Targeted deletion of Actb demonstrates that the β-actin gene, in contrast to the γ-actin gene, is an essential gene uniquely required for cell growth and migration. Cell motility and growth defects in β-actin–knockout primary cells are due to a specific role for β-actin in regulating gene expression through control of the cellular G-actin pool.
Abstract
Ubiquitously expressed β-actin and γ-actin isoforms play critical roles in most cellular processes; however, their unique contributions are not well understood. We generated whole-body β-actin–knockout (Actb−/−) mice and demonstrated that β-actin is required for early embryonic development. Lethality of Actb−/− embryos correlated with severe growth impairment and migration defects in β-actin–knockout primary mouse embryonic fibroblasts (MEFs) that were not observed in γ-actin–null MEFs. Migration defects were associated with reduced membrane protrusion dynamics and increased focal adhesions. We also identified migration defects upon conditional ablation of β-actin in highly motile T cells. Of great interest, ablation of β-actin altered the ratio of globular actin (G-actin) to filamentous actin in MEFs, with corresponding changes in expression of genes that regulate the cell cycle and motility. These data support an essential role for β-actin in regulating cell migration and gene expression through control of the cellular G-actin pool.
INTRODUCTION
Actins are an essential component of the cytoskeleton, with critical roles in a wide range of cellular processes, including cell migration, cell division, and the regulation of gene expression. These functions are attributed to the ability of actin to form filaments that can rapidly assemble and disassemble according to the needs of the cell. There exist six different but highly conserved actin isoforms in vertebrates (Rubenstein, 1990). Four of these isoforms are expressed primarily in striated (αsk and αca) and smooth (αsm and γsm) muscle cells, whereas the two cytoplasmic β-actin and γ-actin isoforms are ubiquitously expressed. Each isoform is the product of a separate gene, with Actb and Actg1 encoding for β-actin and γ-actin, respectively. Conserved from birds to mammals, β-actin and γ-actin differ at only four biochemically similar amino acid residues, suggesting evolutionary pressure to maintain these small sequence differences. In fact, it was recently demonstrated that these amino acid differences confer unique biochemical properties between the two isoforms (Bergeron et al., 2010). How these distinct properties translate into functional specificity within the cell, however, remains elusive.
Despite longstanding ambiguity regarding the significance of the cytoplasmic actin isoforms, it has long been believed that β-actin and γ-actin isoforms confer unique biological functions. Recent genetic models provided considerable support to this idea, as β-actin– and γ-actin–deficient mice have distinct phenotypes. It was first reported that mice hypomorphic for β-actin die during development, of uncharacterized defects (Shawlot et al., 1998; Shmerling et al., 2005). In contrast, γ-actin–null (Actg1−/−) mice are viable; however, they present with some notable phenotypes (Belyantseva et al., 2009). Specifically, γ-actin knockout resulted in developmental delays, reduced postnatal and adult survival, and progressive myopathy (Sonnemann et al., 2006; Belyantseva et al., 2009; Bunnell and Ervasti, 2010). In addition, β-actin– and γ-actin–deficient hair cells display different patterns of progressive hearing loss and distinct stereocilia pathologies (Perrin et al., 2010). The fact that total actin expression levels are maintained in Actg1−/− tissues suggests that these phenotypes are due to changes in actin isoform composition and not the concentration of total actin.
One of the best-studied roles for actin is in cell motility, a fundamental process essential for embryonic development, wound healing, and immune responses. Cell migration is largely driven by the polymerization of actin at the leading edge, which provides the protrusive forces that push the membrane forward (Pollard and Borisy, 2003). We previously demonstrated that γ-actin is not required for cell motility (Bunnell and Ervasti, 2010), which is consistent with evidence that β-actin is the predominant isoform driving cell migration. First, one study demonstrated that overexpression of β-actin leads to an increase in membrane protrusions and cell migration (Peckham et al., 2001). In addition, several studies showed that β-actin is enriched at the leading edge of migrating cells, whereas γ-actin is more uniformly distributed throughout the cell (Otey et al., 1986; Rubenstein, 1990; Hoock et al., 1991; Hill and Gunning, 1993; Bassell et al., 1998; Micheva et al., 1998). Finally, it has been shown that β-actin but not γ-actin is N-terminally arginylated in vivo (Zhang et al., 2010), which is believed to regulate lamella formation in motile cells (Karakozova et al., 2006). A recent study challenged these reports by demonstrating that knockdown of β-actin in fibroblasts had no effect on the speed of cell migration (Dugina et al., 2009). Despite the controversy surrounding the role of β-actin in cell migration, the effects of a targeted deletion of Actb on cell motility have never been examined.
Cell motility is an important process during the entire life span of vertebrates but is especially critical during many stages of development. Using Actg1−/− mice, we previously demonstrated that γ-actin is not required for embryogenesis, suggesting that other actin isoforms can compensate for the loss of γ-actin (Bunnell and Ervasti, 2010). In contrast, we show here that β-actin–null (Actb−/−) mice are early embryonic lethal, indicating that β-actin is an essential gene required for embryonic development. The lethality in Actb−/− mice is likely due to defects in cell growth and migration, as these processes were severely impaired in β-actin–knockout primary mouse embryonic fibroblasts (MEFs). We observed a significant decrease in migration velocity together with reduced leading-edge dynamics in β-actin–knockout MEFs but not Actg1−/− MEFs. Conditional ablation of β-actin in T cells also led to impaired migration, indicating a conserved role for β-actin in cell motility. Of interest, we found that ablation of β-actin in MEFs leads to a decrease in the ratio of globular actin (G-actin) to filamentous actin (F-actin), together with altered expression of genes that regulate the cell cycle and cell migration. These data provide evidence that β-actin may regulate gene expression through specific control of the cellular G-actin pool.
RESULTS
β-Actin is essential for in vivo survival
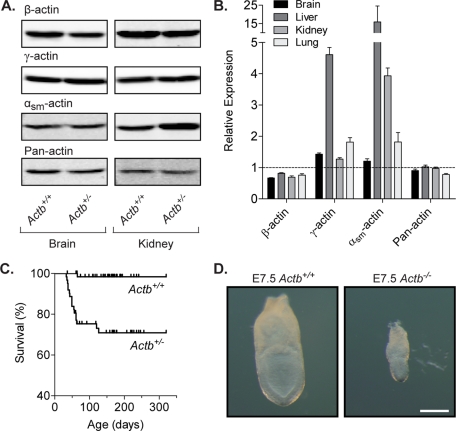
To determine the requirement for β-actin in vivo, we generated a null allele by deleting exons 2 and 3 of the Actb locus, thereby removing the translational start site. Using quantitative Western blot analysis, we found that lysates from several tissues of mice heterozygous for the null allele (Actb+/−) were hypomorphic for β-actin (Figure 1, A and B). β-Actin immunoreactivity was reduced 18% in liver, 24% in lung, 31% in kidney, and 33% in brain lysates from Actb+/− mice compared with identically prepared and loaded lysates from Actb+/+ mice (Figure 1B). Although compensatory up-regulation of γ-actin and αsm-actin was observed in all tissues, total actin expression was restored to control levels only in the liver and kidneys and not in the brain or lungs (Figure 1, A and B). Despite the modest decrease in β-actin across several tissues, ∼30% of Actb+/− mice died between 5 and 18 wk of age (Figure 1C), primarily due to hydrocephalus resulting from unknown causes. Homozygous deletion of Actb resulted in embryonic lethality, as evidenced by the fact that no Actb−/− mice were live born. Genotyping of embryos at various gestational ages from heterozygous crosses revealed lower-than-expected frequencies of Actb−/− mice as early as embryonic day (E) 7.5, with no Actb−/− mice surviving beyond E8.5 (Table 1). Already by E7.5 Actb−/− embryos were considerably smaller than their wild-type counterparts (Figure 1D). The embryonic lethality of Actb−/− mice indicates that β-actin is an essential gene during early stages of development.
