In this paper, we report a new combination of actin inhibitors optimized to rapidly arrest actin dynamics while preserving the actin network. Using this drug combination, we demonstrate that actin dynamics, but not actin structure, are required for the spatial persistence of Rac activation in HL-60 cells.
Abstract
The actin cytoskeleton is regulated by factors that influence polymer assembly, disassembly, and network rearrangement. Drugs that inhibit these events have been used to test the role of actin dynamics in a wide range of cellular processes. Previous methods of arresting actin rearrangements take minutes to act and work well in some contexts, but can lead to significant actin reorganization in cells with rapid actin dynamics, such as neutrophils. In this paper, we report a pharmacological cocktail that not only arrests actin dynamics but also preserves the structure of the existing actin network in neutrophil-like HL-60 cells, human fibrosarcoma HT1080 cells, and mouse NIH 3T3 fibroblast cells. Our cocktail induces an arrest of actin dynamics that initiates within seconds and persists for longer than 10 min, during which time cells maintain their responsivity to external stimuli. With this cocktail, we demonstrate that actin dynamics, and not simply morphological polarity or actin accumulation at the leading edge, are required for the spatial persistence of Rac activation in HL-60 cells. Our drug combination preserves the structure of the existing cytoskeleton while blocking actin assembly, disassembly, and rearrangement, and should prove useful for investigating the role of actin dynamics in a wide range of cellular signaling contexts.
INTRODUCTION
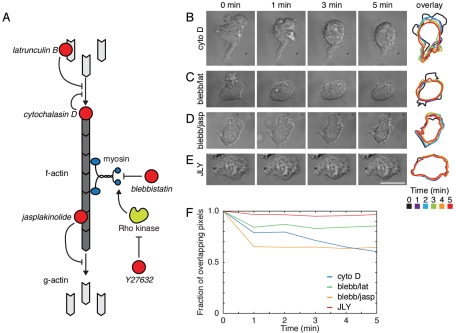
Actin dynamics are essential for many cellular processes, including the control of cell shape, movement, and division (Ridley et al., 2003). To examine the role of actin dynamics in many cellular processes, investigators commonly use pharmacological inhibitors that interfere with polymerization, depolymerization, and rearrangement of the actin cytoskeleton. These inhibitors can be sorted into several general classes. The first and most prevalent class of actin drugs includes those that interfere with actin polymerization. New actin polymerization can be inhibited by either sequestering the free actin monomer pool with drugs like latrunculin (Spector et al., 1983; Coué et al., 1987) or by blocking the fast-growing barbed ends of actin filaments with drugs like cytochalasin (Cooper, 1987; Figure 1A). A second class of actin drugs includes those that interfere with the disassembly of the actin network. For example, jasplakinolide binds actin filaments and interferes with disassembly (Cramer, 1999; Wilson et al., 2010; Figure 1A). Jasplakinolide also lowers the dissociation rate constant for barbed-end actin to essentially zero, resulting in the incorporation of virtually all free actin at high jasplakinolide doses or long posttreatment times (Bubb et al., 2000; Kiuchi et al., 2011). A third class of actin inhibitors includes those that interfere with the rearrangement of the actin network. For example, the molecular motor myosin II is involved in restructuring the actin network from a geometry that favors protrusion to one that favors contraction (Verkhovsky et al., 1999). Myosin II also participates in the disassembly of the actin network (Medeiros et al., 2006; Burnette et al., 2008). Some drugs, such as blebbistatin (Straight et al., 2003), directly inhibit myosin II activity, whereas other compounds, such as the Rho kinase (ROCK) inhibitor Y27632 (Uehata et al., 1997), inhibit myosin II activators (Figure 1A).
FIGURE 1:
The JLY cocktail, but not previously published combinations of actin inhibitors, rapidly arrests cell morphology in HL-60 cells. (A) Schematic of actin inhibitors. Cytochalasin D caps the barbed end of actin filaments, thereby preventing further polymerization. Jasplakinolide binds to the side of actin filaments and inhibits polymer disassembly. Latrunculin B binds to actin monomers and prevents their incorporation into actin polymer. Blebbistatin directly inhibits the ATPase activity of myosin II, and the ROCK inhibitor Y27632 inhibits the signaling cascade that drives phosphorylation and activation of myosin II. (B–E) HL-60 cells were plated on fibronectin-coated slides and imaged with DIC microscopy. At t = −1 min, cells had been preincubated in (B–E) 100 nM fMLP and (C and D) 50 μM blebbistatin or (E) 10 μM Y27632 for 10 min. At t = 0 min, (B) 10 μM cytochalasin D, (C) 500 nM latrunculin B, (D) 1 μM jasplakinolide, or (E) 8 μM jasplakinolide and 5 μM latrunculin B were added. Overlays represent successive cell outlines with black = 0 min, purple = 1 min, blue = 2 min, green = 3 min, orange = 4 min, red = 5 min. Scale bar: 10 μm. Images are representative of a minimum of five cells. (F) Cross-correlation of cell outline between t = 0 min and t = 5 min of cells in (B–E) indicates the ability of each drug or drug cocktail to stabilize cell morphology.
Studies using single actin inhibitors have been successful in identifying cell processes that are dependent on the actin cytoskeleton. For instance, the monomer-sequestering agent latrunculin has been used to show that actin polymerization and morphological rearrangements are not required for directional sensing in Dictyostelium discoideum and neutrophils (Parent et al., 1998; Servant et al., 2000). In contrast, filamentous actin is required for polarization of phosphatidylinositol 3,4,5-trisphosphate (PIP3) in response to uniform chemoattractant in neutrophils (Wang et al., 2002).
