We have identified two new Pot1a-associated telomere proteins, Pat1 and Tpt1, from Tetrahymena. Tpt1 is required to prevent telomere elongation and appears to be the Tetrahymena equivalent of vertebrate TPP1. Pat1 depletion causes gradual telomere shortening, indicating that it is needed for telomerase to gain access to the DNA terminus.
Abstract
We have identified two new telomere proteins, Tpt1 and Pat1, from the ciliate Tetrahymena thermophila. Although Tetrahymena telomerase is well characterized, only one telomere protein had previously been identified. This was the G-overhang binding-protein Pot1a. Tpt1 and Pat1 were isolated as Pot1a binding partners and shown to localize to telomeres. As Tpt1 and Pat1 were both found to be essential, conditional cell lines were generated to explore their function. Tpt1 depletion caused a rapid growth arrest and telomere elongation in the absence of cell division. The phenotype was similar to that seen after Pot1a depletion suggesting that Tpt1 and Pot1a function together to regulate telomere length and prevent telomere deprotection. In contrast, Pat1 depletion had a modest effect on cell growth but caused progressive telomere shortening similar to that observed upon TERT depletion. Thus Pat1 appears to be needed for telomerase to maintain the chromosome terminus. Analysis of Pot1a-Tpt1-Pat1 complex formation using purified proteins indicated that Tpt1 interacts directly with Pot1a while Pat1 interacts with Tpt1. Our results indicate that Tpt1 is the Tetrahymena equivalent of mammalian TPP1, Schizosaccharomyces pombe Tpz1, and Oxytricha nova TEBPβ.
INTRODUCTION
Telomeres have two primary functions: to shield the chromosome terminus from unwanted DNA repair activities and to provide a mechanism for maintaining the chromosome terminus by solving the end replication problem (O'sullivan and Karlseder, 2010). They exist as DNA–protein complexes in which telomeric DNA is packaged by a series of unique telomere proteins. The telomeric DNA generally consists of tandem repeats of a simple G·C-rich sequence that ends in single-stranded overhang on the 3′ G-rich strand. The proteins sequester the DNA terminus from DNA damage sensors, thus preventing the chromosome end from being treated as a DNA double-strand break. At the same time, they render the DNA terminus accessible to telomerase, the enzyme that maintains the chromosome end by synthesizing additional telomeric repeats.
Telomere proteins generally act as part of a multiprotein complex (Linger and Price, 2009). Although all the components are required for the complex to be fully functional, the individual telomere proteins are responsible for specific aspects of telomere maintenance. For example, the mammalian shelterin complex contains the DNA binding proteins TRF1, TRF2, and POT1 in addition to three other proteins – TIN2, TPP1, and RAP1 (Palm and de Lange, 2008; Stewart et al., 2011). TRF1 and TRF2 bind to the double-stranded telomeric DNA whereas POT1 binds to the 3′ G-rich overhang that serves as the substrate for telomerase. TIN2 holds the complex together by interacting with TRF1, TRF2, and TPP1. TPP1 serves as a bridge between the duplex DNA and the G-overhang binding proteins. POT1 binding to the overhang is required to exclude ATR and hence to prevent the overhang from activating a DNA-damage checkpoint (Denchi and de Lange, 2007; Guo et al., 2007). TPP1 serves a regulatory role as it not only enhances POT1 DNA binding affinity but also interacts with telomerase and enhances telomerase processivity (Abreu et al., 2010; Latrick and Cech, 2010).
