Abstract
Fluorescent speckle microscopy (FSM) is a method for measuring the movements and dynamic assembly of macromolecular assemblies such as cytoskeletal filaments (e.g., microtubules and actin) or focal adhesions within large arrays in living cells or in preparations in vitro. The discovery of the method depended on recognizing the importance of unexpected fluorescence images of microtubules obtained by time-lapse recording of vertebrate epithelial cells in culture. In cells that were injected with fluorescent tubulin at ∼10% of the cytosol pool, microtubules typically appeared as smooth threads with a nearly constant fluorescence intensity. One day, when an unusually low concentration of fluorescent tubulin was injected into cells, the images from a sensitive cooled charge-coupled detector camera showed microtubules with an unusual “speckled” appearance—there were fluorescent dots with variable intensity and spacing along the microtubules. A first thought was that the speckles were an artifact. With further thought, we surmised that the speckles could be telling us something about stochastic association of tubulin dimers with the growing end of a microtubule. Numerous experiments confirmed the latter hypothesis. Subsequently the method we call FSM has proven to be very valuable. The speckles turned out not to be a meaningless artifact, but rather a serendipitous find.
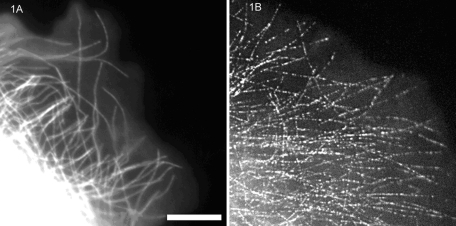
The discovery of the fluorescent speckle microscopy (FSM) technique depended on new advances in cameras with cooled charge-coupled device detectors (CCDs) and their application to fluorescence microscopy in cell biology in the early to mid-1990s. These cameras had significantly higher sensitivity (quantum efficiency), lower noise, and better spatial accuracy than the video cameras with image intensifiers that were commonly used at that time by cell biologists to obtain dynamic images of fluorescently labeled proteins in living cells. In 1996, Clare Waterman-Storer in the Salmon lab was examining how the polymerization and depolymerization of individual microtubules occurred near the leading edge of motile epithelial cells in culture (Waterman-Storer and Salmon, 1997). We were particularly interested in how the assembly of the actin filament cytoskeleton and its retrograde flow inward toward the cell center affected microtubule movement and assembly dynamics near the leading edge. To address this issue, Clare microinjected cells with purified tubulin dimers, the subunit protein of microtubules. The tubulin had been labeled with a red fluorescent fluorophore, X-rhodamine. She tried to inject enough X-rhodamine tubulin so that the labeled tubulin was ∼10% of the total cellular pool of tubulin subunits. After obtaining several time-lapse recordings with our cooled CCD camera of fluorescent microtubule assembly dynamics in her epithelial cell preparations, Clare left the microscope room and pulled Ted Salmon out of the lab to look at her time-lapse images. There was an unexpected feature that concerned her that had not been seen in previous publications in which images were recorded using an intensified video camera. In cells with high levels of injected fluorescent tubulin, the microtubules were brightly labeled, and fluorescence intensity appeared nearly constant along the lengths of microtubules (Figure 1A). However, in dim fluorescent cells containing low levels of injected tubulin, microtubules did not appear continuously labeled along their lengths but appeared as linear arrays of weakly fluorescent “speckles” that had the distribution expected for microtubules near the leading edge (Figure 1B). When Clare played back the time-lapse recording (Supplemental Video S1), it was apparent that the linear speckle arrays extended at their distal ends by adding new speckles with variable intensity and separation. The linear arrays abruptly shortened by loss of the speckles at the distal end—behavior expected for the dynamic instability of microtubule plus ends that face toward the leading edge of the cell.
FIGURE 1:
Comparison of diffraction-limited fluorescent images recorded with a cooled CCD camera and 1.4–numerical aperture objective of microtubules in the lamella of a migrating newt lung epithelial cell injected with X-rhodamine–labeled tubulin. (A) Ten percent labeled tubulin and (B) 0.25% labeled tubulin in the cytosol. Scale bar, 10 μm. (Reproduced with permission from Waterman-Storer CM, Salmon ED (1999). Fluorescent speckle microscopy of microtubules: how low can you go? FASEB J 13(Suppl 2), S225–S230.)