Although actin inhibitors have been a successful tool in identifying actin cytoskeletal-dependent processes, some of these single actin drugs not only block actin polymerization but may also significantly perturb the existing organization of the actin network, potentially complicating their interpretation. For example, latrunculin not only inhibits new assembly but also depolymerizes the existing actin cortex (Spector et al., 1983), which leads to a dramatic alteration in the mechanical properties of cells (Wakatsuki et al., 2001). Other single actin drugs have been shown to perturb the existing structure of the actin network (Cooper, 1987; Bubb et al., 1994; Uehata et al., 1997; Straight et al., 2003). With careful optimization, some single actin drugs can stabilize actin dynamics without altering the overall organization of the actin network in some contexts. For instance, jasplakinolide is able to inhibit chick fibroblast lamellipodium protrusion without significant disruptions to the actin cytoskeleton (Cramer, 1999), and low doses and short periods of treatment with jasplakinolide preserve actin organization in melanophores (Semenova et al., 2008). Additionally, blebbistatin slows actin dynamics in Aplysia bag cell neurons without grossly affecting actin organization (Burnette et al., 2008). We suspected (and later verified) that although previously published actin drugs and drug combinations can preserve actin organization in these slow-moving cells, they would fail to act fast enough to arrest actin dynamics in quickly moving cells, such as neutrophils. Chick fibroblasts and Aplysia bag cell neurons both move at ∼1 μm/min (Lin and Forscher, 1993; Cramer, 1999) compared with 10 μm/min for neutrophils (Hauert et al., 2002).
To investigate the role of actin dynamics in processes like cell polarity and directional sensing, we sought to identify a drug treatment that quickly arrests actin dynamics by stopping polymerization, depolymerization, and reorganization. We wanted the drug treatment to preserve the existing actin organization in a range of cell types, including highly motile cells. In this paper, we report a new combination of actin inhibitors that is optimized to quickly freeze cell morphology. We demonstrate that this cocktail, but none of the previously published combinations of inhibitors, acts within seconds to block assembly, disassembly, and rearrangement of the actin network. Using this drug combination, we demonstrate that actin dynamics, as opposed to morphological polarity or actin accumulation at the leading edge, are required for the spatial persistence of Rac activation in HL-60 cells.
RESULTS
Our drug cocktail, unlike previously published combinations, rapidly arrests morphological changes in HL-60 cells
We sought to identify a combination of actin drugs that inhibits actin dynamics while preserving the structure of the existing cytoskeleton. Using neutrophil-like, differentiated HL-60 cells (Hauert et al., 2002), which exhibit rapid actin dynamics (Weiner et al., 1999), we evaluated several drugs and drug cocktails previously reported to inhibit actin dynamics in other cell types. We reasoned that any cocktail that inhibits actin dynamics while preserving the organization of the existing network should rapidly and persistently preserve cell morphology, as assayed by differential interference contrast (DIC) microscopy.
We first analyzed the ability of cytochalasin D to preserve cell morphology in migrating HL-60 cells. Cytochalasin D not only acts to inhibit actin polymerization by capping barbed ends but also significantly slows disassembly of the actin network in structures such as Listeria actin tails (Kueh et al., 2008). In HL-60 cells, cytochalasin D treatment blocks leading-edge advancement but fails to inhibit significant morphological changes over a 5-min observation window. During this time period, both the leading edge and trailing edge retract, leading to a loss of morphological polarity (Figure 1B and Supplemental Movie S1).
Next we tested two different combinations of actin drugs reported to slow actin rearrangements in other cells. A combination of blebbistatin and latrunculin B has been reported to slow actin dynamics in primary murine dendritic cells (Renkawitz et al., 2009). Differentiated HL-60 cells treated with this drug combination exhibit rapid retraction of the leading edge (Figure 1C and Movie S2). A combination of blebbistatin and jasplakinolide has also been reported to slow actin dynamics in keratocytes (Wilson et al., 2010). However, this drug cocktail fails to rapidly arrest the advancement of the leading edge in HL-60 cells (Figure 1D and Movie S3).
We reasoned that the limitations of previously published cocktails stem from their inability to simultaneously inhibit actin depolymerization, polymerization, and restructuring. To test this hypothesis, we treated cells with a triple-drug cocktail containing jasplakinolide, latrunculin B, and Y27632, which are inhibitors of actin depolymerization, actin polymerization, and myosin II-based contractility, respectively. Cells treated with this triple-drug cocktail (which we refer to as JLY) exhibit a rapid freezing of cell morphology that initiates within seconds and persists for at least 10 min (Figure 1E and Movie S4). Quantitative analysis of changes in cell outline confirmed that JLY maintains HL-60 cell morphology better than previously published methods (Figure 1F). Omission of any member of our three-drug JLY combination impaired the ability of this cocktail to rapidly preserve cell morphology in HL-60 cells (Supplemental Figure S1). With the exception of latrunculin, lower doses of each drug, alternative myosin inhibitors (such as blebbistatin and ML-7), and alternative drug combinations did not preserve cell morphology as efficiently as our optimized concentrations of JLY (Figures S1 and S2). JLY with a concentration of 2.5 μM latrunculin worked in a manner similar to optimized JLY.
