The expression of a small sterol transport protein, STARD4, is regulated by cholesterol levels. We show that the abundance of STARD4 regulates the sensitivity of the SREBP-2 system to changes in cholesterol, providing an additional layer of regulation in the cholesterol homeostatic mechanism.
Abstract
Nonvesicular transport of cholesterol plays an essential role in the distribution and regulation of cholesterol within cells, but it has been difficult to identify the key intracellular cholesterol transporters. The steroidogenic acute regulatory-related lipid-transfer (START) family of proteins is involved in several pathways of nonvesicular trafficking of sterols. Among them, STARD4 has been shown to increase intracellular cholesteryl ester formation and is controlled at the transcriptional level by sterol levels in cells. We found that STARD4 is very efficient in transporting sterol between membranes in vitro. Cholesterol levels are increased in STARD4-silenced cells, while sterol transport to the endocytic recycling compartment (ERC) and to the endoplasmic reticulum (ER) are enhanced upon STARD4 overexpression. STARD4 silencing attenuates cholesterol-mediated regulation of SREBP-2 activation, while its overexpression amplifies sterol sensing by SCAP/SREBP-2. To analyze STARD4's mode of action, we compared sterol transport mediated by STARD4 with that of a simple sterol carrier, methyl-β-cyclodextrin (MCD), when STARD4 and MCD were overexpressed or injected into cells. Interestingly, STARD4 and cytosolic MCD act similarly by increasing the rate of transfer of sterol to the ERC and to the ER. Our results suggest that cholesterol transport mediated by STARD4 is an important component of the cholesterol homeostasis regulatory machinery.
INTRODUCTION
Significant differences in lipid distribution are maintained among intracellular organelles. For example, cholesterol comprises ∼30% of the lipid molecules in the plasma membrane (PM; Ikonen, 2008), and it is also highly enriched in the endocytic recycling compartment (ERC; Hao et al., 2002). The endoplasmic reticulum (ER) is the site of cholesterol synthesis, and it is also the major cellular organelle for regulating cholesterol levels. Cholesterol comprises only ∼5% of the lipid molecules in the ER (Radhakrishnan et al., 2008). This difference in membrane cholesterol content can be explained in part by differences in the stabilization of cholesterol by other lipid components in various organelles (Mesmin and Maxfield, 2009).
There are two major pathways by which levels of unesterified cholesterol are regulated in the ER. The more rapid response to elevated cholesterol is esterification of cholesterol by the enzyme acyl-CoA: cholesterol acyl-transferase (ACAT), an ER enzyme. A major determinant of flux through ACAT is delivery of cholesterol to the ER (Tabas et al., 1988; Lange et al., 1999). The second major regulatory mechanism is mediated by the sterol regulatory element-binding protein (SREBP) transcription factors (Brown and Goldstein, 2009). When the cholesterol content of ER membranes drops below 5 mol%, a membrane protein, SREBP-2, is escorted from the ER to the Golgi by the cholesterol-sensing protein, SREBP cleavage-activating protein (SCAP). SREBP-2 is then proteolytically processed to generate a cytoplasmic form that translocates into the nucleus and activates the expression of genes implicated in sterol biosynthesis, uptake, and metabolism (Horton et al., 2003). Above the 5 mol% threshold, cholesterol-bound SCAP and SREBP-2 are retained in the ER, preventing SREBP-2 processing (Radhakrishnan et al., 2008).
Maintaining appropriate cholesterol levels in various cell membranes is important for many cellular functions, including signal transduction and membrane trafficking (van Meer and Sprong, 2004; Maxfield and Menon, 2006; Simons and Gerl, 2010). For the cholesterol level sensed in the ER to reflect the cholesterol abundance in organelles such as the PM and endosomes, there must be a mechanism for rapid exchange of cholesterol among these organelles. While there is some vesicular transport of lipids from the PM and endosomes to the ER, this is not a major pathway of lipid transport (van Meer et al., 2008), so cholesterol sensing that relies upon vesicular transport to the ER would be very slow.
There is abundant evidence that sterols can be transported among organelles by nonvesicular pathways. Newly synthesized sterol can move from the ER to the PM when secretory membrane traffic is inhibited by genetic or pharmacological manipulations (Urbani and Simoni, 1990; Heino et al., 2000; Baumann et al., 2005). Following photobleaching, the fluorescent sterol dehydroergosterol (DHE) in the ERC recovered with a t1/2 of ∼2.5 min (Hao et al., 2002). The rate of replenishment of sterol in the ERC was not diminished greatly in ATP-depleted cells, indicating that the transport to the ERC was nonvesicular. Such a rapid rate of sterol transport suggests the ERC and the PM, which are the major pools of cellular cholesterol, are maintained with sterol levels close to equilibrium with each other. It is not known if other membranes (e.g., the ER) also maintain sterol concentrations nearly equilibrated with the PM and the ERC.
Since cholesterol is nearly insoluble in water, rapid nonvesicular transport requires carrier proteins. Several candidate proteins have been proposed to be nonvesicular transporters, but it has proven difficult to provide convincing evidence that they play a major role as cholesterol transporters (Alpy and Tomasetto, 2005; Maxfield and van Meer, 2010; Raychaudhuri and Prinz, 2010).
Characterizing the molecular basis for nonvesicular sterol transport is very important for understanding sterol homeostasis. For example, if the abundance of transport carriers was low, this could limit the cell's ability to respond to changes in the sterol content of the PM. This would cause some organelles to be far from the equilibrium distribution of sterol. Since the ER can remove cholesterol from its membrane by esterification, this organelle, in particular, might be held below the equilibrium cholesterol concentration if sterol transport were limiting. In this situation, changes in the abundance of the sterol carrier would alter the cell's sensing of cholesterol in the ER and thus alter steady-state levels of cholesterol.
Demonstration that a protein plays a significant role as a sterol transport carrier would require showing the protein is an efficient sterol transporter and relatively abundant. It should also be possible to show that rates of interorganelle transport in cells depend significantly on the abundance of the transporter. A difficulty is that sterol-binding proteins could regulate sterol transport without being major carriers themselves.
One family of candidate transporters is related to the oxysterol-binding protein (OSBP), which binds 25-hydroxycholesterol (Lehto and Olkkonen, 2003). In yeast, a family of OSBP homology (Osh) proteins has been identified, and several of these proteins have been shown to transport sterol between liposomes (Raychaudhuri et al., 2006). While evidence has been presented indicating that the Osh proteins could be sterol transporters in yeast (Sullivan et al., 2006; Schulz et al., 2009), it is unclear at present whether their abundance and transport kinetics would be sufficient to account for a large fraction of sterol transport in cells (Fairn and McMaster, 2008). A role for OSBP-related proteins in sterol transport in mammalian cells has also been proposed (Jansen et al., 2011).
The family of steroidogenic acute regulatory-related lipid-transfer (START) domain-containing proteins is involved in several aspects of intracellular trafficking of lipids (Soccio and Breslow, 2003; Alpy and Tomasetto, 2005). The START domains bind cholesterol (or other lipids) in a hydrophobic pocket that is covered by a lid, which apparently opens to allow exchange with membranes. The founding member of the family, StAR, is responsible for cholesterol delivery to mitochondria for steroid hormone synthesis (Miller, 2007). Several members of the START family have domains that determine their cellular localization. For example, STARD3/MLN64 is anchored in the membranes of late endosomes by four N-terminal transmembrane domains (Alpy et al., 2001).
Some sterol-binding START proteins contain only the START domain, a protein module of 210 residues; this subgroup includes STARD4, -D5, and -D6. Among these, the widely expressed STARD4 has been shown to increase cholesteryl ester (CE) formation upon overexpression (Rodriguez-Agudo et al., 2008). Interestingly, STARD4 transcription is controlled by SREBP-2 (Soccio et al., 2005). STARD5 gene expression is not regulated by SREBP-2 but by ER stress, and STARD6 gene expression is limited in its tissue distribution (Soccio et al., 2005).
We examined whether STARD4 can serve as a major cholesterol transporter in mammalian cells. An in vitro transport assay showed that STARD4 transfers sterol rapidly between liposomes, and it is more than 1000-fold more potent than methyl-β-cyclodextrin (MCD) in this activity. STARD4 overexpression improves sterol delivery to the ERC and leads to an ACAT-dependent accumulation of sterol esters in lipid droplets, while its silencing dramatically increases free cholesterol levels in the cell. Moreover, in STARD4-silenced cells, SREBP-2 processing in response to changes in cellular cholesterol levels is strongly attenuated. We compared the cellular transport activity of STARD4 with the simple carrier MCD, which was injected into the cytoplasm. Strikingly, MCD mimics STARD4-mediated sterol transport, and reverses the free cholesterol accumulation observed in cells silenced for STARD4, indicating this protein provides a mechanism for cholesterol equilibration among many organelles. We hypothesize that STARD4 expression levels regulate the sensitivity of the cholesterol homeostatic mechanisms by altering the rate of sterol equilibration among organelles.