FIGURE 1:
In vivo characterization of β-actin deficiency. (A) Representative immunoblots of SDS extracts from Actb+/+ and Actb+/− adult brain and kidney tissue probed with antibodies specific for β-actin, γ-actin, αsm-actin, or pan-actin. (B) Protein levels were quantified from brain, liver, kidney, and lung tissues from three separate experiments blotted in triplicate using the LI-COR Odyssey imaging system. Bar graphs represent relative expression levels in Actb+/− tissues as compared with Actb+/+ levels (mean ± SEM). (C) Kaplan–Meier survival curve of Actb+/+ and Actb+/− mice from 0 to 320 d of age. Tick marks represent censored animals. Survival curves are significantly different (p < 0.0001, log-rank test; n ≥ 62 for each genotype). (D) Representative images of E7.5 Actb+/+ and Actb−/− embryos. Scale bar, 200 μm.
TABLE 1:
Early embryonic lethality in Actb−/− mice.
| Age | Number | Actb+/+ | Actb+/− | Actb−/− |
|---|---|---|---|---|
| E7.5* | 51 | 37% | 57% | 6% |
| E8.5* | 58 | 36% | 57% | 7% |
| E9.5* | 26 | 31% | 69% | 0% |
| E13.5* | 27 | 44% | 56% | 0% |
*p < 0.05, chi-square analysis.
Establishing primary cultures of β-actin–knockout MEFs
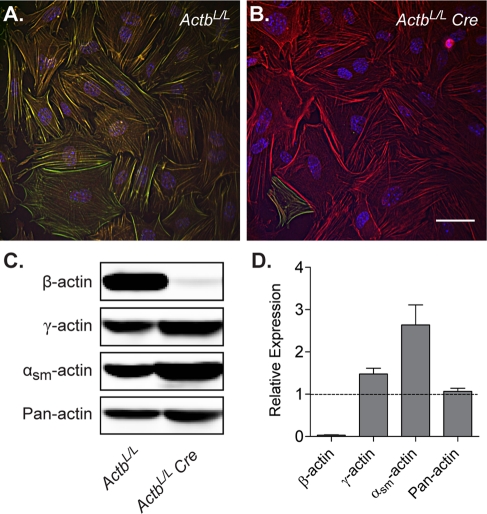
To assess whether there is a unique function for β-actin in cell behavior, we established a system for knocking out β-actin in primary MEFs. Mice carrying floxed alleles of Actb (ActbL/L; Perrin et al., 2010) were crossed to the CAGG-CreERTM transgenic line, which expresses a tamoxifen-inducible Cre recombinase under the control of a ubiquitous promoter (Hayashi and McMahon, 2002). Primary MEFs were generated from ActbL/L and ActbL/L CAGG-CreERTM (herein referred to as ActbL/L Cre) E13.5 embryos and treated with tamoxifen for three consecutive days to induce recombination in Cre-expressing cells. Although shorter treatment times with tamoxifen were also tested, we found that they resulted in less efficient knockout. After tamoxifen treatment, cells were cultured for an additional 4 d to allow for protein turnover. Immunofluorescence analysis with a β-actin–specific antibody demonstrated the absence of β-actin expression in more than 95% of ActbL/L Cre cells (Figure 2, A and B). The small population of β-actin–positive cells likely represents those that did not undergo recombination. Efficient ablation of β-actin was further confirmed by Western blot analysis, which revealed total β-actin protein levels in ActbL/L Cre cells to be <5% of that in ActbL/L cells (Figure 2, C and D). As a result of compensatory up-regulation of other actin isoforms, total actin expression was maintained in ActbL/L Cre cells (Figure 2, C and D), indicating that any phenotypes resulting from knockout of β-actin are due to changes in actin isoform composition and not the concentration of actin.
FIGURE 2:
Efficient knockout of β-actin in MEFs. (A, B) Representative images of ActbL/L (A) and ActbL/L Cre (B) MEFs cultured overnight and costained with antibodies to β-actin (green) and γ-actin (red). Scale bar, 50 μm. (C) Representative immunoblots of cell lysates from ActbL/L and ActbL/L Cre MEFs probed with pan actin or actin isoform–specific antibodies. (D) Relative expression levels in ActbL/L Cre cells as compared with ActbL/L cells using quantitative Western blot analysis (n = 5; mean ± SEM).
β-Actin–knockout MEFs are growth impaired
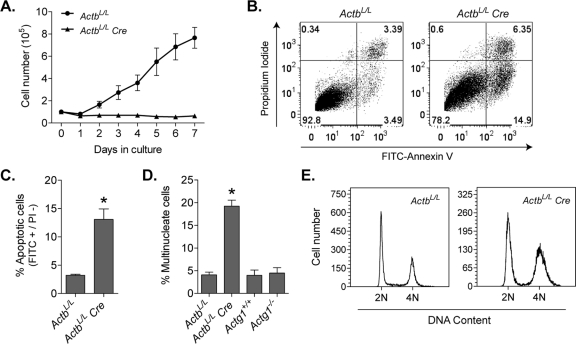
A modest decrease in cell viability was previously observed in Actg1−/− MEFs (Bunnell and Ervasti, 2010); therefore we assessed the growth properties of β-actin–knockout cells. Analysis of growth kinetics revealed ActbL/L Cre cells to be severely growth impaired. Whereas ActbL/L cells exhibited a typical growth pattern, ActbL/L Cre cells failed to increase in number over a 7-d period (Figure 3A). Contributing to this zero growth rate was increased apoptosis, as the percentage of apoptotic cells was 13.1% in ActbL/L Cre MEFs compared with 3.2% in control MEFs (Figure 3, B and C). Multinucleate cells were also significantly increased in ActbL/L Cre MEFs but not in Actg1−/- MEFs (Figure 3D). This result was further corroborated by flow cytometric analysis of DNA content, which revealed a larger percentage of cells with 4N DNA content in β-actin–knockout cells compared with control cells (Figure 3E). These data suggest that cell division may specifically require β-actin.
FIGURE 3:
Severe growth deficiency in the absence of β-actin. (A) Growth curves of ActbL/L and ActbL/L Cre MEFs (n = 4). Cell numbers were significantly decreased in ActbL/L Cre MEFs from day 2 onward as determined by an unpaired t test at each time point. (B) Dot plots from FITC-annexin V flow cytometric analyses. Lower right box, early apoptotic cells; upper right, dead cells. (C) Mean percentage of apoptotic cells, defined as FITC positive and propidium iodide negative (n = 4). (D) Mean percentage of cells containing multiple nuclei from at least three or four independent experiments. More than 250 cells were counted per experiment. (E) Representative traces from flow cytometric analyses of DNA content by propidium iodide staining. Asterisks denote significant differences (p < 0.05); error bars, SEM.