JLY preserves steady-state actin organization
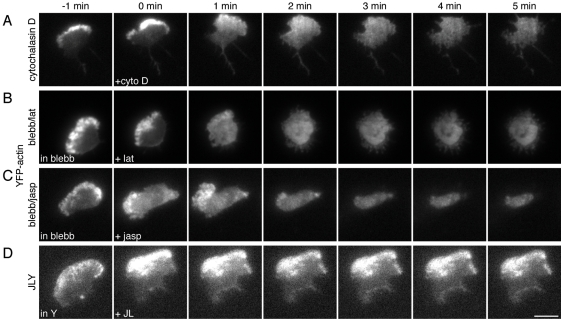
After determining that our triple-drug cocktail JLY effectively preserved cell morphology, we next tested the ability of JLY and other drug combinations to preserve the steady-state actin distribution in HL-60 cells. To visualize filamentous actin in living cells, we used an HL-60 cell line that stably expresses yellow fluorescent protein (YFP)-actin (Van Keymeulen et al., 2006). We selected this probe over other actin-binding proteins, such as Lifeact (Riedl et al., 2008), because, unlike actin-binding proteins, YFP-actin incorporates into the actin network (Westphal et al., 1997) and is compatible with assays of long-term dynamics, such as photobleaching experiments (Lai et al., 2008). We analyzed cells in oblique fluorescence illumination to detect changes in filamentous actin concentration and organization and to reduce background from out-of-focus fluorescence. As expected, the drug combinations that failed to quickly arrest morphological changes, cytochalasin D, blebbistatin–latrunculin, blebbistatin–jasplakinolide (Figure 1, B–D), were unable to maintain the steady-state distribution of filamentous actin (Figure 2, A–C, and Movies S5–S7). In contrast, the JLY drug cocktail, which effectively preserves cell morphology (Figure 1E and Table S1), also maintains steady-state distribution of filamentous actin (Figure 2D and Movie S8).
FIGURE 2:
JLY preserves steady-state actin organization in HL-60 cells. HL-60 cells stably expressing YFP-tagged actin were plated on fibronectin-coated slides and imaged in DIC and oblique fluorescence illumination. Cells were pretreated with 100 nM fMLP and (B and C) 50 μM blebbistatin or (D) 10 μM Y27632 for 10 min before imaging. At t = 0 min, (A) 10 μM cytochalasin D, (B) 500 nM latrunculin B, (C) 1 μM jasplakinolide, or (B) 8 μM jasplakinolide and 5 μM latrunculin B were added. Images are representative of a minimum of five cells. Scale bar: 10 μm.
JLY arrests actin dynamics in multiple cell types
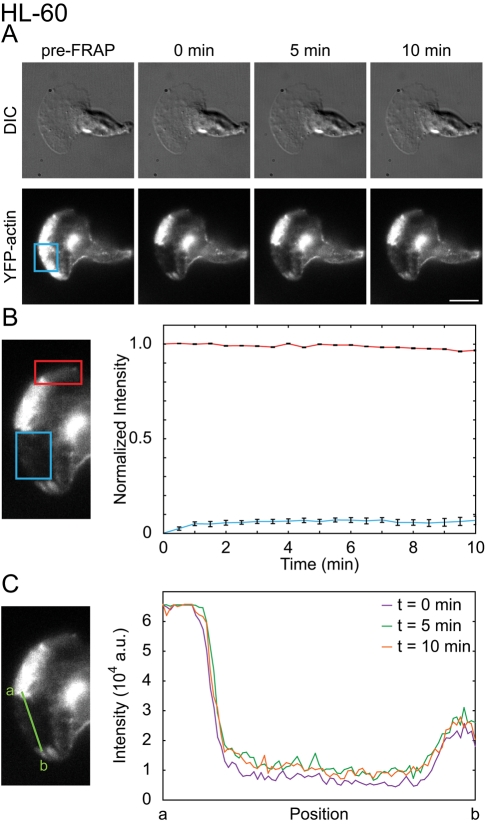
We have demonstrated that the JLY triple-drug cocktail preserves steady-state actin polymer distribution in HL-60 cells. This could either reflect an inhibition of actin polymerization and depolymerization, or could represent significant but balanced rates of actin polymerization and depolymerization with no net change in filament abundance. To discriminate between these possibilities, we assayed fluorescence recovery after photobleaching (FRAP) of YFP-actin. FRAP enables measurement of the rate of actin assembly by quantifying YFP-actin recovery in the bleached region. Our photobleaching setup takes 1–3 s to switch from photobleaching to fluorescence imaging. Because G-actin diffusion is rapid, this time is sufficient for the G-actin pool to equilibrate. As a result, we only assay fluorescence recovery of filamentous actin. JLY-treated cells exhibited only a 7% fluorescence recovery (n = 25) in the 10 min following photobleaching (Figure 3, A and B, and Movie S9), suggesting a lack of significant new actin polymerization. The intensity of an unbleached region of the cell was used to estimate the rate of actin depolymerization after drug treatment. Cells showed only a 3% decrease in actin polymer signal over 10 min (n = 25; Figure 3B), indicating minimal actin depolymerization. Furthermore, we observed persistent boundaries of the bleached region in the 10 min following photobleaching. The sharpness of this boundary suggests that there are minimal actin rearrangements following drug treatment (Figure 3C). Alternative combinations, concentrations, and timing of actin inhibitors were not as effective at arresting actin dynamics (Figures S1, A–I, and S3). We conclude that our optimized combination of JLY effectively arrests actin polymerization, depolymerization, and actin reorganization in HL-60 cells.