RESULTS
Injected MCD and STARD4 overexpression increase the rate of sterol delivery to the ERC
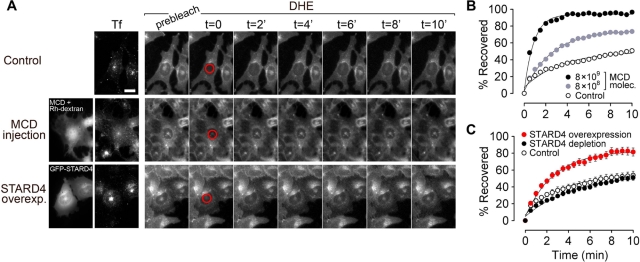
Sterol transfer between the PM and the ERC depends on a rapid, nonvesicular mechanism (Hao et al., 2002). To test whether a larger number of cytosolic sterol carriers would increase the rate of sterol delivery to the ERC, we microinjected cells with different amounts of MCD and assessed the kinetics of DHE transport to the ERC using fluorescence recovery after photobleaching (FRAP; Figure 1). Control U2OS cells, or cells that have been microinjected with MCD, were incubated with extracellular DHE loaded onto MCD for 1 min at 37°C to deliver DHE to the PM. This was followed by a 15-min incubation at 37°C in medium containing Alexa Fluor 633–transferrin (Tf), to achieve a steady-state distribution of DHE and sufficient Tf internalization to label the ERC. After the ERC was photobleach, DHE returned to this organelle with a recovery t1/2 of 3.2 ± 0.3 min in control cells. In MCD-injected cells, the rate of DHE recovery increased in a dose-dependent manner (Figure 1B). When 8 × 109 molecules of MCD were introduced into the cytosol, the recovery t1/2 decreased to 0.9 min. This shows that regulation of the number of sterol carriers in the cytosol can be a rate-determining factor for sterol movement between organelles.
FIGURE 1:
Microinjected MCD and STARD4 overexpression enhance sterol transport to ERC. (A) DHE transport kinetics to the ERC measured by FRAP. U2OS cells were labeled with DHE and incubated with 10 μg/ml Alexa Fluor 633–Tf for 15 min at 37°C in Medium A/glucose to identify the ERC. Where indicated, MCD and rhodamine-dextran were coinjected immediately before labeling cells with DHE. STARD4-overexpressing cells were transfected with GFP-STARD4 for 48 h before the experiment. An image was taken before photobleaching. DHE in the ERC was photobleached (red circles), and images were taken every 30 s. Cells were maintained at 37°C. Scale bar: 20 μm. (B and C) The fluorescence intensity ratio of the photobleached area (ERC) to the entire cell was calculated for each time point and normalized using “prebleach” (100%) and “t = 0” (0%) images. (B) Measurements for single cells. The number of MCD molecules injected was estimated by quantifying the rhodamine fluorescence (see Materials and Methods). MCD (8 × 109): t1/2 = 0.9 min; injected MCD (8 × 108): t1/2 = 2.6 min; control: t1/2 = 3.2 min. (C) FRAP recovery curves for cells transfected with GFP-STARD4 or depleted for STARD4 by shRNA transfection. Each data point is derived from an average of five experiments (± SE). The lines are single exponential fits.
ERC sterol levels mirror the cholesterol content in the PM (Maxfield and Mondal, 2006); however, the cellular proteins used for transport have not been identified. To determine whether STARD4 could play a role in sterol transport between the PM and the ERC, we performed FRAP experiments on green fluorescent protein (GFP)-STARD4–transfected cells, which were labeled with DHE and Tf, as described in the preceding paragraph. After photobleaching, the DHE fluorescence recovered in the ERC with a t1/2 of 2.4 ± 0.5 min (Figure 1C). The rate and extent of sterol recovery in the ERC in these STARD4-overexpressing cells was significantly greater than in control cells. Interestingly, FRAP experiments performed on cells depleted of STARD4 by small interfering RNA (siRNA) knockdown showed little reduction in the rate of DHE delivery to the ERC. This lack of effect may be due, in part, to an increase in cellular cholesterol following STARD4 silencing (see Figure 4 later in the paper). Nevertheless, this suggests that STARD4-independent mechanisms also play a role in sterol exchange to and from the ERC.
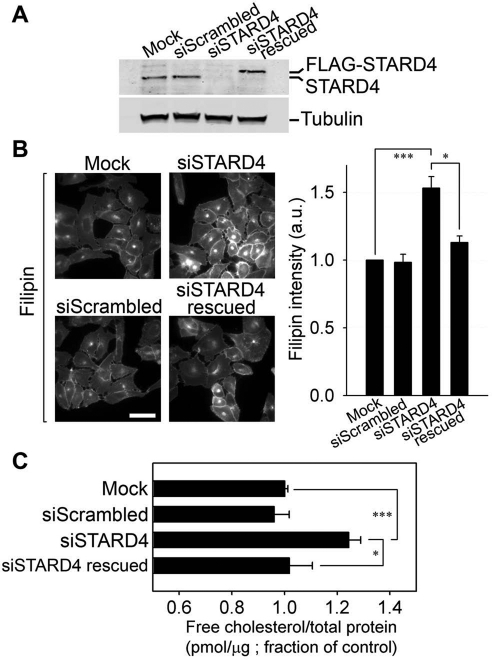
FIGURE 4:
STARD4 silencing causes an increase in free cholesterol level. (A) U2OS cells were transfected with scrambled siRNA or with siSTARD4 and incubated in growth medium for 48 h. Where indicated, 24 h after siRNA transfection, the cells were subjected to another transfection with pCMV-Tag2B-STARD4 having a silent mutation in the siRNA-targeting sequence for 24 h, in order to rescue STARD4 silencing. As a control, the transfection reagent HiPerFect was added alone to the cells (Mock). The immunoblot shows the efficiency of STARD4 silencing. In the rescue experiment (fourth lane), note the appearance of the FLAG-tagged STARD4 band, while endogenous STARD4 is silenced. (B) Epifluorescence microscopy images of cells treated as in (A), which were fixed and stained with filipin. Scale bar: 50 μm. The right panel shows quantification (± SE) of the average cellular filipin fluorescence power in different conditions. Filipin fluorescence was normalized to the control experiment. Intensity measurements were performed on background-subtracted images from an average of 50 fields of cells in each condition. (C) GC/MS measurement of free cholesterol levels in cells transfected or not with the indicated siRNA. The cells were transfected and plated in six-well plates for 48 h. Cellular lipids were extracted and analyzed by GC/MS. Data represent averages (± SE) of three independent experiments normalized to control value. ***, p < 0.001, *, p < 0.02.
STARD4 increases ACAT-dependent sterol esterification
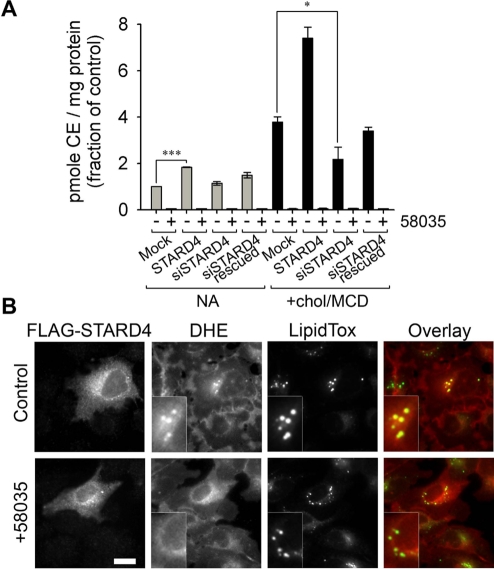
It has been reported that STARD4 overexpression in mouse hepatocytes increases CE formation, whereas no increase was observed with the overexpression of other START domain–only proteins, such as StAR or STARD5 (Rodriguez-Agudo et al., 2008). It is well established that excess free sterol in the ER is converted to sterol ester by ACAT and stored in lipid droplets (Guo et al., 2009). Therefore one hypothesis is that STARD4 could bring sterol molecules to the ER, where the ACAT enzyme resides. To address this possibility, we examined the esterification of cholesterol by ACAT in cells with varying levels of STARD4 expression. We used an esterification assay in which cells were incubated for 6 h with [14C]oleate before lipid extraction. In good agreement with a previous report (Rodriguez-Agudo et al., 2008), a twofold increase in CE level was observed when the STARD4 gene was overexpressed, as compared with control cells (Figure 2A). The STARD4-mediated cholesterol esterification was inhibited significantly by the ACAT inhibitor, 58035.