Migration defects in β-actin–knockout MEFs
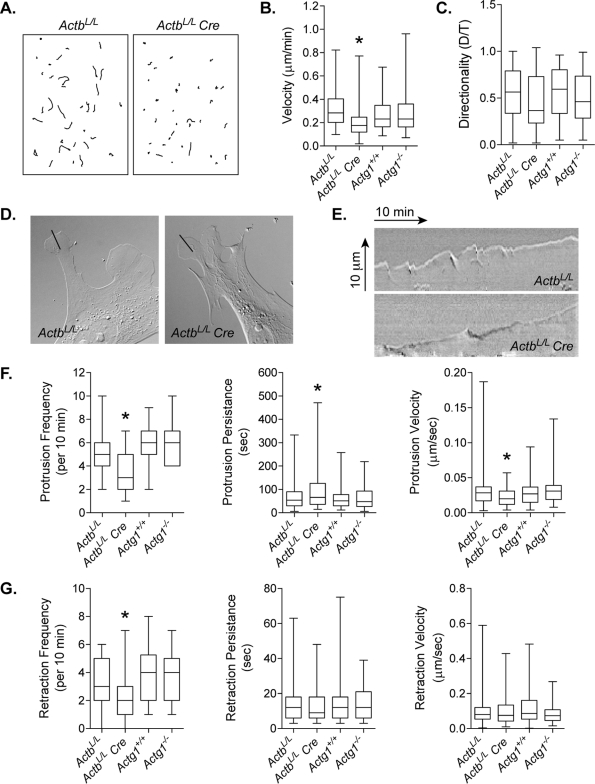
γ-Actin was previously shown to be dispensable for migration in vivo and in vitro (Bunnell and Ervasti, 2010); therefore we investigated whether β-actin might be important for cell migration. Given the growth impairment of ActbL/L Cre cells, confluent cell monolayers for wound-healing assays were difficult to achieve; therefore we used a random cell migration assay. Individual cells were tracked at 10-min intervals for 4 h and the velocity of migration and directional persistence calculated. A clear difference in the migratory ability of β-actin–knockout cells was observed (Figure 4A). In contrast to Actg1−/− cells, migration velocity was significantly decreased in ActbL/L Cre cells compared with controls (Figure 4B). Directional persistence was comparable across all genotypes (Figure 4C).
FIGURE 4:
Impaired migration in β-actin–knockout MEFs. (A) Randomly migrating cells were tracked at 10-min intervals for 4 h. Representative examples of individual migration tracks of ActbL/L and ActbL/L Cre MEFs combined into a single figure. (B) Quantification of migration velocity (n ≥ 88 cells per genotype). (C) Directionality of cell migration was calculated as the linear distance (D) over the total track distance (T) of a cell. (D, E) Individual frames (D) and kymographs (E) from DIC time-lapse movies of ActbL/L and ActbL/L Cre MEFs. Kymographs show lamellipodial activity along the lines in respective DIC images. Images in D represent the last frame of the 10-min movie. Line, 10 μm. (F, G) Quantification of kymographs showing the frequency, persistence, and velocity of protrusion (F) and retraction (G) events. For all box and whisker plots, whiskers indicate maximum and minimum values, the box represents the 25–75th quartile, and the line indicates the median. Asterisks denote significant differences (p < 0.05) from all other genotypes.
It has been suggested that β-actin is the major actin isoform driving membrane protrusions at the leading edge of migrating cells. To test this idea, we analyzed kymographs from active lamellipodia of control, ActbL/L Cre, and Actg1−/− cells in order to assess leading-edge dynamics in the absence of β-actin or γ-actin (Figure 4, D–G). Kymographs generated from ActbL/L Cre lamellipodia revealed that the frequency of protrusion and retraction events was significantly decreased compared with control lamellipodia (Figure 4, F and G). Correspondingly, the average duration of protrusive events was increased in ActbL/L Cre cells (Figure 4F). Most notable, however, is that protrusion velocity was significantly decreased in lamellipodia from ActbL/L Cre cells (Figure 4F). The duration and velocity of retraction events were not affected by the loss of β-actin (Figure 4G). In contrast to ActbL/L Cre cells, leading-edge dynamics in Actg1−/- cells was comparable to that in controls (Figure 4, F and G). Therefore it appears that β-actin has a specific function in membrane protrusion dynamics during cell migration.
Conditional β-actin knockout in CD4-positive T cells results in defective migration
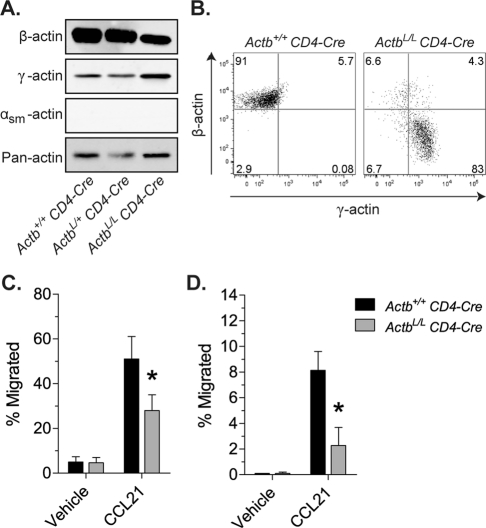
To determine whether β-actin is required for cell migration in other cell types, we used the CD4-Cre driver (Lee et al., 2001) to knock out β-actin from developing thymocytes. Western blot analysis of unfractionated thymocytes from ActbL/L CD4-Cre mice demonstrates decreased β-actin expression and increased γ-actin expression compared with thymocytes from Actb+/+ CD4-Cre animals (Figure 5A). As with fibroblasts, total actin levels were similar to those in control cells (Figure 5A). Of interest, we were unable to detect any of the other actin isoforms in control or β-actin–knockout T cells (Figure 5A). Therefore β-actin–knockout T cells likely express only γ-actin, mitigating potential complications arising from up-regulation of αsm-actin as seen in fibroblasts. The remaining β-actin signal in unfractionated thymocytes is consistent with the caveat that the majority of T cells in the thymus have only recently begun to express CD4 and thus have not had sufficient time for complete β-actin turnover. To compare cells at a later stage of maturation, we used flow cytometry to analyze CD4 single positive/T cell receptor β high (CD4sp/TCRβhi) T cells (Zuniga-Pflucker, 2004). Intracellular colabeling of CD4sp/TCRβhi cells further confirmed a decrease in β-actin expression and a compensatory increase in γ-actin expression in β-actin–knockout T cells (Figure 5B).
FIGURE 5:
Conditional β-actin knockout in CD4-positive T cells leads to impaired migration. (A) Immunoblots of lysates from unfractionated thymocytes probed with pan-actin or actin isoform–specific antibodies. (B) Dot plots from flow cytometric analysis of CD4sp/TCRβhi T cells colabeled with β-actin and γ-actin isoform–specific antibodies showing decreased β-actin expression and increased γ-actin expression in ActbL/L CD4-Cre cells. (C, D) The number of CD4sp/TCRβhi cells migrating through 5-μm-pore (C) or 3-μm-pore (D) transwell filters toward CCL21 as quantified by flow cytometry. Data are expressed as percentage input and reflect an average (± SEM) from three independent experiments, with triplicate values recorded and averaged for each experiment. Asterisks denote significant difference from control cells (p < 0.05).
The migration of developing CD4sp/TCRβhi T cells from the thymic cortex to the medulla is promoted by CC chemokine ligand 21 (CCL21; Takahama, 2006; Yin et al., 2006). Therefore we used an in vitro transwell migration assay to measure basal and chemokine-induced chemotaxis of freshly isolated thymocytes toward CCL21. The percentage of β-actin–knockout CD4sp/TCRβhi T cells responding to CCL21 was significantly reduced compared with control cells (Figure 5, C and D). Therefore, even though fibroblasts and T cells migrate in different ways, both cell types require β-actin, suggesting that β-actin's role in cell motility is universal.