FIGURE 3:
JLY arrests actin depolymerization, polymerization, and myosin-based rearrangements. We used FRAP to analyze the ability of JLY to arrest actin depolymerization, polymerization, and rearrangements in YFP-actin expressing HL-60 cells. (A) Cells imaged in oblique fluorescence illumination were first pretreated with JLY, then photobleached in a rectangular region. Scale bar: 10 μm. (B) FRAP is a measure of actin assembly, and the fluorescence intensity of the nonbleached region estimates depolymerization. HL-60 cells display 7% recovery (blue box) in the 10 min after photobleaching (n = 25). HL-60 cells show 3% loss of filamentous actin signal over 10 min (n = 25), as evaluated by the loss of fluorescence intensity of the unbleached region of filamentous actin (red box). (C) The persistence of the bleached area can assay large-scale actin rearrangements. A line profile (green line) of the borders between unbleached and bleached regions indicates no detectable actin rearrangements in the 10 min after photobleaching for JLY-treated HL-60 cells. Error bars show SEM
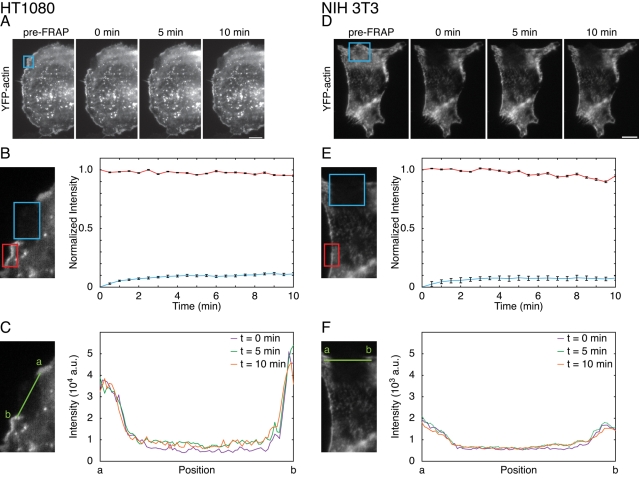
We next evaluated the ability of JLY to block actin dynamics and morphological rearrangements in other mammalian cells, such as HT1080 human fibrosarcoma cells (Rasheed et al., 1974). HT1080 cells were chosen because, like HL-60 cells, they are a model of mammalian cell chemotaxis (Mensing and Czarnetzki, 1984; VanCompernolle et al., 2003). Control HT1080 cells recovered to 60% of their original intensity in approximately 1 min after photobleaching (n = 5; Figure S4A), indicating rapid actin assembly. In contrast, the JLY-treated HT1080 cells exhibited only an 11% fluorescence recovery (n = 20) in the 10 min after photobleaching (Figure 4, A and B, and Movie S10), suggesting an inhibition of actin polymerization. HT1080 cells exhibited only a 5% decrease (n = 20) in fluorescence signal of the non-bleached area of the actin network over 10 min (Figure 4B), suggesting minimal depolymerization. Finally, JLY also blocked actin rearrangements in HT1080 cells, as assessed by the constant boundaries of the bleached region following drug treatment (Figure 4C).
FIGURE 4:
JLY arrests actin dynamics in other mammalian cells. We used FRAP to analyze the ability of JLY to arrest actin depolymerization, polymerization, and rearrangements in YFP-actin expressing (A–C) HT1080 and (D–F) NIH 3T3 cells. Cells imaged in oblique fluorescence illumination were first pretreated with JLY, then photobleached in a rectangular region. (B and E) FRAP is a measure of actin assembly, and the fluorescence intensity of the nonbleached region estimates depolymerization. (C and F) The persistence of the bleached area can assay large-scale actin rearrangements. (B) In HT1080 cells, 11% new actin polymerization (blue box) and 5% depolymerization (red box) occurred in the 10 min following JLY treatment (n = 20). (C) The line profile (green line) between unbleached and bleached regions indicates no detectable actin rearrangements in the 10 min after photobleaching for JLY-treated HT1080 cells. (E) NIH 3T3 cells display 8% recovery (blue box) in the 10 min after photobleaching (n = 14). NIH 3T3 cells show 5% loss of filamentous actin signal over 10 min (n = 14), as evaluated by the loss of fluorescence intensity of the unbleached region of filamentous actin (red box). (F) A line profile (green line) of the borders between unbleached and bleached regions indicates no detectable actin rearrangements in the 10 min after photobleaching for JLY-treated NIH 3T3 cells. Error bars show SEM. Scale bar: 10 μm.
In addition, we tested JLY in NIH 3T3 cells, a widely used cell line for studying mesenchymal cell migration (Ridley et al., 2003; Hakkinen et al., 2011). Control NIH 3T3 cells recovered to 22% of their original intensity in approximately 1 min after photobleaching (n = 13; Figure S4B), indicating rapid actin assembly. In contrast, JLY-treated NIH 3T3 cells showed a 5% fluorescence recovery (n = 14) in the 10 min after photobleaching (Figure 4, D and E, and Movie S11). NIH 3T3 cells showed an 8% decrease (n = 14) in fluorescence signal in the unbleached area of the actin network over 10 min post–drug treatment (Figure 4E). Additionally, JLY also blocked actin rearrangements in NIH 3T3 cells, as indicated by the consistent boundaries of the bleached region following drug treatment (Figure 4F). Together, these data suggest that JLY is effective at inhibiting actin polymerization, depolymerization, and rearrangements in HT1080, NIH 3T3, and HL-60 cells and is likely to be broadly applicable to other mammalian cell lines.
