron V.A. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Migliore C., Petrelli A., Ghiso E., Corso S., Capparuccia L., Eramo A., Comoglio P.M., Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 17.Leivonen S.K., Makela R., Ostling P., Kohonen P., Haapa-Paananen S., Kleivi K., Enerly E., Aakula A., Hellstrom K., Sahlberg N., Kristensen V.N., Borresen-Dale A.L., Saviranta P., Perala M., Kallioniemi O. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 18.Li X.F., Yan P.J., Shao Z.M. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 19.Xu C., Liu S., Fu H., Li S., Tie Y., Zhu J., Xing R., Jin Y., Sun Z., Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Chen J., Radcliffe T., Lebrun D.P., Tron V.A., Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glud M., Klausen M., Gniadecki R., Rossing M., Hastrup N., Nielsen F.C., Drzewiecki K.T. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129:1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 22.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Tang L., Tron V.A., Reed J.C., Mah K.J., Krajewska M., Li G., Zhou X., Ho V.C., Trotter M.J. Expression of apoptosis regulators in cutaneous malignant melanoma. Clin Cancer Res. 1998;4:1865–1871. [PubMed] [Google Scholar]
- 24.Keuling A.M., Felton K.E., Parker A.A., Akbari M., Andrew S.E., Tron V.A. RNA silencing of Mcl-1 enhances ABT-737-mediated apoptosis in melanoma: role for a caspase-8-dependent pathway. PLoS One. 2009;4:e6651. doi: 10.1371/journal.pone.0006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Delft M.F., Wei A.H., Mason K.D., Vandenberg C.J., Chen L., Czabotar P.E., Willis S.N., Scott C.L., Day C.L., Cory S., Adams J.M., Roberts A.W., Huang D.C. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Huang J., Yang N., Greshock J., Megraw M.S., Giannakakis A., Liang S., Naylor T.L., Barchetti A., Ward M.R., Yao G., Medina A., O'brien-Jenkins A., Katsaros D., Hatzigeorgiou A., Gimotty P.A., Weber B.L., Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lujambio A., Calin G.A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., Montuenga L.M., Rossi S., Nicoloso M.S., Faller W.J., Gallagher W.M., Eccles S.A., Croce C.M., Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauhala H.E., Jalava S.E., Isotalo J., Bracken H., Lehmusvaara S., Tammela T.L., Oja H., Visakorpi T. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127:1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 29.Xi Y., Nakajima G., Gavin E., Morris C.G., Kudo K., Hayashi K., Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozopas K.M., Yang T., Buchan H.L., Zhou P., Craig R.W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisvert-Adamo K., Longmate W., Abel E.V., Aplin A.E. Mcl-1 is required for melanoma cell resistance to anoikis. Mol Cancer Res. 2009;7:549–556. doi: 10.1158/1541-7786.MCR-08-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oltersdorf T., Elmore S.W., Shoemaker A.R., Armstrong R.C., Augeri D.J., Belli B.A., Bruncko M., Deckwerth T.L., Dinges J., Hajduk P.J., Joseph M.K., Kitada S., Korsmeyer S.J., Kunzer A.R., Letai A., Li C., Mitten M.J., Nettesheim D.G., Ng S., Nimmer P.M., O'Connor J.M., Oleksijew A., Petros A.M., Reed J.C., Shen W., Tahir S.K., Thompson C.B., Tomaselli K.J., Wang B., Wendt M.D., Zhang H., Fesik S.W., Rosenberg S.H. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 34.Chen S., Dai Y., Harada H., Dent P., Grant S. Mcl-1 down-regulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 2007;67:782–791. doi: 10.1158/0008-5472.CAN-06-3964. [DOI] [PubMed] [Google Scholar]
- 35.Hikita H., Takehara T., Shimizu S., Kodama T., Shigekawa M., Iwase K., Hosui A., Miyagi T., Tatsumi T., Ishida H., Li W., Kanto T., Hiramatsu N., Hayashi N. The Bcl-xL inhibitor, ABT-737, efficiently induces apoptosis and suppresses growth of hepatoma cells in combination with sorafenib. Hepatology. 2010;52:1310–1321. doi: 10.1002/hep.23836. [DOI] [PubMed] [Google Scholar]
- 36.Shin C., Nam J.W., Farh K.K., Chiang H.R., Shkumatava A., Bartel D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O., Becker K.G., Gorospe M., Hide W., Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2: MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braconi C., Valeri N., Gasparini P., Huang N., Taccioli C., Nuovo G., Suzuki T., Croce C.M., Patel T. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caramuta S., Egyhazi S., Rodolfo M., Witten D., Hansson J., Larsson C., Lui W.O. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 41.Segura M.F., Belitskaya-Levy I., Rose A.E., Zakrzewski J., Gaziel A., Hanniford D., Darvishian F., Berman R.S., Shapiro R.L., Pavlick A.C., Osman I., Hernando E. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR-193b levels in melanoma cell lines. Northern blot analysis of miR-193b expression in Malme-3M, MeWo, SK-MEL-2, and SK-MEL-28 cells transfected with either miR-193b or negative control or untreated. Cells were transfected with 5 nmol/L miRNA precursor (miR-193b or negative control) and were harvested 72 hours after transfection. U6 small nuclear RNA was used as an internal control.
The miR-193b expression level was quantified using the TaqMan miRNA assay. RNU6B was used as an internal control. Data are shown as the mean ± SEM of three independent experiments.