β-Actin and γ-actin colocalize in wild-type MEFs
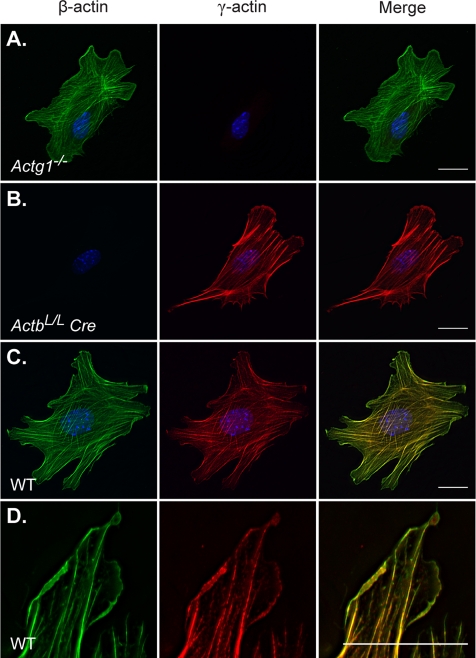
One proposed mechanism for unique functions between the cytoplasmic actin isoforms is differential subcellular localization. Several studies reported distinct localization patterns of β-actin and γ-actin in various cell types (Hoock et al., 1991; Hill and Gunning, 1993; Hill et al., 1994; Bassell et al., 1998; Micheva et al., 1998; Dugina et al., 2009); however, there are discrepancies among studies regarding the precise localization of the two isoforms. Using our β-actin–knockout cells and γ-actin–null cells, we were able to verify isoform specificity of β-actin and γ-actin dye-conjugated antibodies (Figure 6, A and B). Wild-type MEFs, plated under the same culture conditions as used for the migration analysis, demonstrated colocalization of β-actin and γ-actin in all actin-containing structures at the resolution of light microscopy (Figure 6, C and D). Both isoforms were localized to stress fibers, lamellipodial protrusions, and membrane ruffles. Therefore we did not detect differential localization of β-actin and γ-actin in our primary MEFs under the described culture conditions.
FIGURE 6:
β-Actin and γ-actin colocalize in wild-type MEFs. (A–D) Cells were sparsely plated on fibronectin, incubated for 3 h and subsequently costained with antibodies to β-actin (green) and γ-actin (red). Actg1−/− (A) and ActbL/L Cre (B) cells demonstrate the isoform specificity of γ-actin and β-actin antibodies, respectively. In wild-type (WT) cells (C), all actin structures were colabeled with β-actin and γ-actin antibodies. High magnification of membrane protrusions (D) demonstrates colocalization of β-actin and γ-actin in lamellipodia. Cells were maintained in 10% serum. Scale bars, 20 μm.
β-Actin knockout alters gene expression
Actin associates with chromatin-remodeling complexes and all three RNA polymerases, implying a direct role for actin in gene transcription (Zheng et al., 2009). In addition, actin dynamics are known to play a key role in regulating gene expression through the serum response factor (SRF) gene regulatory pathway, target genes of which include actin and actin regulators (Posern and Treisman, 2006). Changes in the expression of proteins that regulate actin would ultimately affect actin-based processes such as cell division and migration; therefore we sought to determine whether gene expression was perturbed in the absence of β-actin or γ-actin, using a Cytoskeleton Regulators quantitative real-time (RT)-PCR array. Of a total 84 genes, 34 were significantly up- or down-regulated in ActbL/L Cre cells, whereas only five were significantly altered in Actg1−/− cells (Supplemental Table S1). In addition, β-actin mRNA levels were decreased 27-fold in ActbL/L Cre cells and increased 1.5-fold in Actg1−/− cells (Supplemental Table S1), consistent with our previously published results demonstrating compensatory up-regulation of β-actin protein in the absence of γ-actin (Bunnell and Ervasti, 2010). Expression of mRNA showing twofold or greater change between control and β-actin–knockout cells notably fell into distinct functional groups. Of the nine genes down-regulated twofold or greater, six are important regulators of the cell cycle (Table 2), which could explain the apparent quiescent state of β-actin–knockout MEFs (Figure 3A). Furthermore, six genes were up-regulated twofold or greater and are important regulators of actin dynamics and myosin activity (Table 2). Three of four transcripts identified as altered by quantitative RT-PCR were also altered when interrogated by Western blot analysis (Supplemental Figure S1).
TABLE 2:
mRNAs showing twofold or greater expression change between ActbL/L and ActbL/L Cre cells fall into distinct functional groups.
| Fold change | p value | ||
|---|---|---|---|
| Cell cycle regulators | |||
| Aurka | Aurora kinase A | −2.0 | 0.00157 |
| Aurkb | Aurora kinase B | −2.7 | 0.00028 |
| Ccnb2 | Cyclin B2 | −3.6 | 0.00044 |
| Cit | Citron | −2.6 | 0.00099 |
| Racgap1 | Rac GTPase-activating protein 1 | −2.7 | 0.00385 |
| Stmn1 | Stathmin 1 | −2.2 | 0.00024 |
| Regulators of actin dynamics | |||
| Dstn | Destrin | 2.8 | 0.00008 |
| Limk2 | LIM motif–containing protein kinase 2 | 2.0 | 0.01898 |
| Regulators of myosin activity | |||
| Cald1 | Caldesmon 1 | 2.1 | 0.01997 |
| Mylk | Myosin, light polypeptide kinase | 4.6 | 0.00023 |
| Mylk2 | Myosin, light polypeptide kinase 2, skeletal muscle | 2.8 | 0.01856 |
| Ppp1r12b | Protein phosphatase 1, regulatory (inhibitor) subunit 12B | 2.9 | 0.00005 |
Data from SABiosciences Mouse Cytoskeleton Regulators RT2 Profiler PCR Array. n = 4.
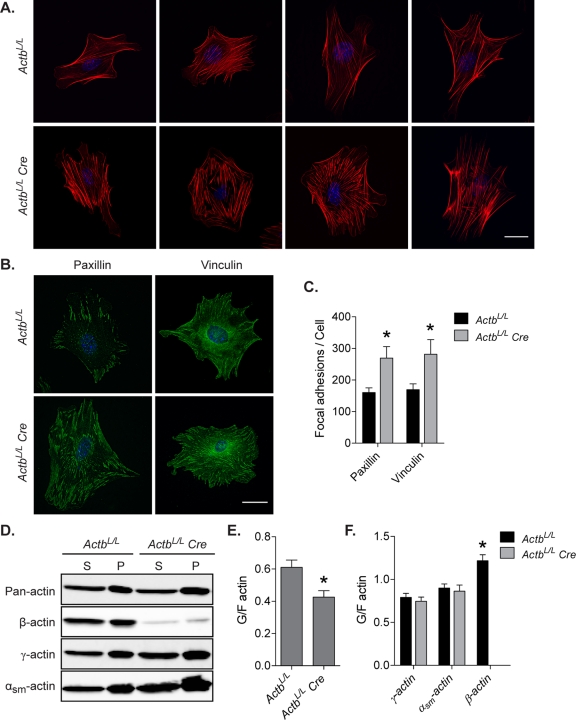
The increased expression of genes that regulate actomyosin contractility is consistent with the observed increase in stress fibers in β-actin–knockout cells labeled with phalloidin (Figure 7A). Because actomyosin contractility and increased stress fiber assembly contribute to focal adhesion formation, we used paxillin and vinculin staining to examine focal adhesions in ActbL/L Cre cells (Figure 7B). As expected, the number of focal adhesions per cell was significantly increased in β-actin–knockout MEFs compared with controls (Figure 7C). Changes in the expression of actin and myosin regulators, together with increased focal adhesion formation, at least in part contribute to the migration defect in β-actin–knockout cells.