FIGURE 2:
STARD4 facilitates ACAT-mediated sterol esterification. (A) Cholesterol esterification by ACAT. Control cells, cells expressing FLAG-STARD4, cells depleted for STARD4 by siRNA, and cells rescued for STARD4 expression after depletion by siRNA were incubated with 1 μCi/ml [14C]oleate for 6 h. Where indicated, the ACAT inhibitor 58035 was added during the incubation. The cholesteryl-[14C]oleate (CE) formed per mg cell protein was measured and normalized to the average control value. Data represent averages (± SE) of three independent experiments. ***, p < 0.001, *, p < 0.05. (B) Epifluorescence images of U2OS cells transfected with FLAG-STARD4 for 18 h. Cells were incubated with DHE-loaded MCD for 1 min at 37°C, rinsed, and incubated 20 min at 37°C. Where indicated, inhibitor 58035 was added to the cells 6 h prior to DHE labeling. Cells were fixed, permeabilized, and stained with Cy3-labeled mouse anti-FLAG M2 antibody and with the lipid-droplet marker LipidTOX Green. Insets: magnifications of lipid-droplet regions. In the overlay images, DHE is red and LipidTOX is green. Yellow dots, indicating colocalization of DHE with LipidTOX, occur only in cells transfected with FLAG-STARD4, not in those treated with 58035. Scale bar: 10 μm.
There is good evidence that sterol moves from the PM to the ER mostly by nonvesicular processes; however, the identity of lipid-transfer proteins responsible for this transport is still unknown (Mesmin and Maxfield, 2009). To test whether STARD4 serves as a sterol carrier between these two compartments, we used U2OS cells in which the expression of STARD4 was silenced by siRNA (Figure 2A). Following siRNA treatment, the cells were incubated for 48 h to allow efficient STARD4 depletion. Unexpectedly, there was no significant effect of STARD4 depletion on cholesterol esterification under these conditions. To see whether levels of STARD4 expression might have greater impact under conditions of cholesterol loading, we incubated the cells with cholesterol-loaded MCD for 2 h before lipid extraction. As expected, cholesterol loading led to a large increase in CE formation by ACAT. In the cholesterol-loaded cells, a significant reduction in cholesterol esterification was observed in STARD4-depleted cells as compared with control cells or cells rescued for STARD4 expression after silencing.
We then analyzed the effect of STARD4 overexpression on the subcellular distribution of DHE. For this, U2OS cells expressing FLAG-tagged STARD4 were incubated with DHE loaded onto MCD for 1 min at 37°C to deliver DHE to the PM. This was followed by a 20-min incubation of the cells at 37°C to allow intracellular DHE transport. The cells were then fixed, permeabilized, and stained with an anti-FLAG antibody. As shown in Figure 2B, the expression of STARD4 had a major effect on the intracellular DHE distribution; DHE becomes strongly enriched in organelles that label with a neutral lipid dye, LipidTOX, which is a lipid-droplet marker (Grandl and Schmitz, 2010). This contrasted with the mainly diffuse DHE distribution found in neighboring untransfected cells. Importantly, DHE colocalization with LipidTOX in cells expressing STARD4 was prevented when the cells were treated with ACAT inhibitor 58035, indicating ACAT activity is required for the DHE incorporation into lipid droplets (Figure 2B, bottom panels). When cells overexpressing STARD4 were incubated for 4 h after DHE labeling, a large fraction of the cellular DHE became concentrated into lipid droplets at the expense of others organelles (Supplemental Figure S1).
We examined whether similar effects on sterol distribution could be observed in cells overexpressing STARD5, a closely STARD4-related protein (Figure S2). In contrast with the effect of STARD4 overexpression, no enrichment of DHE fluorescence was observed in lipid droplets of cells overexpressing FLAG-tagged STARD5. Actually, the expression of STARD5 had no visible effect on intracellular sterol distribution, suggesting that STARD4 and STARD5 have distinct functions in cellular sterol distribution.
The results presented in Figure 2 demonstrate that STARD4 facilitates the transport of sterol to the ER, which is the site of ACAT. The DHE distribution is almost unchanged from untransfected cells when the cells are treated with the inhibitor 58035; this suggests that free sterol reaching the ER may reequilibrate with other organelles when ACAT is not functional.
STARD4 exchanges sterol between membranes
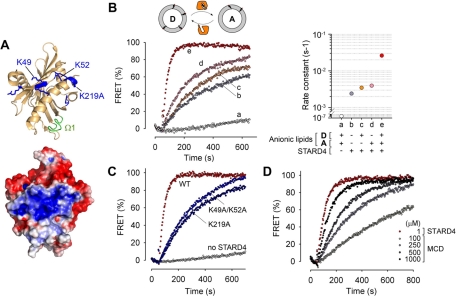
The mechanism of sterol binding to STARD4 is unclear, although modeling based on homologous structures suggests that the hydroxyl is oriented toward the core of the protein (Tsujishita and Hurley, 2000), and molecular dynamics simulations performed on START domains of StAR and STARD3 suggest that a nonpolar loop (Ω1) functions as a hinged lid that transiently opens for sterol uptake and release (Murcia et al., 2006). In the crystal structure of STARD4, there are several positively charged residues exposed near the loop Ω1 of the protein, which form a basic patch (Figure 3A; Romanowski et al., 2002). Given the proximity of the basic patch and Ω1 in STARD4, we suspected this protein might interact with negatively charged lipids, which would facilitate its membrane docking. Therefore we examined the sterol-transfer activity of STARD4 using liposomes of various lipid compositions.
FIGURE 3:
STARD4 transports sterol between negatively charged membranes. (A) Top, ribbon representation of the STARD4 structure (Romanowski et al., 2002), featuring the Ω1 loop (green) and positively charged residues (blue). Bottom, surface representation of STARD4 colored according to electrostatic potential. Structures were created in PyMOL (DeLano Scientific, Palo Alto, CA). (B) DHE transfer assay. The appearance of FRET was used to measure DHE transfer from donor to acceptor liposomes. Donor and acceptor liposomes (100 μM each) were incubated alone (a) or with 1 μM STARD4 (b–e) at 37°C. The donor liposomes contained PC/PE/PS/DHE, and acceptor liposomes contained PC/PE/PI/PS/dansyl-PE. The acidic lipids were replaced with neutral lipids, as indicated in the right panel. Control experiments were performed in the absence of STARD4. (C) Effect of the point mutations K49A/K52A and K219A on the sterol transfer catalyzed by STARD4. (D) Comparison of the sterol-exchange activity of STARD4 (1 μM) with MCD used in a range from 100 to 1000 μM.
To quantify the sterol-exchange activity of STARD4, we used a transport assay in which liposomes containing DHE (donors) and others with dansyl-labeled phosphatidylethanolamine (PE; acceptors) were mixed with purified protein (John et al., 2002). Sterol transfer was measured by the appearance of fluorescence resonance energy transfer (FRET) sensitized emission when these two fluorescent lipids came into proximity (Figure 3B). We used liposomes with compositions similar to the inner leaflet of the PM as donors and ones similar to ER membranes as acceptors (Leventis and Grinstein, 2010). The anionic lipid components in donors and acceptors were phosphatidylserine (PS; 23 mol%) and phosphatidylinositol (PI)/PS (15/5 mol%), respectively. Where indicated, these anionic lipids were replaced by the neutral lipid phosphatidylcholine (PC). We found that the STARD4-catalyzed DHE transfer between negatively charged liposomes was 10-fold faster than between neutral liposomes (Figure 3B). Interestingly, the absence of anionic lipids in either donor liposomes or acceptor liposomes, also resulted in much slower transport. This indicates that both sterol extraction from donors and delivery to acceptors by STARD4 are promoted by negative membrane charges.
To determine the contribution of the basic patch of STARD4 in the sterol-transfer activity, we constructed a double mutant in which two lysines exposed at the surface of the protein, K49 and K52, were both replaced by alanine. Figure 3C shows that this change decreased the rate of sterol transfer approximately eightfold. A K219A mutation within the basic patch of STARD4 also caused a strong decrease in the sterol-transfer rate.
We further assessed the specificity of STARD4 for lipids by replacing C16:0-C18:1-phosphaditylcholine (POPC) with C18:1-C18:1-phosphatidylcholine (DOPC) in liposomes. Figure S3 shows that the sterol-transfer rate increased by up to threefold when acceptor liposomes were enriched in unsaturated lipids. However, when unsaturated lipids were added to donor liposomes, no improvement in the transfer rate was observed.
As shown in Figure S4, the FRET intensity measured in acceptor liposomes was linearly proportional to the DHE concentration, and this allowed us to measure the rate of sterol-transfer per molecule of STARD4 in our in vitro system (Figure S4, B and C). We found that approximately seven molecules of DHE are transferred to acceptor membranes per minute per molecule of STARD4. Moreover, we found that STARD4 was more than 1000-fold more potent in its sterol-transfer activity than MCD (Figure 3D). These results highlight the robustness of STARD4's activity.