JLY does not inhibit the ability of cells to respond to external stimuli
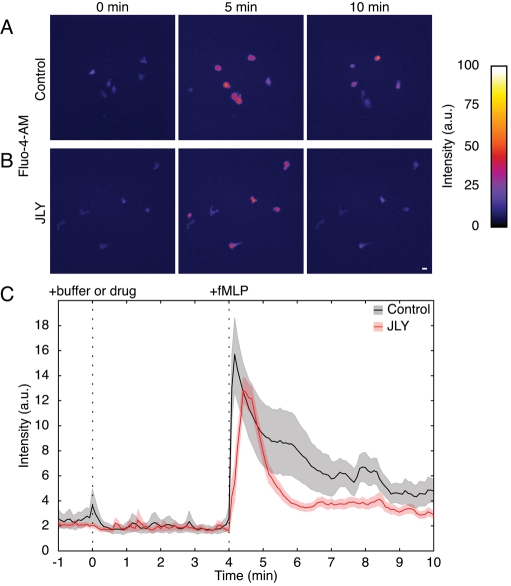
To test whether cells treated with JLY retain their ability to respond to external stimuli, we assayed calcium release of drug-treated HL-60 cells following chemotactic stimulation. Calcium release was evaluated by loading HL-60 cells with the membrane-permeable calcium indicator Fluo-4-AM. Both control cells (n = 9) and JLY-treated cells (n = 20) exhibited chemoattractant-dependent calcium release (Figure 5, A–C, and Movie S12), with a similar proportion of cells responding in both conditions. Calcium fluxes were noted in 100% of control and 95% of JLY-treated cells following chemoattractant stimulation with N-formyl-methionine-leucine-phenylalanine (fMLP). JLY treatment did not significantly alter either the basal calcium levels or the magnitude or kinetics of calcium release (Figure 5C). These data indicate that arresting actin dynamics with JLY does not prevent cell response to external stimuli.
FIGURE 5:
Cells retain their responsivity to external signals following JLY treatment. JLY-treated HL-60 cells retain their ability to activate intracellular calcium release in response to chemoattractant. Both untreated cells (A, n = 9) and JLY-treated cells (B, n = 20), release calcium in response to stimulation with 100 nM fMLP, as assayed with the calcium indicator dye Fluo-4-AM. The proportion of cells that release calcium in response to chemoattractant is similar for control and JLY-treated cells (100 and 95%, respectively). Calibration bar indicates intensity (a.u.); scale bar: 20 μm. (C) A representative time course of calcium release following stimulation with 100 nM fMLP at t = 4 min in control and JLY-treated cells. Quantitative analysis was performed using a region of interest (ROI) that was chosen to be large enough to contain the cell in all time points. The average intensity was background-subtracted, and this corrected intensity value (average ROI intensity – background intensity) was plotted over time. Control cell time course is shown in black and JLY-treated cell time course is shown in red. Light gray and red shading indicate SEM for control and JLY-treated cells, respectively.
To interpret results of experiments using JLY to study cell signaling, it is important to know whether JLY competes with signaling proteins for binding to the actin network. Although jasplakinolide can interfere with some actin-binding molecules, such as phalloidin (Bubb et al., 1994), the doses of jasplakinolide used in JLY did not interfere with the actin-binding proteins Lifeact (Riedl et al., 2008) and Coronin3 (Spoerl et al., 2002) or the Arp2/3 complex (Mullins et al., 1998; Figure S5).
Actin dynamics are essential for the spatial persistence of Rac activity
We next examined the relationship between actin dynamics and the polarization of Rac activity, a key chemotactic signal in HL-60 cells. Actin inhibitors have previously been used to investigate how various chemotactic signals depend on actin dynamics in HL-60 cells. Some polarized responses are still observed in the absence of the actin cytoskeleton. For example, latrunculin B–treated HL-60 cells are able to polarize PIP3 in response to external gradients of chemoattractant (Servant et al., 2000). In contrast, latrunculin B–treated HL-60 cells lose their ability to polarize PIP3 in response to uniform chemoattractant stimulation (Wang et al., 2002). Moreover, latrunculin B–treated HL-60 cells are defective in their ability to recycle the actin nucleation–promoting complex SCAR/WAVE (Suppressor of cyclical AMP Receptor/Wiskott–Aldrich syndrome protein verprolin-homologous protein) from the membrane (Weiner et al., 2007; Millius et al., 2009).
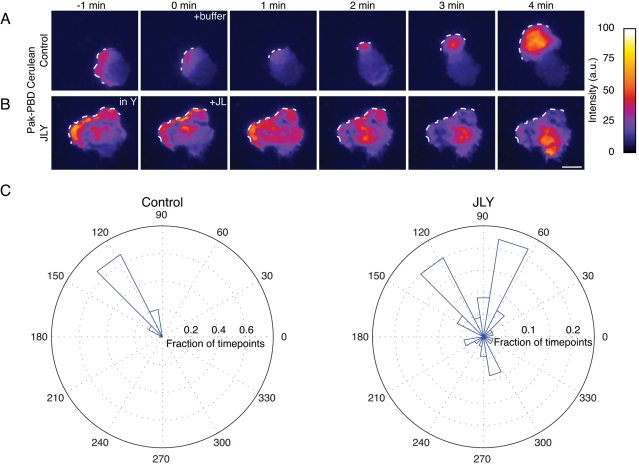
The role of the actin cytoskeleton in the spatial dynamics of Rac activation has not previously been investigated in HL-60 cells. Are actin rearrangements required to maintain polarity of Rac activity in uniform chemoattractant (actin plays this role for PIP3) or to terminate Rac activation (actin plays this role for WAVE complex recycling), or do actin rearrangements serve another role? Using population assays, we previously showed that latrunculin treatment does not alter the magnitude of chemoattractant-induced Rac activation in HL-60 cells (Weiner et al., 2006). However, whether actin plays a role in the spatial organization of Rac activity in these cells is unknown. We used total internal reflection fluorescence (TIRF) imaging of a fluorescently tagged Rac-GTP–binding domain, Pak-PBD-Cerulean (Weiner et al., 2007), to follow the dynamics of Rac activation in living HL-60 cells. In control HL-60 cells stimulated with uniform chemoattractant, Pak-p21-binding domain (Pak-PBD) persistently accumulates at the leading edge (Figure 6, A and C, and Movie S13). However, after actin dynamics are arrested with JLY, Rac activity loses its restriction to the morphological leading edge (Figure 6, B and C, and Movie S14). The loss of actin dynamics does not extinguish Rac activation, but rather causes the Pak-PBD signal to wander throughout the cell. We conclude that actin dynamics are essential for the spatial persistence of Rac activity at the leading edge during random migration.