Although telomeres from different organisms are packaged by proteins that share similar functions, the overall organization of the protein complexes is variable. In budding yeast, the duplex DNA and the G-overhang are packaged by separate complexes with the double-stranded DNA (dsDNA) bound by Rap1, Rif1, and Rif2 and the overhang bound by Cdc13 Stn1 and Ten1 (Linger and Price, 2009). In contrast, fission yeast telomeres are packaged by a single complex that is structurally similar to shelterin. The complex contains a duplex DNA binding protein, Taz1, a G-overhang binding protein, Pot1, and a series of bridging proteins, Rap1, Poz1, and Tpz1, that link Taz1 to Pot1 (Miyoshi et al., 2008). Pot1 is part of a three-protein subcomplex that contains Pot1, Tpz1, and Ccq1. In some ways, Tpz1 appears to be a functional homologue of mammalian TPP1 as it dimerizes with Pot1, and depletion of Tpz1 or Pot1 causes a similar loss of telomere protection (Wang et al., 2007; Kibe et al., 2010). Ccq1, however, is responsible for telomerase recruitment and interacts with Tpz1 rather than Pot1 (Miyoshi et al., 2008; Tomita and Cooper, 2008). As described later in the text, the ciliate Tetrahymena also has a Pot1 complex that contains proteins responsible for telomere protection and telomere length regulation.
Studies with ciliated protozoa have contributed a wealth of information about telomere biology because these organisms have a unique nuclear organization that results in each cell containing thousands of telomeres and an abundance of telomerase (Price, 2006). Tetrahymena thermophila has been particularly useful because it is both genetically tractable and amenable to biochemical analysis (Turkewitz et al., 2002). Indeed, many of the fundamental paradigms for telomerase biogenesis and catalytic mechanism have come from studies with Tetrahymena, and the telomerase holoenzyme from Tetrahymena is arguably the best characterized from any species (Jacobs et al., 2006; Zaug et al., 2008; Min and Collins, 2009, 2010; Berman et al., 2010; Robart et al., 2010; Zaug et al., 2010). The holoenzyme consists of five unique telomerase-specific subunits, in addition to the catalytic subunit TERT and the telomerase RNA TER that are each essential for telomere maintenance (Min and Collins, 2009). The various subunits contribute to different aspects of telomerase assembly, catalytic mechanism, and telomere association.
Tetrahymena telomeres consist of 250- to 350-base-pair G4T2·C4A2 repeats that are packaged into a non-nucleosomal DNA–protein complex (Blackburn and Chiou, 1981; Cohen and Blackburn, 1998; Price, 2006). The G-rich strand terminates in a 3′ overhang of ∼14 or 20 nt that is bound by the telomerase subunit Teb1 and then extended through the action of TERT and TER (Jacob et al., 2001; Min and Collins, 2010). Although Tetrahymena telomerase is well studied, Pot1a is the only Tetrahymena telomere protein to be described thus far (Jacob et al., 2007). We now report the identification of two additional telomere proteins Tpt1 and Pat1 that associate with Pot1a. Tpt1 binds directly to Pot1a and appears to be equivalent to the Pot1/TEBPα interaction partners from mammalian cells (TPP1), fission yeast (Tpz1), and Oxytricha (TEBPβ). Pat1 is a novel protein that is required to prevent progressive shortening and likely functions by promoting telomerase access to the chromosome terminus.
RESULTS
Identification of Tpt1 and Pat1
To identify new telomere proteins from Tetrahymena, we used mass spectrometry (MS) to analyze proteins that copurified with Pot1a. Nuclear extracts were made from a Tetrahymena cell line expressing TAP-tagged Pot1a (Jacob et al., 2007), and the tagged protein was isolated on immunoglobulin G (IgG) Sepharose. Bound protein complexes were eluted with TEV protease and the released proteins analyzed by MS against the predicted proteome of the T. thermophila macronuclear genome (Eisen et al., 2006; Coyne et al., 2008). Two TAP-tagged Pot1a samples and two control samples (mock purification from a Tetrahymena cell line not expressing TAP-Pot1a) were prepared. Proteins identified in the control samples were subtracted from the TAP-Pot1a samples.