Taken together, these results demonstrate that STARD4 is a sterol transporter that operates preferentially between negatively charged membranes. Because STARD4 exhibits faster exchange rates when acceptor liposomes are enriched in unsaturated lipids, it is well adapted to deliver sterol to membranes, such as the ER, that have a large fraction of unsaturated acyl chains.
Cellular free cholesterol level increases upon STARD4 silencing
To measure the contribution of STARD4 to intracellular sterol sensing, we used U2OS cells in which the expression of STARD4 was silenced by siRNA. Following siRNA treatment, the cells were incubated in growth medium for 48 h to allow efficient STARD4 depletion (Figure 4A). We then fixed and stained the cells with the cholesterol-binding dye, filipin, in order to estimate their free cholesterol content. Strikingly, a large increase in filipin intensity was observed in STARD4-deficient cells compared with control cells or cells transfected with scrambled siRNA, reflecting an increase in the free cholesterol content (Figure 4B). Qualitatively, the filipin labeling pattern in STARD4 knockdown cells resembled untreated cells with a pronounced staining of the PM and the ERC, but the labeling was much brighter in silenced cells.
To exclude possible off-target activity of the siRNA that might contribute to the increase of cellular free cholesterol, we performed a rescue experiment in which siRNA-treated cells were transfected with a FLAG-STARD4 RNA interference (RNAi)-resistant construct 24 h before fixation and staining with filipin. As shown in Figure 4A, while endogenous STARD4 was silenced, FLAG-STARD4 was expressed to a comparable level in these conditions. The right panel in Figure 4B shows that the increase of free cholesterol was reversed in cells expressing FLAG-STARD4. We also used gas chromatography/mass spectrometry (GC/MS) to determine the free cholesterol content in siRNA-transfected cells, and this confirmed that cholesterol content was increased significantly in STARD4-deficient cells (Figure 4C). This increase could also be reversed by expressing the STARD4 construct.
The SREBP-2 response to changes in cholesterol levels is attenuated in STARD4-depleted cells
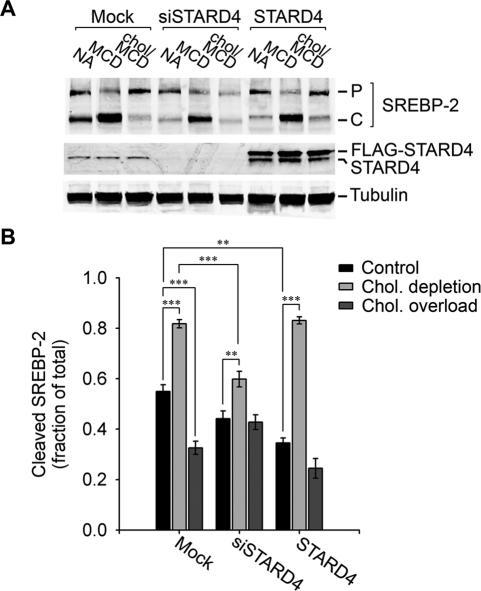
The increase in cholesterol levels when STARD4 was silenced suggests that levels of STARD4 might influence sterol sensing in the ER. To examine this, we compared SREBP-2 processing in cells silenced for STARD4 with control cells exposed to diverse cholesterol conditions (Figure 5). To reduce cellular free cholesterol, U2OS cells were transferred to lipoprotein-deficient medium supplemented with 1 mM MCD for 2 h. As expected, we observed a large increase in production of the cleaved form of SREBP-2 as compared with control cells (Figure 5A). When we raised cellular free cholesterol by adding cholesterol in complex with MCD (500 μM) to the cell medium for 2 h, there was a buildup of the precursor form of SREBP-2 and a decrease in the cleaved form (Figure 5B).
FIGURE 5:
STARD4 silencing attenuates cholesterol-mediated regulation of SREBP-2 processing. (A) Cells were transfected with or without siSTARD4 and plated in 10-cm dishes for 48 h in growth medium. Alternatively, cells were transfected with FLAG-STARD4 cDNA for 24 h. The cells were then washed and incubated for 2 h in McCoy's medium supplemented with: 1%FBS (NA); 1%LPDS, 1 mM MCD, 10 μM mevinolin (MCD); 1%LPDS, 500 μM cholesterol complexed to MCD, 10 μM mevinolin (Chol/MCD). ALLN (10 μM) was present in all conditions. The cells were then lysed, separated by 16% SDS–PAGE, and immunoblotted with anti-SREPB-2, anti-STARD4, and anti-α-tubulin antibodies. P = precursor form of SREBP-2; C = cleaved form of SREBP-2. (B) Quantification (± SE) of three independent experiments in which cleaved SREBP-2 is expressed as a fraction of the total. ***, p < 0.001, **, p < 0.01.
In STARD4-silenced cells, in spite of their higher free cholesterol levels, the cleaved fraction of SREBP-2 was comparable to what we found in control, nontreated cells. We did observe a limited increase of the SREBP-2 cleaved fraction when cholesterol was depleted from cells silenced for STARD4 (Figure 5B). When we loaded these cells with cholesterol for 2 h using MCD-cholesterol complexes, there was no decrease in SREBP-2 cleavage, pointing toward a reduced sensitivity to change in cellular cholesterol levels.
The overexpression of STARD4 on SREBP-2 processing was also tested. As expected, cholesterol depletion caused a large increase in SREBP-2 processing in cells overexpressing STARD4. Both the control cells and the cholesterol-overloaded cells showed somewhat reduced SREBP-2 processing compared with their mock-transfected counterparts.
Taken together, these results suggest that STARD4 has an important role in relaying information about the cellular sterol status to the ER-based sterol sensors.
MCD injected into the cytosol enhances sterol transport to the ER
Our results indicate that STARD4 rapidly transfers sterol to the ER, presumably because its structure is well adapted for targeting the surface of this organelle. It may be that STARD4 is targeted to transfer cholesterol among only a few membranes (e.g., PM, ERC, and ER). Alternatively, STARD4 may function as a relatively nonselective sterol exchanger. Enhanced delivery to the ER might then be detected, as the ER can act as a cholesterol sink by esterifying cholesterol via ACAT.
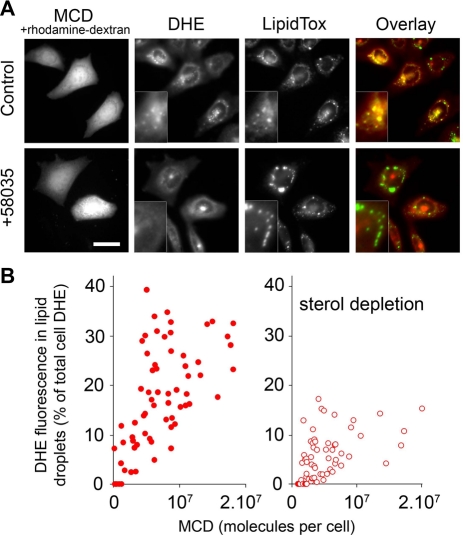
To test the hypothesis that nonselective transport could enhance sterol delivery to the ER, we introduced MCD into the cytosol by microinjection and compared its effects with STARD4. We microinjected DHE-labeled CHO-derived TRVb1 cells with MCD together with rhodamine-dextran, and then incubated them for 90 min at 37°C to allow intracellular DHE transport. Strikingly, DHE became enriched in lipid droplets stained with LipidTOX in injected cells, while noninjected cells showed a typical DHE distribution at the cell periphery and the ERC (Figure 6A). As shown in the bottom panels of Figure 6A, the DHE colocalization with LipidTOX was inhibited in cells treated with ACAT inhibitor 58035, demonstrating the requirement for ACAT in this process. The rate of DHE incorporation into lipid droplets of noninjected cells was much slower, with a t1/2 of 7–8 h (Figure S5).
FIGURE 6:
MCD microinjection increases ACAT-dependent sterol incorporation into lipid droplets. (A) Wide-field fluorescence microscopy images of living CHO cells. Cells were incubated with DHE-loaded MCD for 1 min at 37°C, rinsed, and injected or not with MCD together with rhodamine-dextran. This was followed by an incubation of 90 min at 37°C in Medium A/glucose. Where indicated, ACAT inhibitor 58035 was added to the cells 6 h before the experiment. Before imaging, the cells were stained for 15 min with the lipid-droplet marker LipidTOX. Magnifications of lipid-droplet regions are shown in insets. Panels on far right show pseudocolored overlay images, in which DHE is red and LipidTOX is green. Scale bar: 20 μm. (B) Per-cell quantification of the DHE fluorescence expressed as the percentage of DHE present in lipid droplets relative to the total DHE fluorescence, as a function of the estimated number of MCD molecules injected (see Materials and Methods). The right panel shows the same experiment performed with cells incubated for 2 h in Ham/F12 medium supplemented with 5% LPDS, 1 mM MCD, and 10 μM mevinolin before the steps described in (A). The fluorescence ratio of DHE in lipid droplet to the whole-cell DHE was measured in MetaMorph, as described previously (Majumdar et al., 2007), using a binary mask generated after thresholding the LipidTOX-stained area, and another by thresholding the rhodamine-dextran area, which represents the whole cell area.