FIGURE 7:
Increased focal adhesion formation and F-actin content in β-actin–knockout MEFs. (A) Phalloidin-stained MEFs showing increased stress fibers in ActbL/L Cre cells. (B) Representative images of MEFs labeled with paxillin or vinculin antibodies. (C) Quantification of focal adhesions per cell using integrated morphometry analysis from paxillin- and vinculin-stained images (n ≥ 7 cells per antibody). (D) Immunoblots probed with pan-actin or actin isoform–specific antibodies of supernatant (S) and pellet (P) fractions from cell lysates after high-speed centrifugation. G-actin and F-actin pools are found in the supernatant and pellet fractions, respectively. (E, F) Quantitative Western blot results of ratios of G- to F-actin from immunoblots labeled with a pan-actin antibody (E) or actin isoform–specific antibodies (F) (n = 7, mean ± SEM). Asterisks denote significant differences (p < 0.05). Scale bars, 30 μm.
β-Actin knockout leads to decreased availability of G-actin
Given the observed increase in stress fibers in ActbL/L Cre cells (Figures 6B and 7A) without a change in total actin expression (Figure 2D) and the fact that changes in actin dynamics can affect gene transcription (Posern and Treisman, 2006), we investigated whether the ratio of G- to F-actin was altered in ActbL/L Cre cells. We used high-speed centrifugation to separate the G- and F-actin pools from cell lysates, followed by quantitative Western blot analysis to assess the ratio of G- to F-actin (Figure 7D). In ActbL/L Cre cells the ratio of G- to F-actin was significantly decreased compared with control cells (Figure 7E), consistent with the observed increase in stress fibers in the absence of β-actin. Using actin isoform–specific antibodies, we discovered that in wild-type cells the ratio of G- to F-actin was significantly higher for β-actin than for either γ-actin or αsm-actin isoforms (Figure 7F). In ActbL/L Cre cells neither γ-actin nor αsm-actin shifted their ratio of G- to F-actin to compensate for the loss of β-actin (Figure 7F), resulting in the significant decrease in percentage of G-actin in β-actin–knockout cells (Figure 7E). We previously showed that ratios of G- to F-actin were not altered in Actg1−/− MEFs (Bunnell and Ervasti, 2010); therefore β-actin has a specific role in maintaining the G-actin pool.
DISCUSSION
Actb−/− mice were early embryonic lethal, indicating that β-actin is an essential gene. Our data are consistent with the previously reported embryonic lethality of mice homozygous for hypomorphic alleles of Actb (Shawlot et al., 1998; Shmerling et al., 2005). We observed lower-than-expected frequencies of Actb−/− mice as early as E7.5, with those recovered at this stage appearing to have already begun degenerating. In contrast, Actg1−/− mice are fully viable throughout development, with some lethality occurring postbirth (Belyantseva et al., 2009; Bunnell and Ervasti, 2010). Taken together, the results indicate that β-actin, but not γ-actin, is an essential gene required for early embryogenesis. Rapid cell growth and dynamic cell movements are critical processes during early stages of embryonic development, both of which were shown to be severely impaired in β-actin–knockout cells. Therefore we speculate that the lethality of Actb−/− mice is due largely to impaired cell growth and migration.
We previously demonstrated that Actg1−/− MEFs are mildly growth impaired, resulting from reduced cell survival (Bunnell and Ervasti, 2010). In contrast, β-actin–knockout MEFs were severely growth impaired and exhibited a significant increase in apoptosis. Thus, whereas γ-actin confers advantages for cell growth and survival, β-actin is absolutely essential for maintaining cell growth potential. In addition, we observed an increase in the percentage of multinucleated cells in ActbL/L Cre MEFs but not in Actg1−/− MEFs, indicating a possible unique function for β-actin in cell division. Our data are consistent with a recent report demonstrating that β-actin is important for proper cell shape changes during mitosis (Luxenburg et al., 2011). In addition, it has been shown that β-actin is enriched at the contractile ring in dividing cells (Dugina et al., 2009). Because β-actin is the more dynamic of the two cytoplasmic actin isoforms (Bergeron et al., 2010), it may be specifically required for the rapid reorganization of the actin cytoskeleton during cell division.
We demonstrated here for the first time that targeted deletion of β-actin in two different primary cell types leads to dramatic migration defects, indicating a distinct requirement for β-actin in cell motility. Cell migration results from a coordinated effort between protrusive forces generated by actin polymerization at the leading edge, adhesive forces with the underlying substratum, and actomyosin contractile forces (Mitchison and Cramer, 1996; Lammermann and Sixt, 2009). We found evidence that all three of these migratory components are perturbed in the absence of β-actin. Specifically, in β-actin–knockout MEFs we observed 1) decreased leading-edge dynamics, 2) increased focal adhesion formation, and 3) increased expression of genes that regulate myosin activity. Together, these data indicate that β-actin is important for multiple aspects of cell migration.
Our data from ActbL/L CD4-Cre T cells is consistent with an essential role for β-actin in migration that is highly dependent on protrusion and contraction events. Whether adhesive forces are perturbed in β-actin–knockout T cells remains to be investigated; however, leukocyte migration is believed to be largely independent of underlying adhesiveness (Sixt, 2011). Leukocyte movement is thus primary governed by actin-dependent protrusive and contractile forces (Lammermann and Sixt, 2009; Jacobelli et al., 2010), and we suspect that both of these forces specifically require β-actin in T cells.
One possible explanation for the differential requirement of the two isoforms during cell migration is inherent biochemical differences between β-actin and γ-actin. It was recently demonstrated that β-actin polymerizes and depolymerizes more rapidly than γ-actin in vitro, with the differences between the isoforms more pronounced when in the calcium-bound form (Bergeron et al., 2010). Of note, calcium flickers at the leading edge of migrating cells are believed to promote fibroblast migration (Wei et al., 2009). Therefore our observations of reduced leading-edge dynamics in β-actin–knockout cells could reflect the fact that β-actin is more dynamic than γ-actin in the context of increased calcium at the leading edge.
Another proposed mechanism for the unique role of β-actin in cell motility is differential subcellular localization of the two cytoplasmic actin isoforms. Several studies reported preferential localization of β-actin at the leading edge in migrating cells (Otey et al., 1986; Hoock et al., 1991; Hill and Gunning, 1993; Bassell et al., 1998; Micheva et al., 1998). Furthermore, zipcode-binding protein 1 has been shown to regulate the asymmetric targeting of β-actin mRNA to lamellipodia (Kislauskis et al., 1994; Ross et al., 1997; Farina et al., 2003). Under our cell culture conditions, however, we did not observe differential localization patterns of β-actin and γ-actin in wild-type cells using actin isoform–specific antibodies verified in knockout MEFs. Nonetheless, it remains possible that the ratio of β-actin to γ-actin is elevated at the leading edge of migrating cells. Of note, Bergeron et al. (2010) demonstrated that β-actin and γ-actin could readily copolymerize in vitro, with the polymerization properties of the copolymer reflecting the relative ratio of the two isoforms. It is therefore tempting to speculate that our observations of colocalization indicate copolymers of actin isoforms and that β-actin's incorporation into leading-edge filaments may be necessary to generate copolymers with the proper biochemical properties required for cell motility.