Through this analysis, we identified two proteins that specifically copurified with TAP-Pot1a in both experimental samples but in neither control sample (Supplemental Figure 1). For each protein, we identified multiple peptides that mapped to single predicted open reading frames (Supplemental Table 1 and Supplemental Figure 2). RT-PCR was used to verify the corresponding full-length cDNA sequence. The encoded proteins have predicted molecular weights of 61.4 and 44.7 kDa and are subsequently referred to as Tpt1 (TPP1/Tpz1 in Tetrahymena thermophila) and Pat1 (Pot1 associated Tetrahymena), respectively. Secondary structure prediction and tertiary structure threading failed to identify any known folds or obvious functional motifs in Pat1 or Tpt1. Preliminary analysis of Pat1 and Tpt1 (described later in the text) suggested that Pat1 is a novel protein and Tpt1 appears to be equivalent to the primary POT1 binding partner previously identified in Schizosaccharomyces pombe (Tpz1), vertebrates (Tpp1), and other ciliates (TEBPβ) (Wang et al., 2007; Miyoshi et al., 2008; Baumann and Price, 2010).
Verification that Tpt1 and Pat1 are telomere proteins
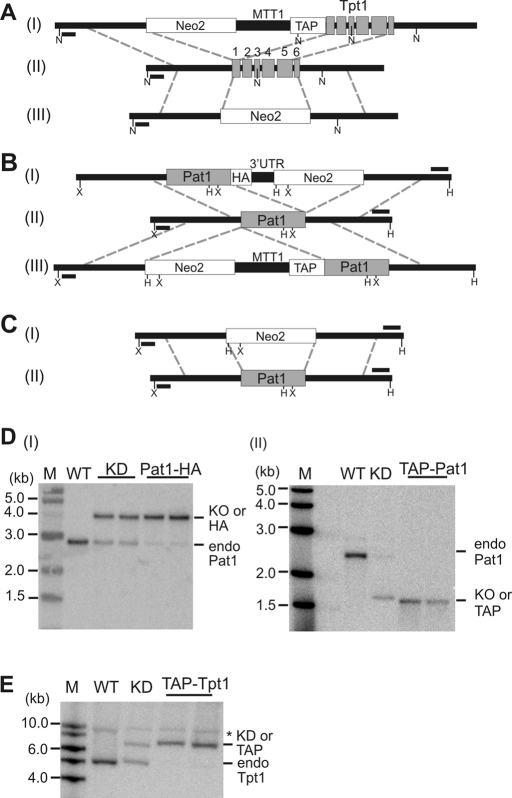
To confirm that Tpt1 and Pat1 are telomere proteins, we sought to verify the interaction with Pot1a and to demonstrate telomere localization. To this end, we generated cell lines expressing either N-terminal TAP-tagged Tpt1 (TAP-Tpt1) or Pat1 (TAP-Pat1), or C-terminal hemagglutinin-tagged Pat1 (Pat1-HA). In each case, the cDNA encoding the tagged version of the protein was recombined into the endogenous gene locus to replace a segment of the native gene (Figure 1). The genes encoding the TAP-tagged proteins were placed under control of a cadmium-regulated metallothionein (MTT1) promoter whereas the Pat1-HA gene retained the endogenous promoter. The TAP affinity tag contained a 6-His motif followed by two protein A motifs and a TEV cleavage site. For Pat1, we obtained full replacement of the endogenous gene indicating that the TAP and HA tags did not interfere with protein function (Figure 1D and Supplemental Figure 3A). For Tpt1, replacement was almost complete with the TAP-tagged allele (Figure 1E and Supplemental Figure 3B); however, only partial replacement was obtained with a C-terminal HA-tagged allele, suggesting that the C-terminal tag could not be tolerated (unpublished data).