To probe the effectiveness of intracellular MCD, we measured the DHE fluorescence associated with lipid droplets in cells in which the amount of injected MCD was determined by coinjection of a fluorescent tracer. As shown in Figure 6B, DHE is incorporated into lipid droplets of MCD-injected cells in a dose-dependent manner. We observed that MCD-mediated sterol trafficking to lipid droplets was reduced significantly when the cells were subjected to sterol depletion, indicating that sterol trafficking relies to a large extent on the chemical activity of cholesterol.
An increase in cytosolic carriers improves sterol sensing by SCAP
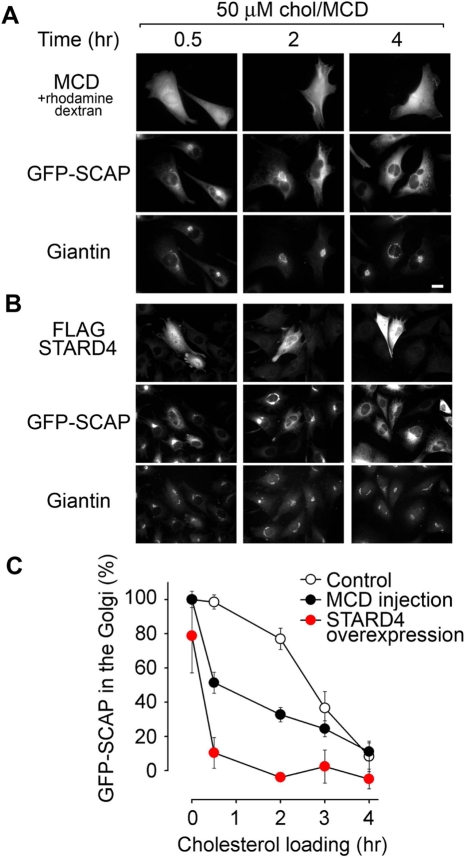
To further demonstrate the ability of STARD4 and MCD to transport cholesterol to the ER, STARD4 was overexpressed or MCD was injected into cells expressing the sterol sensor SCAP fused with GFP at the N-terminus. While GFP-SCAP colocalized with the Golgi marker Giantin in cholesterol-depleted cells, it exhibited more diffuse localization, characteristic of the ER, in cholesterol-rich conditions (Figure S6) in agreement with an earlier study (Nohturfft et al., 2000). We first induced cholesterol depletion by incubating the cells with extracellular MCD to induce GFP-SCAP transport to the Golgi. We then monitored GFP-SCAP distribution after adding back cholesterol with extracellular MCD-cholesterol complexes (Figure 7). As shown in Figure 7A, GFP-SCAP in the Golgi was reduced more rapidly in cells injected with MCD than noninjected cells when cholesterol was supplied to the cells by exchange into the PM. Figure 7B shows that the overexpression of STARD4 had an even more dramatic effect, as the ratio of GFP-SCAP in the Golgi area dropped much faster. These results are quantified in Figure 7C.
FIGURE 7:
GFP-SCAP distribution in relation to sterol content in control, MCD-injected cells, and cells overexpressing STARD4. (A) Wide-field microscopy images of CHO cells stably transfected with GFP-SCAP. Cells grown on coverslips were washed in Hank's balanced salt solution and incubated in growth medium (1% LPDS) supplemented with 2 mM MCD and 10 μM mevinolin for 2 h at 37°C. Cells were injected with MCD along with rhodamine-dextran and then switched for the indicated time to growth medium supplemented with 500 μM cholesterol complexed with MCD and 10 μM mevinolin in order to reload cells with cholesterol. Cells were then fixed, permeabilized, and stained with rabbit anti-Giantin antibody followed by a fluorescently conjugated secondary antibody (anti–rabbit Alexa Fluor 633). Scale bar: 20 μm. (B) Experiment performed as in (A), except that the cells were transfected with FLAG-STARD4 for 48 h prior to the experiment instead of being microinjected. (C) Quantification of the percentage of GFP-SCAP in the Golgi as a function of the cholesterol-loading time. Each point in the graph represents the average (± SE) of measurements obtained from 15–40 cells. GFP-SCAP in the Golgi at 0 h in control cells was normalized to 100%. The zero line represents the relative amount of GFP-SCAP localized to the Golgi in cells treated with 100 μM chol/MCD for 1 h (see Figure S6).
The experiments presented in Figures 6 and 7 show that an untargeted sterol exchanger, such as MCD, enhances sterol delivery to the ER. In addition, changes in the abundance of sterol carriers alter cholesterol sensing by the homeostatic regulatory machinery.
MCD injection in cells silenced for STARD4 reverses free cholesterol accumulation and transport defects
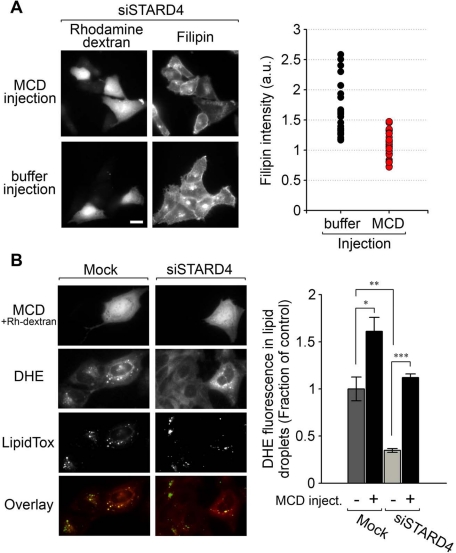
Finally, we microinjected MCD into cells silenced for STARD4 to determine if this could reverse the increase in free cholesterol observed in these cells (Figure 8A). We performed a control experiment in which STARD4-silenced cells were injected with the tracer only. After injection, the cells were fixed and stained with filipin to allow assessment of their free cholesterol content. As expected, the filipin fluorescence intensity declined in cells injected with MCD, while buffer-injected cells conserved their elevated intensity (Figure 8A, left panel). The average filipin intensity measured for 30 cells injected with MCD was comparable to the filipin level measured in cells not transfected with STARD4 siRNA (Figure 8A, right panel).
FIGURE 8:
Cytosolic injection of MCD relieves the increase in free cholesterol observed in cells silenced for STARD4, and rescues sterol transport defects. (A) Left panel, wide-field fluorescence microscopy images of U2OS cells transfected with siSTARD4 for 48 h, then subjected to microinjection of MCD along with rhodamine-dextran, or with the tracer rhodamine-dextran alone as a control (buffer injection). Following injections, the cells were incubated for 2 h at 37°C. Cells were then fixed, stained with filipin, and imaged. The right panel shows quantification of filipin fluorescence power per microinjected cell. Fluorescence power was normalized to a control experiment in which the transfection reagent alone was added to the cells. Measurements were performed on 30 injected cells for each condition. Scale bar: 20 μm. (B) Control or STARD4-depleted cells were labeled with DHE and microinjected (or not) with MCD/rhodamine-dextran. Cells were placed in growth medium (1% LPDS) supplemented with 500 μM cholesterol complexed with MCD and 10 μM mevinolin for 2 h at 37°C. Before imaging, cells were labeled with the lipid-droplet marker LipidTOX Green. Right panel represents quantification (± SE) of three independent experiments. Measurements were performed on a total of 70 injected cells for each condition.
To examine whether MCD could rescue sterol transport to the ER in cells silenced for STARD4, we analyzed the subcellular distribution of DHE in cholesterol-loaded STARD4-silenced cells injected with MCD. In Figure 8B, the comparison between injected versus noninjected cells indicates that MCD rescues the STARD4 function of transferring DHE to the ER, where it can be esterified and incorporated into lipid droplets. Moreover, in these conditions, a significant reduction of DHE fluorescence in lipid droplets is observed in STARD4-silenced cells, as compared with control cells, supporting biochemical results presented in Figure 2A. We conclude that cholesterol transport to the ER can be achieved by MCD, an untargeted sterol carrier, and this compensates for the lack of STARD4.