FIGURE 6:
Actin rearrangements are required for the spatial persistence of Rac activity in randomly migrating HL-60 cells. Rac activity was assayed by TIRF imaging of a fluorescent Rac-GTP–binding domain (Pak-PBD-Cerulean). (A) Control cells are polarized and maintain a persistent field of Rac activity at the leading edge. (B) In JLY-treated cells, the Rac activity field loses its persistent localization to the leading edge. White dotted line indicates the morphological leading edge. Images are representative of a minimum of five cells. Calibration bar indicates intensity (a.u.); scale bar: 10 μm. (C) Quantification of the spatial persistence of the Rac activity in control and JLY-treated HL-60 cells. Spatial persistence is measured by analyzing the vector between the center of mass of the cell and the center of mass of the Rac activity field over 11 min, then plotting the angle of those vectors over the fraction of time points in a rose plot.
DISCUSSION
Pharmacological inhibitors have been broadly used for studying the role of the actin cytoskeleton in a wide array of signaling contexts. One of the most commonly used actin drugs is latrunculin, which blocks new actin assembly by sequestering the monomeric pool of actin (Spector et al., 1983; Coué et al., 1987). Inhibiting new polymerization results in the rapid disassembly of the existing cytoskeleton, because cells rely on new actin polymerization to balance the rapid depolymerization of the existing actin network. Latrunculin-treated cells have a much smaller pool of polymerized actin than control cells (Spector et al., 1983; Ayscough et al., 1997), leading to significantly altered mechanical properties of the cell (Wakatsuki et al., 2001).
To overcome these drawbacks and to more specifically test the role of actin dynamics in cellular function, we have developed a pharmacological cocktail that halts actin polymerization and depolymerization, while also maintaining the organization of the existing actin network. In this study, we systematically analyze the ability of several combinations of actin drugs to preserve cell morphology and assay the dynamic organization of the actin cytoskeleton. We find that previously published drug combinations are unable to preserve cell morphology and arrest actin dynamics in rapidly migrating HL-60 cells (Figures 1 and 2). We identify a drug combination that simultaneously inhibits actin disassembly, actin polymerization, and myosin II-based rearrangements through the concerted activities of jasplakinolide, latrunculin B, and Y27632, respectively (Figure 3). Our combination of drugs acts within seconds to preserve morphological changes and arrest actin dynamics in HL-60, HT1080, and NIH 3T3 cells (Figures 1, 3, and 4). This cocktail will enable researchers to analyze how cell function depends on actin dynamics without the complication of perturbing the existing organization of the actin network.
Actin dynamics are intimately involved in directed cell migration in amoeboid cells, such as neutrophils and Dictyostelium. The cellular signals that drive morphological changes are normally tightly coupled to actin rearrangements in space and time. Several groups have used actin inhibitors to investigate how cell polarity relates to the actin cytoskeleton and morphological changes. Directional sensing is not dependent on morphological changes: latrunculin-treated Dictyostelium and neutrophils maintain their ability to polarize signals such as PIP3 in response to external gradients of chemoattractant (Parent et al., 1998; Servant et al., 2000). However, several chemotactic signaling events depend on the actin cytoskeleton. Latrunculin-treated neutrophils lose their ability to generate PIP3 asymmetry in uniform chemoattractant (Wang et al., 2002). Furthermore, actin polymerization is essential for the recycling of nucleation-promoting factors such as the SCAR/WAVE2 complex in neutrophils (Weiner et al., 2007; Millius et al., 2009). In this study, we use JLY to show that actin dynamics are not essential for global Rac activation but are required for the spatial persistence of Rac activity at the morphological leading edge during random migration (Figure 6).