The presence of arginylated β-actin but not γ-actin in vivo (Zhang et al., 2010) might also confer a specific function for β-actin in cell migration. Evidence from one study suggests that arginylation of β-actin helps to regulate lamella formation and motility in fibroblasts (Karakozova et al., 2006). Whether the motility defects observed in β-actin–knockout cells are specifically due to the loss of arginylated actin, however, will require further investigation.
The altered gene expression of cytoskeleton regulators in β-actin–knockout MEFs at least in part contributes to the observed cell growth and motility defects in fibroblasts and suggests a specific role for β-actin in regulating gene expression. Actin has been suggested to play a direct role in gene transcription through its interaction with chromatin-remodeling complexes and all three RNA polymerases; however, the exact function of actin in transcription remains elusive (Zheng et al., 2009). Nonetheless, a recent study demonstrated that a decrease in nuclear β-actin levels mediates laminin 111–induced quiescence in epithelial cells (Spencer et al., 2011), consistent with our observations of a quiescent state in β-actin–knockout MEFs.
A more defined role for actin in regulating gene expression is in the SRF signaling pathway (Posern and Treisman, 2006). Specifically, monomeric actin acts negatively on the SRF pathway by directly binding to and inhibiting the SRF coactivator MAL. Many of the MAL-SRF target genes include actin and actin regulators, and thus it appears that actin itself is part of an autoregulatory feedback circuit that links actin dynamics with transcriptional regulation. We demonstrated here that in wild-type fibroblasts a greater proportion of β-actin is in the monomeric form relative to the other actin isoforms and that β-actin is important for maintaining the G-actin pool. Presumably, the decreased availability of monomeric actin in β-actin–knockout MEFs would lead to increased expression of MAL-SRF target genes. Of note, five of the six known SRF target genes in the Cytoskeleton Regulators RT-PCR array were significantly up-regulated in β-actin–knockout cells (Miano et al., 2007; Table S1). γ-Actin and αsm-actin are also known SRF target genes (Miano et al., 2007), and the expression of both was shown to be elevated in β-actin–knockout cells, suggesting a possible mechanism for the maintenance of total actin. Together, these data provide the first evidence that β-actin might regulate the SRF signaling pathway by specifically controlling the cellular G-actin pool.
Given the lethality of Actb−/− mice, it is not surprising that β-actin homozygous mutations have never been identified in humans. However, there are two independent reports of β-actin heterozygous mutations (Nunoi et al., 1999; Procaccio et al., 2006). In one case the mutation was identified in identical twins and associated with multiple developmental abnormalities (Gearing et al., 2002; Procaccio et al., 2006). In the second case the mutation was associated with recurrent infection resulting from neutrophil dysfunction, including depressed chemotactic responses (Nunoi et al., 1999). In addition, significant increases in β-actin expression levels were observed in highly invasive variants of several different tumor cell lines (Le et al., 1998; Nowak et al., 2005; Popow et al., 2006), indicating a potential role for β-actin in tumor metastasis. These reports agree with our evidence that β-actin is essential for embryogenesis and plays a critical role during cell migration. Analysis of β-actin conditional knockouts will be necessary to further study the in vivo requirement of β-actin during specific developmental processes and in different migratory cell types.
MATERIALS AND METHODS
Animals
Animals were housed and treated in accordance with the standards set by the University of Minnesota Institutional Animal Care and Use Committee. The floxed Actb allele was described previously (Perrin et al., 2010). Mice carrying the floxed allele were crossed to mice expressing EIIa-cre (Holzenberger et al., 2000) to generate Actb+/− mice. Actb+/− mice were backcrossed to C57Bl/6 for 10 generations before intercross breedings were arranged. CAGGCre-ERTM transgenic mice (Hayashi and McMahon, 2002) (obtained from Jackson ImmunoResearch Laboratories, West Grove, PA, on the C57Bl/6 background), Actg1+/− mice (Belyantseva et al., 2009), and CD4-Cre mice (Lee et al., 2001) were previously described. Genotypic analyses were performed by standard PCR methods.
Timed matings were established between heterozygous pairs and vaginal plugs checked in the morning as an indication that copulation had occurred. The day a plug was detected was considered E0.5 of development. At the designated embryonic age, pregnant females were killed and embryos liberated from the uterine muscle and deciduum. Yolk sac tissue was recovered from later-stage embryos and standard DNA isolation and PCR methods used for genotype assessment. At E7.5–8.5 the entire embryonic tissue was placed in 10 μl of lysis buffer (500 mM KCl, 100 mM Tris-HCl, 0.45% NP-40, 0.45% Tween 20, and 500 μg/ml proteinase K) and incubated at 55°C for 3 h. Samples were boiled for 10 min to inactivate the proteinase K and used directly for PCR.
Cell culture and treatment
Primary MEF cultures were established from E13.5 embryos as described (Bunnell and Ervasti, 2010). Cells from each embryo were grown to confluency on a 10-cm plate and subsequently frozen at 1 × 106 cells/ml in MEF freezing medium (DMEM supplemented with 30% fetal bovine serum [FBS] and 10% dimethyl sulfoxide) until ready for use. Cells were then thawed and plated with MEF culture medium (DMEM supplemented with 10% FBS, 50 μg/ml streptomycin, 50 U/ml penicillin, and 100 μM nonessential amino acids). The following day, ActbL/L and ActbL/L CAGGCre-ERTM cells were dosed with 1 μM tamoxifen (Sigma-Aldrich, St. Louis, MO) to initiate recombination. After 3 consecutive days of tamoxifen treatment, MEF cultures were maintained for an additional 4 d to allow for protein turnover. Cells were passaged by trypsinization (0.05% trypsin-EDTA) on a consistent basis, and all analyses were carried out at the third passage. Cells were incubated in a humidified 37°C, 5% CO2 incubator.
Immunoblot analyses
Tissues were collected from 9-wk-old Actb+/+ and Actb+/− animals, snap frozen in liquid nitrogen, and ground into powder using a liquid nitrogen–cooled mortar and pestle. Pulverized tissue was boiled in 1% SDS, 5 mM ethylene glycol tetraacetic acid, and protease inhibitors for 2 min, followed by brief centrifugation to pellet out insoluble material. MEF cell lysates were prepared as previously described (Bunnell and Ervasti, 2010). Protein concentration of tissue and cell lysates was determined using the DC Protein Assay Kit (Bio-Rad, Hercules, CA). Equal amounts of total protein were separated by SDS–PAGE and immunoblotted with the following antibodies: pan-actin (C4; Seven Hills Bioreagents, Cincinnati, OH), β-actin (AC15; Sigma-Aldrich), γ-actin (mAb 2-4; Sonnemann et al., 2006), αsm-actin (1A4; Sigma-Aldrich), aurora kinase B (Cell Signaling, Beverly, MA), destrin (AE-14; Sigma-Aldrich), caldesmon-1 (Cell Signaling), and myosin phosphatase 1 and 2 (YE336; Abcam, Cambridge, MA). Fluorescently labeled secondary antibodies were detected and quantified from at least three separate experiments using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Data are presented as a fraction of wild-type protein levels.