FIGURE 1:
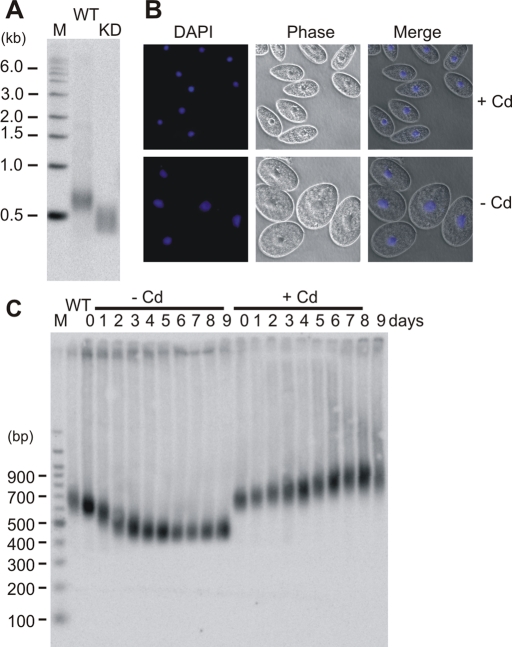
Generation of cell lines with Tpt1 or Pat1 gene replacements. (A–C) Gene-targeting constructs used to make modified Tpt1 or Pat1 cell lines. In each case, the gene-targeting construct was recombined into the native gene locus (panel II). Exons are in gray. Southern blot probes are shown as black bars. X, XbaI; H, HindIII; N, NdeI restriction sites. (A) Constructs used to make TAP-Tpt1 (I) and Tpt1 KD cells (III). (B) Pat-HA (I) and TAP-Pat1 (III) cells. (C) Pat1 KD cells (I). (D and E) Southern blots showing Pat1 or Tpt1 gene replacement. Bands from endogenous, knockout, or HA- or TAP- tagged genes are marked. (D) (I) Pat1 KD or Pat1-HA (HA), HindIII digest. (II) Pat1 KD or TAP-Pat1, XbaI digest. (E) WT, KD, or TAP-Tpt1 (TAP), NdeI digest.
We verified that endogenous Pot1a interacts with Tpt1 and Pat1 through pull-down experiments with Ni-Sepharose and extracts from TAP-Tpt1– or TAP-Pat1–expressing cells (Figure 2A) or Pot1a antibody (Jacob et al., 2007, and Supplemental Figure 3C) and extracts from Pat1-HA–expressing cells (Figure 2B). Western blot analysis indicated that in each case the tagged protein copurified with Pot1a. Note that TAP-Tpt1 and TAP-Pat1 can be visualized with secondary antibody alone because of the protein A motif in the tag (Figure 2A), thus both Pot1a and Tpt1 or Pat1 can be observed in blots with the Pot1a antibody. To ensure that the interaction with Pot1a was not mediated by DNA or RNA, nuclear extracts were treated with micrococcal nuclease or ethidium bromide before TAP-Tpt1 or TAP-Pat1 pull-down (Figure 2A, panels I and II, lane 8; Supplemental Figure 3D and unpublished data). Neither treatment disrupted copurification of Pot1a with Tpt1 or Pat1, indicating that the interaction between these proteins is not dependent on nucleic acid.