The JLY cocktail is a useful tool for revisiting previous studies in the chemotactic signaling field and for refining our understanding of the complex feedback loops that underlie cell polarity. Cells that lack actin polymer can still polarize signals such as PIP3 in response to external gradients of chemoattractant (Parent et al., 1998; Servant et al., 2000). However, these cells show a number of qualitatively different behaviors compared with control cells. If latrunculin-treated Dictyostelium are exposed to a point source of chemoattractant and are challenged with a second input, they rapidly reorient polarity, if the second source is of significantly higher concentration (Devreotes and Janetopoulos, 2003), or establish two crescents of PIP3 accumulation, if the second source is equal to the first input (Janetopoulos et al., 2004). In contrast, untreated Dictyostelium challenged with two point sources of chemoattractant maintain a single axis of PIP3 accumulation and often preserve their existing leading edge (Devreotes and Janetopoulos, 2003). These experiments suggest uniform sensitivity for unpolarized latrunculin-treated cells, whose internal signaling state mirrors the external chemoattractant gradient. In contrast, cells with a functional actin cytoskeleton exhibit localized sensitivity at the leading edge, possibly due to actin-based scaffolding of polarity components at the cell front (Devreotes and Janetopoulos, 2003). If the primary role of actin is to be a passive scaffold for signaling components, then JLY-treated cells (which maintain the existing organization of the actin cytoskeleton but lose actin dynamics) should phenocopy control cells in the dynamics of polarity signals. However, JLY-treated cells are unable to maintain the spatial persistence of Rac activity at the morphological leading edge (Figure 6B), unlike control cells (Figure 6A), which are able to do so. These data suggest that actin dynamics, and not only morphological polarity or actin accumulation at the leading edge, are required for the proper spatial dynamics of Rac activation in HL-60 cells. Our combination of actin inhibitors should prove useful in future experiments aimed at discriminating the role of actin dynamics and global morphological rearrangements for other signals in cell polarity and chemotaxis.
MATERIALS AND METHODS
Cell culture
HL-60 cells were cultured as previously described (Weiner et al., 2007). Briefly, cells were cultured in RPMI 1640 (10-041-CM, Cellgro; Mediatech, Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) and antibiotic/antimycotic (Invitrogen). HL-60 cells were differentiated by adding 1.5% dimethyl sulfoxide (DMSO; D2650; Sigma-Aldrich, St. Louis, MO). HT1080 cells were grown in DMEM (11995, Invitrogen) with 10% heat-inactivated FBS (Invitrogen) and antibiotic/antimycotic (Invitrogen). NIH 3T3 cells were grown in DMEM (11995; Invitrogen) with 10% bovine calf serum (BCS; UCSF Cell Culture Facility, San Francisco, CA).
Prior to imaging, slides were coated with fibronectin at 0.1 mg/ml for 45 min. Then, slides were washed three times with Dulbecco's phosphate-buffered saline. Next slides were blocked with media, and HL-60 cells were plated for 15 min at 37°C in complete growth media. HT1080 and NIH 3T3 cells were plated for 1 h at 37°C in complete growth media. Prior to imaging, slides were washed once or twice with media to remove floating cells and debris. NIH 3T3 cells were imaged in modified Hank's buffered saline solution (mHBSS) with 10% BCS.
Imaging
Images were acquired on a Nikon Ti-E inverted microscope with a 40× PlanApo 0.95 numerical aperture (NA), 60× Apo TIRF 1.49 NA, or 100× Apo TIRF 1.49 NA objective and an electron-multiplying charge-coupled device (EM-CCD) camera (Evolve; Photometrics, Tucson, AZ) controlled by NIS-Elements (Nikon, Melville, NY). Sample drift was minimized using an autofocus system (Perfect Focus; Nikon). Laser lines (440, 488, 514 nm; all 200 mW) were supplied from a Spectral Applied Research LMM5 Laser Merge Module (Richmond Hill, Ontario, Canada). This laser launch uses acousto-optic tunable filters (AOTFs) to control laser output to a TIRF fiber for imaging or a Mosaic Digital Micromirror device (Andor, Belfast, UK) for spatially controlled photobleaching. Bleaching experiments were performed with full laser power (488 nm; 200 mW), and TIRF or oblique imaging was performed with 50 mW or less (through AOTF and neutral density–based laser attenuation).
Calcium assays were imaged using a CCD camera (Cool Snap HQ; Photometrics) and a Nikon TE-2000 inverted microscope with a 20× PlanFluor 0.5 NA objective in an In Vivo Scientific microscope incubator (St. Louis, MO) to create a 37°C climate.
NIS-Elements was used for image acquisition, and NIS-Elements, ImageJ, MATLAB (MathWorks, Natick, MA), and Microsoft Excel (Redmond, WA) were used for data analysis. Pak-PBD images were denoised in collaboration with John Sedat (University of California, San Francisco, CA), using software developed by Jerome Boulanger (Kervrann and Boulanger, 2006). Default parameters were chosen for a two-dimensional time series.
Reagents
The following reagents were used: pig fibronectin (purified in our lab from pig plasma), fMLP (F3506; Sigma), cytochalasin D (250255; Calbiochem, San Diego, CA), blebbistatin (203389; Calbiochem), latrunculin B (428020; Calbiochem), jasplakinolide (420127; Calbiochem), Y27632 (688001; Calbiochem), and calcium dye, Fluo-4-AM (F14201; Invitrogen).
To test existing methods of arresting actin dynamics, the published concentrations of each drug were used (Kueh et al., 2008; Renkawitz et al., 2009; Wilson et al., 2010). For cytochalasin D experiments, cells were preincubated with 100 nM fMLP for a minimum of 2 min, and then a solution containing cytochalasin D and fMLP was added for a final concentration of 10 μM cytochalasin D and 100 nM fMLP. In experiments containing blebbistatin, cells were preincubated in 100 nM fMLP and 50 μM blebbistatin for 10 min. Cells were then treated with either solutions of latrunculin B or jasplakinolide to achieve final concentrations of 100 nM fMLP, 50 μM blebbistatin, and 500 nM latrunculin B, or 100 nM fMLP, 50 μM blebbistatin, and 1 μM jasplakinolide, respectively
For JLY treatment, HL-60 cells were preincubated in 100 nM fMLP and 10 μM Y27632 for 10 min. Next jasplakinolide and latrunculin B were added for a final concentration of 100 nM fMLP, 10 μM Y27632, 8 μM jasplakinolide, and 5 μM latrunculin B. In HT1080 and NIH 3T3 cells, the concentration of Y27632 in the cocktail was 20 μM Y27632. HT1080 cells were preincubated in 100 nM fMLP and 20 μM Y27632 for 10 min, and NIH 3T3 cells were preincubated in 20 μM Y27632 for 10 min before the addition of jasplakinolide and latrunculin B.