DISCUSSION
It has been very difficult to develop convincing evidence that any lipid-transfer protein plays a major role in cholesterol transport through the cytoplasm (Maxfield and van Meer, 2010). One of the challenges is that many proteins may transport sterols through the cytoplasm, and they may have overlapping functions. For example, there are seven members of the Osh family of proteins in budding yeast that may be sterol carriers (Raychaudhuri and Prinz, 2010). Deletion strains for any of these show only partial reductions in nonvesicular transport of sterol between the PM and the ER, and even a strain with deletions of six Osh proteins and a conditional allele for the seventh shows significant sterol transport under nonpermissive conditions (Maxfield and Menon, 2006; Sullivan et al., 2006). If a protein is an important sterol carrier, changes in expression level of the transporter should have measurable effects on sterol transport in cells, as we have shown here for STARD4.
We became interested in STARD4, as STARD4 is the only gene encoding a sterol carrier among all the genes for which transcription is increased by activation of SREBP-2 (Horton et al., 2003; Soccio et al., 2005). Additionally, overexpression of STARD4 leads to an increase in esterification of cholesterol that might be attributed to increased transport to the ER, the site of the esterifying enzyme, ACAT (Rodriguez-Agudo et al., 2008). STARD4 belongs to the START domain superfamily of proteins, which has been implicated in several cholesterol-trafficking pathways in cells (Strauss et al., 2003; Prinz, 2007).
As shown in Figure 3, STARD4 has the ability to transport sterol between membranes, with a rate of transfer per molecule more than 1000-fold greater than the rate exhibited by MCD. Under the conditions of our assays, we observed transfer rates up to seven molecules per minute per molecule of STARD4. For comparison, the yeast Osh4p transfers ∼0.5 molecule of sterol per minute in a similar assay (Raychaudhuri et al., 2006). It is possible that faster rates could be obtained with higher liposome concentrations. In U2OS cells, there are ∼105 STARD4 molecules per cell (Figure S7). The in vitro transfer rates measured in our assay would not be sufficient to account for a large fraction of the nonvesicular transport in cells, but conditions in the cytoplasm, such as the very high concentration of membranes, may accelerate transport rates significantly.
Changes in STARD4 expression have significant effects on sterol transport in cells. Overexpression of STARD4 increases the rate of sterol delivery to the ERC (Figure 1). Similarly, overexpression of STARD4 increases the rate of cholesterol transport to the ER, as assayed by increased cholesterol esterification by ACAT, and this effect was especially pronounced in cholesterol-loaded cells (Figure 2). The increased sterol esterification upon STARD4 overexpression is also shown by greater incorporation of DHE into lipid droplets (Figure 2). Overexpression also increases the sensitivity of SCAP to changes in cholesterol concentration (Figure 7), presumably as a consequence of increased sterol in the ER. It is noteworthy that statin treatment increases expression of STARD4 approximately threefold in cell culture (Soccio et al., 2005) and in mice (Riegelhaupt et al., 2010). Increased STARD4 expression would increase cholesterol delivery to the ER, which would reduce the SREBP-induced expression of low-density lipoprotein receptors. This would attenuate the serum cholesterol–lowering effects of statins. This hypothesis will need to be tested further in animal studies.
The effects of silencing STARD4 expression are more complex. Reduction of STARD4 expression leads to an increase in cellular free cholesterol (Figure 3). This may be related to the reduction in the SREBP-2 response in cells silenced for STARD4 when they are challenged with either low or high levels of cholesterol (Figure 5). That is, there is a desensitization of the cholesterol homeostatic machinery when STARD4 levels are reduced. Similarly, ACAT-dependent esterification of cholesterol as a response to cholesterol loading was less effective in cells depleted for STARD4 (Figures 2 and 8B). However, depletion of STARD4 does not measurably reduce the rate of DHE delivery to the ERC, and it does not greatly affect sterol esterification (Figure 2) or SREBP-2 processing (Figure 5) in normal cell growth conditions. One hypothesis is that the cellular cholesterol increases in normal growth conditions when STARD4 levels are reduced, until the steady-state rate of delivery of cholesterol to the ER is the same as in untreated cells. That is, there are fewer transporters, but this is counterbalanced by more cholesterol in the PM and the ERC at steady state. These complex effects of reduction in STARD4 levels also may reflect the fact that there are other sterol transporters. The increased free cholesterol in cells depleted for STARD4 may also compensate for slower rate constants for transport. The effects of reduced STARD4 seem to be most pronounced when there is an excess of cholesterol, which might be expected to stress the capacity of sterol transport systems.
STARD4 does not have any obvious organelle-targeting motifs, although it does have a basic patch near the sterol-exchange site that confers a preference for interactions with membranes containing lipids with acidic head groups (Figure 3). The cytosolic leaflets of the PM and endosomes are enriched in anionic lipids. PS, in particular, represents ∼30 mol% of the phospholipids in the PM inner leaflet (Leventis and Grinstein, 2010). Anionic lipids are also abundant in the ER (van Meer et al., 2008). ER membranes contain more unsaturated lipids than other organelles, and this could enhance STARD4-mediated delivery of sterol to the ER (Figure S3).
The lack of a highly specific organelle-targeting motif raises the possibility that STARD4 could act as a relatively nonselective sterol transporter. Such a general sterol carrier would effectively serve to equilibrate sterol chemical activity among many organelles. As discussed in detail elsewhere (Lange and Steck, 2008; Mesmin and Maxfield, 2009), sterol chemical activity may be nearly equilibrated among organelles, even if the concentration of sterols is quite different. We compared the effects of overexpressing STARD4 with microinjection of MCD, a sterol exchanger that should be quite nonselective in its interactions with membranes. Like STARD4 overexpression, injection of MCD into the cytoplasm increased the rate of transport of sterol to the ERC (Figure 1), and it increased the esterification of sterols by ACAT—indicating an increased rate of sterol delivery to the ER (Lange et al., 1999). Strikingly, injection of MCD into cells can restore normal cholesterol levels in cells with reduced expression of STARD4 (Figure 8), and it restored the esterification of DHE in cholesterol-loaded cells that were STARD4-silenced. These results show that MCD can substitute for STARD4 in important aspects of cholesterol homeostasis.
It is interesting to note that the relative distribution of sterol between the PM and the ERC does not change noticeably when STARD4 levels are elevated or when MCD is injected into cells. This is consistent with the idea that these organelles are nearly equilibrated in their cholesterol content. In contrast, the amount of ACAT-dependent esterification increases when the abundance of sterol transporters increases. This suggests that the ER may be held below its equilibrated cholesterol concentration by the activity of ACAT, which removes free cholesterol from the system by converting it into cholesteryl esters, which are stored in lipid droplets.
While our results suggest that STARD4 is an important sterol transporter, it is clear that there must be other transporters that may have distinct or overlapping transport roles. These transporters might include other START-domain proteins, as well as the oxysterol-binding protein–related protein family (Raychaudhuri and Prinz, 2010; Jansen et al., 2011). Recently, STARD4−/− mice were created (Riegelhaupt et al., 2010), and these mice were smaller than wild-type mice. However, the survival of the mice indicates that STARD4 is not essential. Intriguingly, of the six START domain–containing proteins that have been knocked out in mice, only StAR has given a pronounced lipid phenotype (Caron et al., 1998). This suggests that there is functional redundancy among the START-domain proteins and possibly overlapping functions with other families of sterol transporters.
We propose that STARD4 helps to equilibrate cholesterol among organelles, including the ER, which contains the main homeostatic regulators of cholesterol levels. Because the expression of STARD4 is governed by SREBP-2, there is a negative feedback loop in which cholesterol transported by STARD4 to the ER contributes to its own down-regulation (Figure S8). Cholesterol levels in cells depend on a delicate balance among sterol uptake, synthesis, esterification, and deesterification. STARD4 abundance can have effects on sterol sensing by SCAP and also on esterification by ACAT. As a result, controlling the sterol-carrier abundance is a crucial component of the sterol homeostatic machinery.
MATERIALS AND METHODS
Reagents and antibodies
All tissue culture media were from Life Technologies (Carlsbad, CA). MCD, mevinolin, filipin, and the ACAT inhibitor, 58035, were purchased from Sigma-Aldrich (St. Louis, MO). The neutral lipid stain, LipidTOX Green, and Dextran (10,000 MW) conjugated with tetramethyl-rhodamine were from Invitrogen (Carlsbad, CA). Alexa Fluor 633 was conjugated to iron-loaded Tf (Sigma) following the manufacturer's instruction. Mouse anti–α-tubulin and anti–FLAG M2-Cy3 monoclonal antibodies were from Sigma. Goat anti-STARD4 (T-17) polyclonal antibody and mouse anti–SREBP-2 (1C6) monoclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-Giantin polyclonal antibody (Covance, Princeton, NJ) was used as a marker for the Golgi. [14C]oleate was from Perkin Elmer-Cetus (Waltham, MA).