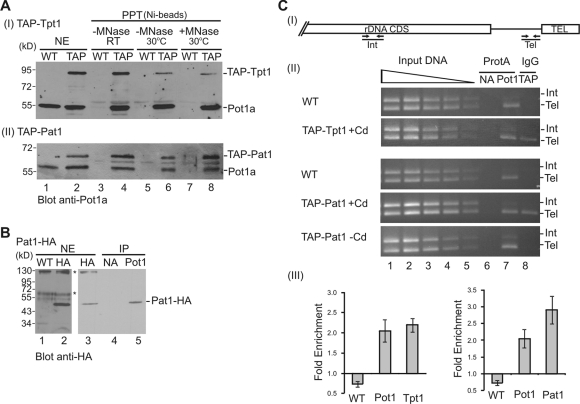
FIGURE 2:
Tpt1 and Pat1 bind Pot1a and associate with telomeres. (A and B) Tpt1 and Pat1 copurify with Pot1a. (A) Ni-Sepharose precipitation (PPT) of nuclear extracts (NE) from WT or conditional TAP-Tpt1 (TAP) cells (panel I) or TAP-Pat1 (TAP) cells (panel II). Western blot with Pot1a antibody. Lanes 7 and 8, extract was treated with MNase before pull down. (B) Immunoprecipitation (IP) of nuclear extracts (NE) from Pat1-HA cells with no antibody (NA) or Pot1a antibody (Pot1). Western blot with HA antibody. *, Cross-reacting bands. (C) Tpt1 and Pat1 are present at rDNA telomeres. (I) Schematic of rDNA telomere. Arrows indicate positions of PCR primers used to amplify subtelomeric (Tel) and internal (Int) regions. (II) Multiplex PCR of ChIP products from WT or conditional TAP-Tpt1 or TAP-Pat1 cells grown ± cadmium. Lanes 1–5, fourfold dilutions of input DNA; lane 6, protein A beads, no antibody (NA); lane 7, Protein A beads + Pot1a antibody (Pot1); lane 8, IgG Sepharose to precipitate Tap-tagged Tpt1 or Pat1 (TAP). (III) Quantification of multiplex PCR to show fold enrichment of telomeric DNA. Minimum of three experiments.
We tested for Tpt1 and Pat1 localization to macronuclear telomeres using chromatin immunoprecipitation (ChIP). Cells expressing TAP-tagged Tpt1 or Pat1 were formaldehyde cross-linked and fragmented by sonication, and chromatin associated with the TAP-tagged protein was precipitated with IgG Sepharose. The precipitated DNA was then amplified by multiplex PCR using primer pairs corresponding either to a region immediately adjacent to the telomere or to an internal region of the ribosomal DNA (rDNA) mini-chromosome (Figure 2C and Jacob et al., 2007). As shown in Figure 2C, precipitation of either TAP-Tpt1 or TAP-Pat1 enriched for telomeric DNA (Tel) relative to the internal region (Int). The level of enrichment was similar to that seen in the positive control with Pot1a antibody. In contrast, ChIP with wild-type (WT) cells or TAP-Pat1 cells grown in the absence of cadmium to repress Pat1 expression (see Figure 5 later in this paper) did not result in enrichment of the telomeric fragment. These results confirm that both Tpt1 and Pat1 localize to telomeres.
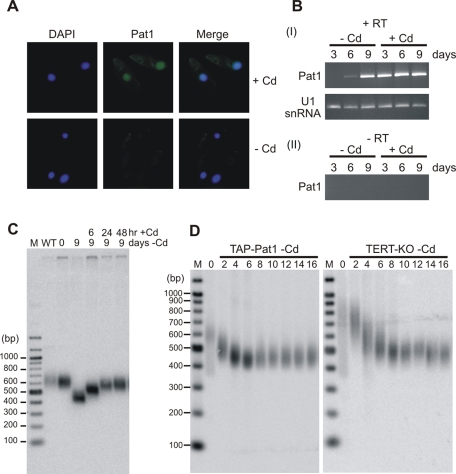
FIGURE 5:
Telomere shortening is reversible and similar to that seen after TERT depletion. (A) Localization of Pat1 in conditional TAP-Pat1 cells grown ± Cd for 3 d. Cells were stained with Pat1 antibody and DAPI. (B) RT-PCR showing level of TAP-Pat1 expression. (I) mRNA was isolated from TAP-Pat1 cells grown ± Cd for 3, 6, or 9 d. (II) as for (I) but without reverse transcriptase. (C) Southern blot showing the length of the rDNA telomere in TAP-Pat1 cells after growth without (–Cd) cadmium for 9 d followed by addition of cadmium for 6, 24, or 48 h. (D) Southern blots showing rDNA telomere length in conditional TAP-Pat1 cells (TAP-Pat1) and conditional TERT knockout cells (TERT-KO) after growth without cadmium for 2–16 d.