Analysis of cell growth
To generate the growth curve, equal numbers (105) of MEFs were plated in each well of six-well plates and cells counted every 24 h in duplicate using a hemocytometer. On the final day of the growth curve cells were replated on fibronectin-coated coverslips and stained with phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) as described later to quantify the percentage of multinucleate cells. Cellular DNA content was analyzed by flow cytometry after staining ethanol-fixed cells with a solution of 20 μg/ml propidium iodide containing 200 μg/ml ribonuclease A. Apoptosis was detected by flow cytometry using a fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit (BD Pharmingen, San Diego, CA) as previously described (Bunnell and Ervasti, 2010). Fluorescent signals were collected on a FACSCalibur (BD Biosciences, San Diego, CA) flow cytometer at the University of Minnesota's Masonic Cancer Center Flow Cytometry Core Facility, and data analysis was carried out using FlowJo software (TreeStar, Ashland, OR).
Live-cell imaging
Live-cell imaging was performed on a Delta Vision personalDV (Applied Precision, Issaquah, WA) using differential interference contrast (DIC) imaging with a 40× (numerical aperture [NA] 1.35) oil objective or by phase contrast illumination with a 10× (NA 0.25) objective. Cells were plated at a density of 2 × 103 cells/cm2 on glass-bottom dishes (MatTek Corporation, Ashland, MA) precoated with 5 μg/ml fibronectin (BD Biosciences) and allowed to adhere for 3 h before the start of imaging. For optical clarity, the dish was sealed with vacuum grease and a glass coverslip. MEF culture media containing 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was used to stabilize the pH, and the cells were maintained at 37°C by an environmental chamber enclosing the microscope. For random migration, images were captured at 10-min intervals and cells tracked using the Manual Tracking plugin for ImageJ software (National Institutes of health, Bethesda, MD). Cells that divided or made contact with other cells during the experiment were not used for data analysis. Velocity was calculated as the total track distance divided by the total time (240 min), and directionality (D/T) was calculated as the linear distance (D) divided by the total track distance (T). More than 85 cells from at least three independent experiments were analyzed. For kymography, cells were imaged for 10 min at 3-s intervals using the 40× objective. Kymographs were generated and analyzed as previously described (Hinz et al., 1999) using the ImageJ Kymograph plugin. At least 74 protrusion and 54 retraction events were quantified from between 15 and 40 lamellipodial protrusions for each genotype. Only lamellipodia that protruded consistently during the entire 10-min movie were analyzed.
Immunofluorescence microscopy
MEFs were plated on glass coverslips precoated with 5 μg/ml fibronectin (BD Biosciences) and cultured for 3 h or overnight as indicated. Cells were fixed in 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA) in phosphate-buffered saline (PBS) for 10 min at 37°C and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Treatment with cold methanol at −20°C for 10 min was necessary for use with β-actin and γ-actin antibodies. Cells were blocked for 1 h in 3% BSA and incubated with the directly conjugated actin isoform–specific antibodies β-actin (FITC-conjugated AC15; Abcam) and γ-actin (Alexa 568–conjugated mAb 1-37; Invitrogen, Carlsbad, CA; Perrin et al., 2010). Focal adhesions were detected using primary antibodies to paxillin (clone 349; BD Biosciences) and vinculin (clone hVIN-1; Sigma-Aldrich), followed by an Alexa 488 goat anti–mouse secondary antibody (Invitrogen). Focal adhesions were quantified with Image J software using anti-paxillin and anti-vinculin staining to threshold objects 0.5–20 μm2 in size. Subsequently, the analysis of particles command was used to count the number of thresholded objects as a measure of focal adhesions per cell. F-actin was labeled with phalloidin–Alexa 488 (Invitrogen). Coverslips were mounted using ProLong Gold antifade reagent with DAPI (Invitrogen). Images were acquired using a 20× (NA 0.75), 40× oil (NA 1.35), or 100× oil (NA 1.40) objective on a Delta Vision personalDV microscope using softWoRx 3.7.1 software (Applied Precision). Stacks of images were collected at 0.20-μm intervals, deconvolved using Resolve3d software (Applied Precision), and processed using ImageJ 1.43 software.
Quantitative RT-PCR gene expression array
Cells were harvested by trypsinization and total RNA isolated from cell lysates using an RT2 qPCR-Grade RNA Isolation Kit (SABiosciences, Frederick, MD). RNA concentration and sample purity was determined using a NanoDrop (Wilmington, DE) spectrophotometer. cDNA was generated from equivalent amounts of total RNA using an RT2 First-Strand Kit (SABiosciences). The real-time PCR reaction was prepared using an SABiosciences RT2 qPCR Master Mix and loaded into each well of the 96-well Mouse Cytoskeleton Regulators RT2 Profiler PCR Array (SABiosciences). Real-time PCR detection was performed on a calibrated Bio-Rad MyiQ Real-Time PCR Detection System using PCR array protocol and plate setup template files from SABiosciences. Baseline and threshold values were set manually at the same level across all runs, and threshold cycle data were analyzed using the SABiosciences Excel-based PCR Array Data Analysis Template.
Ratios of G- to F-actin
Ratios of G- to F-actin were determined using a commercial kit (Cytoskeleton, Denver, CO) as previously described (Bunnell and Ervasti, 2010). Samples were analyzed by Western blotting with actin isoform–specific or pan-actin antibodies.
T-cell analysis
Whole thymi were harvested from 6- to 12-wk-old mice and T cells dispersed by gently mashing the tissue through 70-μm cell strainers (BD Falcon) using a rubber syringe plunger while rinsing with harvesting buffer (PBS containing 2% calf serum). The single-cell suspension was washed in excess harvesting buffer, and trypan blue–negative viable cells were enumerated using a hemocytometer. For analysis of T cell migration, thymocytes were resuspended at a density of 2 × 107 cells/ml into migration medium (RPMI containing 10 mM HEPES and 1% BSA, pH 7.0), preincubated at 37°C for 30 min, and chemotaxis assessed as previously described (Martin et al., 2008; Shiow et al., 2008). Briefly, 100 μl (2 × 106 cells) were placed in the upper portion of 5- or 3-μm transwell 6.5-mm chemotaxis inserts (Costar #3421 or #3415, respectively; Corning, Corning, NY), whereas the lower chamber contained either vehicle (migration medium alone) or migration medium with 1 μg/ml recombinant mouse CCL21 (R&D Systems, Minneapolis, MN). Triplicate samples of each condition were incubated at 37°C for 90 min (5-μm inserts) or 4 h (3-μm inserts), at which time the cells in the lower chamber were collected and washed with FACS buffer (ice-cold Hank's balanced salt solution containing 2% calf serum). Samples were stained with anti–CD4-PE (RM4-5), anti–CD8-PerCP-Cy5.5 (clone 53-6.7), and anti–TCR-β (clone H57-597) (all from eBioscience, San Diego, CA) for 30 min on ice, washed with cold FACS buffer, and finally resuspended in exactly 200 μl of FACS buffer containing 20,000 PKH67 reference beads (Sigma-Aldrich). A FACSCalibur flow cytometer (BD Biosciences) was used to collect an equal fraction (represented by 5000 reference beads) from each sample, and the number of CD4-positive/CD8-negative/TCR-βhi cells was counted and divided by input number in this gate to obtain the percentage migration.