Cell culture and transfection
Cells were grown as a monolayer in a humidified incubator at 37°C in a 5% CO2 atmosphere. The cell culture media were supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. The human osteosarcoma cell line U2OS was grown in McCoy's medium supplemented with 10% fetal bovine serum (FBS). The CHO-derived TRVb1 cells (McGraw et al., 1987) were grown in bicarbonate-buffered Ham's F12 medium containing 200 μg/ml G418 and 5% FBS. CHO cells stably expressing GFP-SCAP were grown in DMEM/Ham's F12 (1:1) with 400 μg/ml G418 and 1% lipoprotein-deficient serum (LPDS). Cells for wide-field microscopy were plated on 35-mm plastic dishes, the bottoms of which were replaced with poly-d-lysine-coated coverslips. For STARD4 expression, U2OS cells were transiently transfected with pCMV-Tag2B plasmid (Stratagene, Santa Clara, CA) containing FLAG-tagged hSTARD4, or with pEGFP-C1-hSTARD4 by electroporation using the Gene Pulser II system (Bio-Rad, Hercules, CA), or, alternatively, with HiPerFect transfection reagent (Qiagen, Valencia, CA). The same vectors containing STARD5 cDNA were used to obtain FLAG-tagged hSTARD5 and GFP-STARD5 expression. Empty vector served as a negative control. Studies were performed 1 or 2 d after transfection.
Fluorescence microscopy
Wide-field fluorescence microscopy and digital image acquisition were carried out using a Leica DMIRB microscope equipped with an Andor iXonEM Blue EMCCD camera driven by MetaMorph Imaging System software (Universal Imaging/Molecular Devices, Sunnyvale, CA). All images were acquired using oil-immersion objectives (40×, 1.25 NA, or 63×, 1.36 NA) with 2 × 2 pixel binning. Cells being imaged were in PBS when fixed, or Medium A (150 mM NaCl, 20 mM HEPES, pH 7.55, 1 mM CaCl2, 5 mM KCl, 1 mM MgCl2) when alive. When indicated, Medium A was supplemented with 2 mg/ml glucose (Medium A/glucose). DHE was imaged using a filter cube obtained from Chroma Technology (Bellows Falls, VT; 335-nm [20-nm band pass] excitation filter, 365-nm-long pass dichromatic filter, and 405-nm [40-nm band pass] emission filter]. Standard TRITC, FITC, and Cy5 cubes were obtained from Chroma. To detect expression of FLAG-tagged protein constructs, cells were fixed and permeabilized with 0.05% saponin, and then subjected to immunofluorescence using either mouse anti–FLAG M2-Cy3 or rabbit anti-FLAG (Sigma). Filipin staining was performed as previously described (Pipalia et al., 2006), and was imaged using an A4 filter cube (Leica, Wetzlar, Germany). Fluorescence cross-over from one channel to another was measured using single-labeled samples of each probe and found to be insignificant. Images were background-corrected as previously described (Hao et al., 2002).
RNAi
For STARD4 RNAi, siRNA with target sequence 5′-TGGTCAGCTTTGGAATATAA-3′ was mixed with HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions, and added to cells in suspension before plating. To generate a STARD4 RNAi–resistant construct, a synonymous substitution in the siRNA-targeting sequence, in which CTTTGG (corresponding to amino acids LW) is replaced by CTATGG (also LW), was produced by QuickChange Site-Directed Mutagenesis (Stratagene). For STARD4 RNAi in defined cells, pGFP-V-RS vectors (OriGene, Rockville, MD) having a short-hairpin RNA (shRNA) expression cassette with the target gene–specific sequence mentioned above, were transfected in U2OS cells according to manufacturer's instructions for 48 h.
ACAT assay
U2OS cells were plated in 24-well plates after transfection and cultivated in growth medium for 24 h for STARD4 overexpression or for 48 h for STARD4 silencing. Cells were then incubated with fresh medium containing [14C]oleate (1 μCi/ml) bound to bovine serum albumin, prepared as previously described (Goldstein et al., 1983), with or without ACAT inhibitor 58035 (30 μg/ml) for 6 h. When indicated, 500 μM MCD/cholesterol was added to the culture medium during the final 2 h of the incubation. Lipids were then extracted twice with hexane/2-propanol (3:2) and resolved on normal-phase thin-layer chromatography plates using the solvent system: hexane /diethyl ether/glacial acetic acid (80:20:1). The plates were exposed to phosphor-imaging screens (GE Healthcare, Piscataway, NJ), and scanned using a Typhoon Trio imager (GE Healthcare). The spots corresponding to cholesteryl [14C]oleate were background-corrected and quantified using MetaMorph. Protein concentration after solubilization with 0.5 M NaOH was determined by the Bio-Rad DC protein assay.
STARD4 expression and purification
STARD4 and STARD4 mutants (K49A/K52A and K219A) were purified essentially as previously described (Romanowski et al., 2002), with some modifications. Briefly, mouse STARD4 cloned in pGEX-6P-1 vector was expressed in Escherichia coli BL21(DE3) cells. After expression, bacteria were lysed by sonication in 10 mM Tris (pH 8), 140 mM NaCl, 5 mM dithiothreitol (DTT), 1 mM EDTA, supplemented with 1 mM phenylmethylsulfonyl fluoride, a cocktail of anti-proteases (Roche), 0.1% Triton X-100, and 0.25 mg/ml lysozyme. After a short incubation, the disrupted bacteria were further homogenized using a Dounce homogenizer. The supernatant was loaded on a glutathione Sepharose 4B column, and STARD4 in fusion with glutathione S-transferase (GST) was eluted in 50 mM Tris (pH 8), 140 mM NaCl, and 5 mM DTT, supplemented with 10 mM reduced glutathione. The N-terminal GST tag was removed by digestion with PreScission protease (GE Healthcare), which was followed by another passage through a glutathione Sepharose 4B column. STARD4-containing fractions were further purified by gel filtration on a Superose 6 column (GE Healthcare).
Liposomes
Lipids in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL), except DHE (powder, Sigma). A dried film was prepared by evaporation of a mixture of the indicated lipids in chloroform and resuspended in 50 mM HEPES (pH 7.2) and 120 mM potassium acetate. After five cycles of thawing and freezing in liquid nitrogen, the liposome suspension was extruded sequentially through 0.4- and 0.1-μm (pore size) polycarbonate filters using a hand extruder (Avanti) at a final lipid concentration of 1 mM. Liposomes were stored at room temperature and used the same day. Liposomes referred to as “donors” or “acceptors” were used in a sterol-transfer assay (described in the following section). The composition of donor liposomes was 31 mol% POPC, 23 mol% POPE, 23 mol% POPS, and 23 mol% DHE. The composition of acceptor liposomes was 70 mol% POPC, 7 mol% POPE, 15 mol% liver PI, 5 mol% POPS, and 3 mol% Dansyl-PE. In some experiments, anionic lipids were replaced by the neutral lipid POPC. Donor and acceptor liposome compositions were intended to approach that of the PM (inner leaflet) and ER membranes, respectively.
Sterol-transfer assay
The sterol transport activity of STARD4 was measured by a FRET assay. Experiments were performed in quartz cuvettes (100 μl) in a buffer (50 mM HEPES, pH 7.2, 120 mM potassium acetate) equilibrated at 37°C on a SpectraMax M2 fluorometer (MDS Analytical Technologies, Sunnyvale, CA). The assay required donor liposomes containing DHE and acceptor liposomes containing a low percentage of dansyl-labeled lipid (dansyl-PE; John et al., 2002). Sterol transfer was monitored by the appearance of FRET emission measured at 500 nm with excitation at 340 nm.
Cell microinjection
Cytosolic microinjections were performed using back-loaded borosilicate glass capillaries and a Narishige micromanipulator. The microinjection solution was 10 mM HEPES (pH 6.9), 140 mM KCl, with the addition of 0.5 mg/ml rhodamine-dextran and, when indicated, 2.5 mM MCD. To determine the number of MCD molecules injected in a single cell, the cellular integrated fluorescence intensity of the coinjected dye, rhodamine-dextran, was compared with a standard curve obtained by imaging rhodamine-dextran in aqueous solutions of known concentrations placed in a hemocytometer chamber. Therefore the number of rhodamine-dextran molecules injected could be calculated, and the number of cellular MCD molecules was determined based on this. Images of the standard solutions were acquired using imaging parameters identical to those used in microinjection experiments.
Free cholesterol measurement by GC/MS
Cellular lipids were extracted twice with hexane/2-propanol (3:2). During the first extraction, β-sitosterol was added as an internal standard for quantification. Dried lipids were resuspended in hexane and separated on a Varian Factor Four capillary column, using a Varian 400 GC/MS/MS system. The system settings were used as described previously (Rosenbaum et al., 2010). The protein concentration after solubilization with 0.5 M NaOH was determined by the Bio-Rad DC protein assay.