Depletion of Tpt1 leads to growth arrest and telomere elongation
To learn more about Tpt1 function, we attempted to generate a simple gene replacement by substituting the ∼45 copies of the macronuclear TPT1 gene with a selectable marker cassette (Figure 1A). Selection for drug resistance, however, yielded cells with only ∼50% replacement of the WT gene locus (Figure 1E). This inability to replace all endogenous copies of TPT1 indicated that it is essential. Despite the incomplete gene replacement, we isolated genomic DNA to examine the length of the rDNA telomeric restriction fragments. Southern hybridization revealed that Tpt1 knockdown (KD) resulted in longer telomeres, suggesting that the protein plays a role in telomere length regulation (Figure 3A).
FIGURE 3:
Tpt1 depletion causes a cell-cycle checkpoint and telomere elongation. (A) Southern blot showing length of rDNA telomere in WT and Tpt1 KD cells. (B) Growth curves for TAP-Tpt1 conditional cells grown with (+Cd) or without (–Cd) cadmium. Cd was readded to a portion of the –Cd culture after 24 h. (C) Phase contrast (PC) images of conditional TAP-Tpt1 cells grown with (+Cd) or without (–Cd) cadmium for 24 h. Nuclei were stained with DAPI. (D) Southern blot showing length of rDNA telomeres in TAP- Tpt1 cells grown ± Cd for 0–96 h. (E) Localization of Pot1a in TAP-Pat1 cells grown ± Cd for 24 h. Cells were stained with Pot1a antibody and DAPI.
Because the TPT1 gene is essential, we turned to the conditional TAP-Tpt1 cell line in which the endogenous promoter is replaced with the MTT1 promoter. By growing the cells in cadmium to allow TAP-Tpt1 expression, we were able to obtain almost complete replacement of the endogenous TPT1 gene (Figure 1E and Supplemental Figure 3B). We then examined the effect of Tpt1 depletion by growing the cells in the absence of cadmium. Tpt1 depletion caused a rapid growth arrest such that the population underwent at most two population doublings (Figure 3B). The arrested cells then became large and round (Figure 3C). Both the growth arrest and abnormal morphology were reversed if cadmium was added back to the culture after 24 h (Figure 3B and unpublished data), suggesting that Tpt1 depletion had activated a cell-cycle checkpoint. When we isolated DNA and used Southern hybridization to examine the size of telomeric restriction fragments from the rDNA, we found that Tpt1 depletion had caused them to become much longer and more heterogeneous in size (Figure 3D).
Because the apparent increase in telomere length caused by Tpt1 depletion occurred in the absence of continued division (unpublished data, but see Figure 3B), it was unclear whether the increase in size of the telomeric restriction fragments reflected actual telomere growth or merely extension of the G-overhang in the arrested cells. To address this possibility, we examined G-overhang length by using a ligation-mediated primer extension protocol previously developed for this purpose (Jacob et al., 2001). This approach indicated that G-overhang length was unchanged by Tpt1 depletion (Supplemental Figure 4A). As the primer extension step used in this protocol is inadequate to measure overhang length if the overhangs are long (>75 nt; C. Price and N. Jacob, unpublished results), we also examined overhang length by comparing the migration of telomeric restriction fragments in agarose gels before and after treatment with Exo1 to degrade the overhangs (Supplemental Figure 4B). Again, the Exo1 treatment did not cause a noticeable change in migration pattern, indicating that Tpt1 depletion had little effect on G-overhang length. Thus Tpt1 depletion causes telomere growth, indicating that Tpt1 functions in telomere length regulation.