As we discussed these observations, several other scientists joined us, including our colleague Michael Caplow at the University of North Carolina, who is an internationally recognized expert on the biochemistry of microtubule assembly in vitro, and Tim Mitchison, the discoverer of microtubule dynamic instability, who happened to be visiting for a seminar. Mike's first impression of the data was that we had a problem with our fluorescently labeled tubulin. Rather than the normal dimers, he believed that our preparation might contain oligomers of fluorescent dimers and these oligomers were responsible for the speckles seen in weakly fluorescent cells. As an alternative, he suggested that these oligomers could be formed by microtubule-binding proteins in the cell or attachment to vesicles and should be considered as artifacts to be eliminated from our experiments by making better fluorescently labeled tubulin.
One of these possibilities initially seemed to be the likely answer, but we also thought of another, much more interesting possibility: that fluorescent speckles were the product of the stochastic association of a low concentration of labeled tubulin dimers with the growing end of a microtubule. This idea was stimulated by studies in the Kinosita lab in Japan, where they were polymerizing actin filaments that had single red fluorophores along the length of a filament separated by a distance resolvable in the light microscope (Sase et al., 1995). This was achieved by including a very low concentration (∼0.01%) of fluorescent actin subunits among the unlabeled subunit pool. We speculated that something similar might be happening to make the fluorescent speckles on microtubules when cells were microinjected with very low amounts of fluorescent tubulin. However, the fluorescent speckles probably contained multiple fluorophores because the concentration of fluorescent tubulin subunits was higher than 0.01%, so the pattern was more like a “fluorescent bar code” rather than widely spaced single fluorophores. We also doubted whether our imaging system was sensitive enough to see individual fluorophores above the background autofluorescence within cells. Indeed, Tim Mitchison noticed in time-lapse movies of the speckled microtubules (Supplemental Video S1) that the “bar code” pattern of speckles along the microtubule lattice stayed constant over time but changed only if the microtubule depolymerized and repolymerized, indicating that the pattern was formed during polymerization. All of these ideas for the source of the uneven fluorescent microtubules in bright cells and the fluorescent speckles in dim cells created much discussion.
Over the next few weeks we put together the following mechanism to explain both the low-contrast speckle distribution along microtubules in cells injected with 10% concentration of labeled tubulin (Figure 1A) and the high-contrast speckle distribution along microtubules in cells injected with low concentration of labeled tubulin (Figure 1B; e.g., 1%). What our eyes detect as speckle contrast along a microtubule is the SD between adjacent resolvable regions divided by the mean. For the resolution of a 1.4–numerical aperture objective, the resolvable region along a microtubule is 0.27 μm (for our camera, which did not limit optical resolution). Because there are ∼1024 tubulin dimers per micrometer length of a microtubule, 0.27 μm corresponds to N = 440 dimers. If the fraction of labeled tubulin in the subunit pool is f, then for stochastic (random) dimer incorporation into an end during growth, the mean number of labeled tubulin dimers within a resolvable region along a microtubule is M = fN, whereas the SD (fluctuation in intensity) is given by [fN(1 – f)]0.5 ∼ (M)0.5. For f = 0.1 as in Figure 1A, M = 44 fluorophores in a resolvable region, whereas SD = 6.5, which is small compared with the mean. This explained the low contrast of speckles along microtubules in the bright fluorescent cells injected with 10% labeled tubulin (Figure 1A). In contrast, for f = 0.01, as might have occurred in Figure 1B, M = 4.4 fluorophores and SD = 2.15, which is large compared with the mean. This explained the high contrast of speckles along microtubules in cells injected with 1% labeled tubulin or lower.