Calcium assay
Cells were incubated for 0.5–1 h at 37°C in media containing 1 μM Fluo-4-AM. Next cells were plated as described in the Cell Culture section. Neutral density filters were used to attenuate the intensity of the fluorescence excitation light to prevent spontaneous release of calcium. Cells were loaded and imaged at an intensity signal between one to two times the background intensity. Positive calcium release was scored for cells exhibiting at least a threefold increase in fluorescence of intensity.
Supplementary Material
Acknowledgments
We thank John Sedat for image denoising collaboration; Jared Toettcher for assistance with data analysis; and Angelika Noegel, Roland Wedlich-Soldner, and Matt Welch for constructs. We also thank Scott Hansen, Mark Von Zastrow, and members of the Weiner lab for helpful discussion and a critical reading of the manuscript. This work was supported by a National Institutes of Health grant (R01-GM084040) to O.D.W.
Abbreviations used:
- AOTF
acousto-optic tunable filters
- BCS
bovine calf serum
- DIC
differential interference contrast
- DMSO
dimethyl sulfoxide
- EM-CCD
electron-multiplying charge-coupled device
- FBS
fetal bovine serum
- fMLP
N-formyl-methionine-leucine-phenylalanine
- FRAP
fluorescence recovery after photobleaching
- JLY
jasplakinolide–latrunculin B–Y27632 cocktail
- mHBSS
modified Hank's buffered saline solution
- NA
numerical aperture
- PBD
p21-binding domain
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- ROCK
Rho kinase
- ROI
region of interest
- SCAR
Suppressor of cyclical AMP receptor
- TIRF
total internal reflection fluorescence
- WAVE
Wiskott-Aldrich syndrome protein verprolin-homologous protein
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0379) on August 31, 2011.
REFERENCES
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Senderowicz AM, Sausville EA, Duncan KL, Korn ED. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J Biol Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Bubb MR, Spector I, Beyer BB, Fosen KM. Effects of jasplakinolide on the kinetics of actin polymerizationAn explanation for certain in vivo observations. J Biol Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- Burnette DT, Ji L, Schaefer AW, Medeiros NA, Danuser G, Forscher P. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev Cell. 2008;15:163–169. doi: 10.1016/j.devcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coué M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- Cramer LP. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr Biol. 1999;9:1095–1105. doi: 10.1016/s0960-9822(99)80478-3. [DOI] [PubMed] [Google Scholar]
- Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauert AB, Martinelli S, Marone C, Niggli V. Differentiated HL-60 cells are a valid model system for the analysis of human neutrophil migration and chemotaxis. Int J Biochem Cell Biol. 2002;34:838–854. doi: 10.1016/s1357-2725(02)00010-9. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervrann C, Boulanger J. Optimal spatial adaptation for patch-based image denoising. IEEE Trans Image Process. 2006;15:2866–2878. doi: 10.1109/tip.2006.877529. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Nagai T, Ohashi K, Mizuno K. Measurements of spatiotemporal changes in G-actin concentration reveal its effect on stimulus-induced actin assembly and lamellipodium extension. J Cell Biol. 2011;193:365–380. doi: 10.1083/jcb.201101035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai FPL, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TEB, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Forscher P. Cytoskeletal remodeling during growth cone-target interactions. J Cell Biol. 1993;121:1369–1383. doi: 10.1083/jcb.121.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Mensing H, Czarnetzki BM. Leukotriene B4 induces in vitro fibroblast chemotaxis. J Invest Dermatol. 1984;82:9–12. doi: 10.1111/1523-1747.ep12258678. [DOI] [PubMed] [Google Scholar]
- Millius A, Dandekar SN, Houk AR, Weiner OD. Neutrophils establish rapid and robust WAVE complex polarity in an actin-dependent fashion. Curr Biol. 2009;19:253–259. doi: 10.1016/j.cub.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Schumann K, Weber M, Lämmermann T, Pflicke H, Piel M, Polleux J, Spatz JP, Sixt M. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. 2009;11:1438–1443. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova I, Burakov A, Berardone N, Zaliapin I, Slepchenko B, Svitkina T, Kashina A, Rodionov V. Actin dynamics is essential for myosin-based transport of membrane organelles. Curr Biol. 2008;18:1581–1586. doi: 10.1016/j.cub.2008.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Spoerl Z, Stumpf M, Noegel AA, Hasse A. Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J Biol Chem. 2002;277:48858–48867. doi: 10.1074/jbc.M205136200. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Uehata M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- VanCompernolle SE, Clark KL, Rummel KA, Todd SC. Expression and function of formyl peptide receptors on human fibroblast cells. J Immunol. 2003;171:2050–2056. doi: 10.4049/jimmunol.171.4.2050. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Wong K, Knight ZA, Govaerts C, Hahn KM, Shokat KM, Bourne HR. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J Cell Biol. 2006;174:437–445. doi: 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Self-polarization and directional motility of cytoplasm. Curr Biol. 1999;9:11–20. doi: 10.1016/s0960-9822(99)80042-6. [DOI] [PubMed] [Google Scholar]
- Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR. Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Mühlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, Ji L, Keren K, Danuser G, Theriot JA. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465:373–377. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.