Interestingly, the effect of Tpt1 depletion was strikingly similar to what we observed after Pot1a depletion, which also causes growth arrest within two population doublings, appearance of large round cells, and comparable telomere elongation in the absence of cell division with little change in G-overhang length (Jacob et al., 2007). This similarity in phenotypes led us to ask whether, as observed for mammalian TPP1 and POT1 (Liu et al., 2004), Tpt1 is required for localization of Pot1a to the macronucleus. Immunolocalization studies to analyze Pot1a distribution after Tpt1 depletion indicated that this is indeed the case. In cells expressing Tpt1, Pot1a localized to the macronucleus as previously observed (Figure 3E; Jacob et al., 2007). After Tpt1 depletion, however, Pot1a was no longer apparent in the macronucleus, indicating that Tpt1 is required for either Pot1a stability or nuclear localization. Thus our results indicate that Tpt1 and Pot1a function together to regulate telomere length and hide the chromosome terminus from the DNA damage–sensing machinery.
Depletion of Pat1 leads to progressive telomere shortening
When we attempted to generate a simple PAT1 gene disruption, we were again unsuccessful, indicating that Pat1 is also essential. This time we were able to replace ∼80% of the endogenous copies of the macronuclear gene with the selectable marker cassette (Figure 1C and D). As before, we isolated DNA from the KD cell line and examined the length of the rDNA telomeric restriction fragments. In contrast to what was observed after Tpt1 depletion, Pat1 KD resulted in shorter telomeres (Figure 4A).
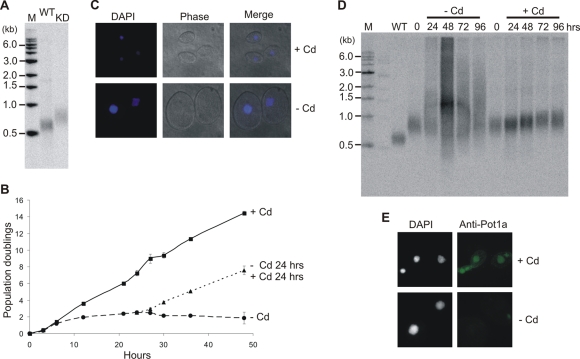
FIGURE 4:
Pat1 depletion results in shorter telomeres. (A) Southern blot showing length of rDNA telomere in WT and Pat1 KD cells. (B) Phase contrast (PC) images of conditional TAP-Pat1 cells grown with (+Cd) or without (–Cd) cadmium for 6 d. Nuclei were stained with DAPI. (C) Southern blot showing length of rDNA telomeres in conditional TAP-Pat1 cells grown ± Cd for 1–9 d.
To examine the effect of more complete Pat1 depletion, we again turned to a conditional cell line. In this case, the PAT1 gene was replaced by a TAP-Pat1 allele under control of the MTT1 promoter (Figure 1, B and D). By selecting for drug resistance in the presence of cadmium, we were able to obtain full replacement of the endogenous PAT1 gene locus (Figure 1D and Supplemental Figure 3A). When we then removed cadmium to examine the effect of Pat1 depletion on cell growth, we did not observe an immediate phenotype. When the cells were kept in continuous log phase culture in the absence of cadmium, however, the growth rate slowed slightly such that by day 6 the cells divided every 3.4 h instead of every 3 h (unpublished data). The slower growth was accompanied by an enlargement of both the cells and the macronuclei, suggesting a problem with cell division (Figure 4B). On subsequent days these changes gradually reversed so that by day 9 ∼95% of the cells had returned to their original size and nuclear morphology. The growth rate also increased by day 9, but it did not return to that observed when the cells were grown in cadmium.