To prove this hypothesis correct, we had to do many tests (Waterman-Storer and Salmon, 1998, 1999; Waterman-Storer et al., 1998), which all were supportive (in particular, we had to satisfy the critical eye of Michael Caplow!). Our experimental results showed that fluorescent tubulin sediments as a 6S dimer in an analytical ultracentrifuge and thus was not forming oligomers. We showed that microtubules exhibit the expected fluorescent speckle patterns when assembled in vitro from pure tubulin and increasing concentrations of labeled dimer; speckle patterns are random and randomly change after microtubule shortening and regrowth; speckle contrast depends on the fraction of labeled tubulin dimer as predicted by the aforementioned equations; fluorescent tubulin speckle contrast does not depend on microtubule-associated proteins; the number of predicted fluorophores within a speckle at low fractions of labeled tubulin matches the number measured by steps in photobleaching; and, finally, the speckle intensity distributions along microtubules match predictions from computer simulations based on the foregoing model (Waterman-Storer and Salmon, 1999).
Our initial publications documenting the FSM technique showed how plus-end microtubule polymerization and depolymerization kinetics can be separated from motor-driven microtubule translocation velocity for individual microtubules in interphase cytoplasm, the treadmilling of individual severed microtubule fragments near the leading edges of motile cells, and the two-dimensional kinetics of poleward flux of spindle microtubules in tissue cells and cytoplasmic extracts (Waterman-Storer and Salmon, 1997, 1998, 1999; Waterman-Storer et al., 1998). Low concentrations of microinjected fluorescent actin subunits were also used to demonstrate the advantages of FSM for analyzing the two-dimensional pattern of actin polymerization at the leading edge of migrating cells and the retrograde flow of actin filaments toward the cell center (Waterman-Storer et al., 1998).
Since its discovery, FSM has been used by numerous investigators of microtubule and actin polymerization and depolymerization dynamics and polymer motility in living cells and reconstituted preparations (Danuser and Waterman-Storer, 2006; Cameron et al., 2011) and has been applied to study the dynamics of other macromolecular ensembles, such as integrin-based focal adhesions (Hu et al., 2007). Indeed, a recent Google search on FSM turned up over 68,000 hits. The technology has been greatly enhanced by the work of Gaudenz Danuser and coworkers, who have developed sophisticated computer programs for automatically detecting speckles and measuring their translocation and lifetimes to convert observational FSM into quantitative FSM (Danuser and Waterman-Storer, 2006). This has revealed a multitude of dynamic information about microtubule and actin cytoskeletal dynamics and function during cell motility and microtubule assembly dynamics and poleward flux in mitotic spindles (Cameron et al., 2011). More recently, with the introduction of better cooled CCD cameras, including those with electron multiplication, measurement of polymer dynamics with single fluorophore speckles has become easier (e.g., Watanabe and Mitchison, 2002; Yang et al., 2007). Thus, our careful consideration of an imaging “artifact” serves as a powerful example of how paying attention to the results of even apparently failed experiments can lead to fortuitous discoveries.
Acknowledgments
We thank Tim Mitchison, Gaudenz Danuser, and Michael Caplow for their editorial contributions.
Abbreviations used:
- CCD
charge-coupled device detector
- FSM
fluorescent speckle microscopy
Footnotes
REFERENCES
- Cameron LA, Houghtaling BR, Yang G. Fluorescent speckle microscopy. Cold Spring Harb Protoc. 2011;1 doi: 10.1101/pdb.top106. 2011(5) [DOI] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Sase I, Miyata H, Corrie JE, Craik JS, Kinosita K Jr. Real time imaging of single fluorophores on moving actin with an epifluorescence microscope. Biophys J. 1995;69:323–328. doi: 10.1016/S0006-3495(95)79937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. How microtubules get fluorescent speckles. Biophys J. 1998;75:2059–2069. doi: 10.1016/S0006-3495(98)77648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Fluorescent speckle microscopy of microtubules: how low can you go? FASEB J. 1999;13(2):S225–S230. doi: 10.1096/fasebj.13.9002.s225. [DOI] [PubMed] [Google Scholar]
- Yang G, Houghtaling BR, Gaetz J, Liu JZ, Danuser G, Kapoor TM. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]