SREBP-2 processing and immunoblot analysis
U2OS cells transfected with STARD4 siRNA and untransfected control were plated and maintained in growth medium with 5% FBS for 48 h. Cells transfected with STARD4 cDNA were maintained in the same medium for 24 h. Cells were then incubated for 2 h at 37°C in McCoy's medium with 1% LPDS, 10 μM mevinolin, 10 μM N-acetylleucylleucylnorleucinal (ALLN), which was supplemented with 1 mM MCD or 500 μM MCD/cholesterol to deplete or load the cells with cholesterol, respectively. Cells were subsequently harvested at 0°C and lysed in 10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS, complemented with an anti-protease cocktail (leupeptin, pepstatin A, aprotinin, ALLN, and Pefabloc) for 30 min at 4°C. The samples were cleared by centrifugation (16,000 × g, 20 min) and subjected to SDS–PAGE. After SDS–PAGE, the proteins were transferred to nitrocellulose membrane (Whatman, Piscataway, NJ) and incubated for 16 h at 4°C with anti-SREBP-2, anti-STARD4, or anti–α-tubulin antibodies. Infrared fluorescent dye-coupled secondary antibodies (LI-COR Biosciences, Lincoln, NE) were incubated following the manufacturer's instructions. Immunoblots were visualized and quantified with the Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Rebecca Juliano for making CHO cells stably expressing GFP-SCAP. We thank J. David Warren and the Milstein Synthetic Chemistry Core Facility for sharing their GC/MS/MS system and other instruments, and Geri Kreitzer for sharing a Flaming/Brown micropipette puller. This work was supported by National Institutes of Health grants R37-DK27083 to FRM and R37-AG019391 to D.E.
Abbreviations used:
- ACAT
cholesterol acyl-transferase
- ALLN
N-acetylleucylleucylnorleucinal
- CE
cholesteryl ester
- DOPC
C18:1-C18:1-phosphatidylcholine
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERC
endocytic recycling compartment
- FBS
fetal bovine serum
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- LPDS
lipoprotein-deficient serum
- MCD
methyl-β-cyclodextrin
- OSBP
oxysterol-binding protein
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PM
plasma membrane
- POPC
C16:0-C18:1-phosphaditylcholine
- PS
phosphatidylserine
- RNAi
RNA interference
- SCAP
SREBP cleavage-activating protein
- shRNA
short-hairpin RNA
- siRNA
small interfering RNA
- SREBP
sterol regulatory element binding protein
- START
steroidogenic acute regulatory-related lipid transfer
- START
StAR-related lipid transfer
- Tf
transferrin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0372) on September 7, 2011.
REFERENCES
- Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC. The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein. J Biol Chem. 2001;276:4261–4269. doi: 10.1074/jbc.M006279200. [DOI] [PubMed] [Google Scholar]
- Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- Baumann NA, Sullivan DP, Ohvo-Rekila H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 2009;50:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron KM, Soo SC, Parker KL. Targeted disruption of StAR provides novel insights into congenital adrenal hyperplasia. Endocr Res. 1998;24:827–834. doi: 10.3109/07435809809032693. [DOI] [PubMed] [Google Scholar]
- Fairn GD, McMaster CR. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell Mol Life Sci. 2008;65:228–236. doi: 10.1007/s00018-007-7325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Grandl M, Schmitz G. Fluorescent high-content imaging allows the discrimination and quantitation of E-LDL-induced lipid droplets and Ox-LDL-generated phospholipidosis in human macrophages. Cytometry A. 2010;77:231–242. doi: 10.1002/cyto.a.20828. [DOI] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV Jr, Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M, Lin SX, Karylowski OJ, Wustner D, McGraw TE, Maxfield FR. Vesicular and non-vesicular sterol transport in living cells. The endocytic recycling compartment is a major sterol storage organelle. J Biol Chem. 2002;277:609–617. doi: 10.1074/jbc.M108861200. [DOI] [PubMed] [Google Scholar]
- Heino S, Lusa S, Somerharju P, Ehnholm C, Olkkonen VM, Ikonen E. Dissecting the role of the Golgi complex and lipid rafts in biosynthetic transport of cholesterol to the cell surface. Proc Natl Acad Sci USA. 2000;97:8375–8380. doi: 10.1073/pnas.140218797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- John K, Kubelt J, Muller P, Wustner D, Herrmann A. Rapid transbilayer movement of the fluorescent sterol dehydroergosterol in lipid membranes. Biophys J. 2002;83:1525–1534. doi: 10.1016/S0006-3495(02)73922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Steck TL. Cholesterol homeostasis and the escape tendency (activity) of plasma membrane cholesterol. Prog Lipid Res. 2008;47:319–332. doi: 10.1016/j.plipres.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Ye J, Rigney M, Steck TL. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J Lipid Res. 1999;40:2264–2270. [PubMed] [Google Scholar]
- Lehto M, Olkkonen VM. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim Biophys Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Cruz D, Asamoah N, Buxbaum A, Sohar I, Lobel P, Maxfield FR. Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol Biol Cell. 2007;18:1490–1496. doi: 10.1091/mbc.E06-10-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Menon AK. Intracellular sterol transport and distribution. Curr Opin Cell Biol. 2006;18:379–385. doi: 10.1016/j.ceb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Mondal M. Sterol and lipid trafficking in mammalian cells. Biochem Soc Trans. 2006;34:335–339. doi: 10.1042/BST0340335. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J Cell Biol. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Maxfield FR. Intracellular sterol dynamics. Biochim Biophys Acta. 2009;1791:636–645. doi: 10.1016/j.bbalip.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL. Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim Biophys Acta. 2007;1771:663–676. doi: 10.1016/j.bbalip.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Murcia M, Faraldo-Gomez JD, Maxfield FR, Roux B. Modeling the structure of the StART domains of MLN64 and StAR proteins in complex with cholesterol. J Lipid Res. 2006;47:2614–2630. doi: 10.1194/jlr.M600232-JLR200. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Pipalia NH, Huang A, Ralph H, Rujoi M, Maxfield FR. Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J Lipid Res. 2006;47:284–301. doi: 10.1194/jlr.M500388-JLR200. [DOI] [PubMed] [Google Scholar]
- Prinz WA. Non-vesicular sterol transport in cells. Prog Lipid Res. 2007;46:297–314. doi: 10.1016/j.plipres.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Prinz WA. The diverse functions of oxysterol-binding proteins. Annu Rev Cell Dev Biol. 2010;26:157–177. doi: 10.1146/annurev.cellbio.042308.113334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegelhaupt JJ, Waase MP, Garbarino J, Cruz DE, Breslow JL. Targeted disruption of steroidogenic acute regulatory protein D4 leads to modest weight reduction and minor alterations in lipid metabolism. J Lipid Res. 2010;51:1134–1143. doi: 10.1194/jlr.M003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Agudo D, Ren S, Wong E, Marques D, Redford K, Gil G, Hylemon P, Pandak WM. Intracellular cholesterol transporter StarD4 binds free cholesterol and increases cholesteryl ester formation. J Lipid Res. 2008;49:1409–1419. doi: 10.1194/jlr.M700537-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum AI, Zhang G, Warren JD, Maxfield FR. Endocytosis of beta-cyclodextrins is responsible for cholesterol reduction in Niemann-Pick type C mutant cells. Proc Natl Acad Sci USA. 2010;107:5477–5482. doi: 10.1073/pnas.0914309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Adams RM, Maxwell KN, Breslow JL. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J Biol Chem. 2005;280:19410–19418. doi: 10.1074/jbc.M501778200. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Breslow JL. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem. 2003;278:22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- Strauss JF III, Kishida T, Christenson LK, Fujimoto T, Hiroi H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol Cell Endocrinol. 2003;202:59–65. doi: 10.1016/s0303-7207(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Sullivan DP, Ohvo-Rekila H, Baumann NA, Beh CT, Menon AK. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem Soc Trans. 2006;34:356–358. doi: 10.1042/BST0340356. [DOI] [PubMed] [Google Scholar]
- Tabas I, Rosoff WJ, Boykow GC. Acyl coenzyme A:cholesterol acyl transferase in macrophages utilizes a cellular pool of cholesterol oxidase-accessible cholesterol as substrate. J Biol Chem. 1988;263:1266–1272. [PubMed] [Google Scholar]
- Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- Urbani L, Simoni RD. Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J Biol Chem. 1990;265:1919–1923. [PubMed] [Google Scholar]
- van Meer G, Sprong H. Membrane lipids and vesicular traffic. Curr Opin Cell Biol. 2004;16:373–378. doi: 10.1016/j.ceb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.