To assess actin levels, 1 × 107 cells from unfractionated thymus were washed with PBS and then lysed for 30 min on ice with T cell lysis buffer (PBS containing 1% NP-40, 50 mM Tris pH 7.6, 2 mM EDTA, 10 mg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). Insoluble material was removed by centrifugation at 14,000 rpm for 10 min. Determination of protein concentration and immunoblotting of these clarified lysates were carried out as described earlier. Flow cytometry analysis of actin isoform expression was performed by fixation of 2 × 106 cells with 2% PFA (Electron Microscopy Sciences) for 20 min at room temperature, permeabilization for 5 min at room temperature with 0.05% Triton X-100, and antigen retrieval with 80% ice-cold methanol for 5 min. Cells were then washed in FACS buffer and stained with anti–CD4-Pacific Blue, anti–CD8-allophycocyanin, anti–TCR-β eF780 (all from eBioscience), together with anti–β-actin FITC and anti–γ-actin AF568, and analyzed on an LSRII flow cytometer (BD Biosciences).
Statistical analysis
Unless otherwise noted, comparisons between two groups were performed by an unpaired t test. Comparisons between more than two groups were performed by one-way analysis of variance, followed by Tukey's post hoc test. A value of p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Ben Perrin for assistance with live-cell imaging, Theodore Sanborn and True Lee for CD4-Cre mouse genotyping and colony maintenance, and Kris Hogquist for helpful discussions. This work was supported by National Institutes of Health Grants AR049899 to J.M.E. and AI031126 and AI038474 to Y.S.
Abbreviations used:
- αsm-actin
α smooth-muscle actin
- CCL21
CC chemokine ligand 21
- CD4sp/TCRβhi
CD4 single positive/T cell receptor β high
- E
embryonic day
- F-actin
filamentous actin
- G-actin
globular actin
- MEF
mouse embryonic fibroblast
- SRF
serum response factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0582) on September 7, 2011.
REFERENCES
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc Natl Acad Sci USA. 2009;106:9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron SE, Zhu M, Thiem SM, Friderici KH, Rubenstein PA. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J Biol Chem. 2010;285:16087–16095. doi: 10.1074/jbc.M110.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell TM, Ervasti JM. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton (Hoboken) 2010;67:564–572. doi: 10.1002/cm.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugina V, Zwaenepoel I, Gabbiani G, Clement S, Chaponnier C. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing M, et al. Aggregation of actin and cofilin in identical twins with juvenile-onset dystonia. Ann Neurol. 2002;52:465–476. doi: 10.1002/ana.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hill MA, Gunning P. Beta and gamma actin mRNAs are differentially located within myoblasts. J Cell Biol. 1993;122:825–832. doi: 10.1083/jcb.122.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MA, Schedlich L, Gunning P. Serum-induced signal transduction determines the peripheral location of beta-actin mRNA within the cell. J Cell Biol. 1994;126:1221–1229. doi: 10.1083/jcb.126.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Alt W, Johnen C, Herzog V, Kaiser HW. Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Bouc YL. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoock TC, Newcomb PM, Herman IM. Beta actin and its mRNA are localized at the plasma membrane and the regions of moving cytoplasm during the cellular response to injury. J Cell Biol. 1991;112:653–664. doi: 10.1083/jcb.112.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobelli J, Friedman RS, Conti MA, Lennon-Dumenil AM, Piel M, Sorensen CM, Adelstein RS, Krummel MF. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat Immunol. 2010;11:953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammermann T, Sixt M. Mechanical modes of “amoeboid” cell migration. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Le PU, Nguyen TN, Drolet-Savoie P, Leclerc N, Nabi IR. Increased beta-actin expression in an invasive Moloney sarcoma virus-transformed MDCK cell variant concentrates to the tips of multiple pseudopodia. Cancer Res. 1998;58:1631–1635. [PubMed] [Google Scholar]
- Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AL, Schwartz MD, Jameson SC, Shimizu Y. Selective regulation of CD8 effector T cell migration by the p110 gamma isoform of phosphatidylinositol 3-kinase. J Immunol. 2008;180:2081–2088. doi: 10.4049/jimmunol.180.4.2081. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Vallee A, Beaulieu C, Herman IM, Leclerc N. beta-Actin is confined to structures having high capacity of remodelling in developing and adult rat cerebellum. Eur J Neurosci. 1998;10:3785–3798. doi: 10.1046/j.1460-9568.1998.00391.x. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Nowak D, Skwarek-Maruszewska A, Zemanek-Zboch M, Malicka-Blaszkiewicz M. Beta-actin in human colon adenocarcinoma cell lines with different metastatic potential. Acta Biochim Pol. 2005;52:461–468. [PubMed] [Google Scholar]
- Nunoi H, Yamazaki T, Tsuchiya H, Kato S, Malech HL, Matsuda I, Kanegasaki S. A heterozygous mutation of beta-actin associated with neutrophil dysfunction and recurrent infection. Proc Natl Acad Sci USA. 1999;96:8693–8698. doi: 10.1073/pnas.96.15.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, Lessard JL, Bulinski JC. Immunolocalization of the gamma isoform of nonmuscle actin in cultured cells. J Cell Biol. 1986;102:1726–1737. doi: 10.1083/jcb.102.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham M, Miller G, Wells C, Zicha D, Dunn GA. Specific changes to the mechanism of cell locomotion induced by overexpression of beta-actin. J Cell Sci. 2001;114:1367–1377. doi: 10.1242/jcs.114.7.1367. [DOI] [PubMed] [Google Scholar]
- Perrin BJ, Sonnemann KJ, Ervasti JM. beta-Actin and gamma-actin are each dispensable for auditory hair cell development but required for stereocilia maintenance. PLoS Genet. 2010;6:e1001158. doi: 10.1371/journal.pgen.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Popow A, Nowak D, Malicka-Blaszkiewicz M. Actin cytoskeleton and beta-actin expression in correlation with higher invasiveness of selected hepatoma Morris 5123 cells. J Physiol Pharmacol. 2006;57((Suppl 7),):111–123. [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Procaccio V, et al. A mutation of beta-actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am J Hum Genet. 2006;78:947–960. doi: 10.1086/504271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein PA. The functional importance of multiple actin isoforms. Bioessays. 1990;12:309–315. doi: 10.1002/bies.950120702. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Deng JM, Fohn LE, Behringer RR. Restricted beta-galactosidase expression of a hygromycin-lacZ gene targeted to the beta-actin locus and embryonic lethality of beta-actin mutant mice. Transgenic Res. 1998;7:95–103. doi: 10.1023/a:1008816308171. [DOI] [PubMed] [Google Scholar]
- Shiow LR, et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat Immunol. 2008;9:1307–1315. doi: 10.1038/ni.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmerling D, et al. Strong and ubiquitous expression of transgenes targeted into the beta-actin locus by Cre/lox cassette replacement. Genesis. 2005;42:229–235. doi: 10.1002/gene.20135. [DOI] [PubMed] [Google Scholar]
- Sixt M. Interstitial locomotion of leukocytes. Immunol Lett. 2011;138:32–34. doi: 10.1016/j.imlet.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Sonnemann KJ, Fitzsimons DP, Patel JR, Liu Y, Schneider MF, Moss RL, Ervasti JM. Cytoplasmic gamma-actin is not required for skeletal muscle development but its absence leads to a progressive myopathy. Dev Cell. 2006;11:387–397. doi: 10.1016/j.devcel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci. 2011;124:123–132. doi: 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- Wei C, Wang X, Chen M, Ouyang K, Song LS, Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Chtanova T, Ladi E, Robey EA. Thymocyte motility: mutants, movies and migration patterns. Curr Opin Immunol. 2006;18:191–197. doi: 10.1016/j.coi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Han M, Bernier M, Wen JK. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276:2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga-Pflucker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.