Examination of the rDNA telomeres from cells harvested each day during continuous growth without cadmium revealed that telomere length decreased in parallel with the decline in growth rate. For the first 5–6 d after cadmium removal, the telomeres became progressively shorter and less heterogeneous in length (Figure 4C). They then stopped shortening, however, and reached a new length set point. Southern blot analysis verified that the TAP-PAT1 allele had been retained and the endogenous PAT1 allele was undetectable even after 9 d without cadmium (unpublished data). The telomeres of conditional TAP-Pat1 cells grown with cadmium showed a slight increase in length over the 9 d of log phase growth. This telomere elongation was expected as WT cells also exhibit telomere elongation when maintained in log phase culture (Jacob et al., 2004). Analysis of G-overhang length by ligation-mediated primer extension indicated that this was unaltered by Pat1 depletion (Supplemental Figure 4A).
We next used indirect immunofluorescence to monitor Pat1 distribution within the TAP-Pat1 cells. When the cells were grown in cadmium, we observed macronuclear staining with antibody to Pat1 (Figure 5A and Supplemental Figure 3A). This staining, however, was completely gone 3 d after cadmium removal. As was previously reported for Pot1a (Jacob et al., 2007), we failed to see staining in the micronucleus, suggesting that either Pat1 protein is absent or its levels are too low for detection. ChIP analysis of conditional TAP-Pat1 cells grown without cadmium for 3 d verified that Pat1 had been lost from the telomere (Figure 2C, panel II). Taken together, these results indicate that loss of Pat1 from the telomere results in progressive telomere shortening.
Because our initial attempts to disrupt PAT1 indicated that the gene is essential, we suspected that the eventual stabilization of telomere length and the return of the conditional TAP-Pat1 cells to a near WT growth rate might be due to up-regulation of the MTT1 promoter in cells with critically short telomeres. Such promoter upregulation had previously been observed in a conditional TERT knockout cell line in which the rescuing allele was also expressed from the MTT1 promoter (Jacob et al., 2003). Indeed, RT-PCR confirmed that, although the level of TAP-Pat1 mRNA was greatly decreased after 3 d growth without cadmium, it became slightly elevated after 6 d and further elevated by 9 d (Figure 5B). Thus PAT1 expression becomes reactivated at approximately day 6, but the level of expression is insufficient for telomeres to return to their original length. Re-addition of cadmium after 9 d resulted in substantial telomere elongation within just 6 h (approximately two cell divisions) with the telomeres achieving WT length within 24 h (Figure 5C). This finding indicates that the effect of Pat1 depletion is readily reversible.
As progressive telomere shortening is characteristic of telomerase-deficient cells, we next asked whether Pat1 and TERT depletion affect telomere lengths in a similar manner. To do this, we compared the telomere shortening profile in conditional TAP-Pat1 and conditional TERT knockout cells. Although the TERT conditional cells started with longer telomeres, the overall rate and extent of telomere shortening were strikingly similar when the two cell lines were grown in continuous culture without cadmium (Figure 5D). The similarity in telomere shortening profile after Pat1 and TERT depletion, together with the lack of a sudden growth arrest, indicates that Pat1 probably functions by regulating telomerase action on the chromosome terminus rather than by protecting the telomere from DNA repair activities.
Interactions between Pat1 or Tpt1 and telomerase
One explanation for why Pat1 is needed for telomerase action at the telomere would be if Pat1 is a component of the telomerase holoenzyme that is needed for catalytic activity. To test for this possibility, we isolated telomerase from TAP-Pat1 cells grown with and without cadmium and compared the level of enzyme activity. Depletion of Pat1 did not alter the level of activity relative to that observed from WT and TAP-Pat1–expressing cells (Figure 6A). This result was markedly different from the dramatic drop in activity observed on TERT depletion. Thus Pat1 does not appear to be an essential component of the telomerase holoenzyme.
FIGURE 6:

Lack of interaction between Pat1 or Tpt1 and telomerase. (A) Telomerase activity in extracts made from WT cells and TAP-TERT or TAP-Pat1 conditional cells grown ± cadmium. RC, recovery control for telomerase product isolation. (B) Telomerase activity precipitated with IgG beads and extracts from WT cells or cells expressing TAP-tagged TERT (TT), TAP-Pat1 (Pat1), or TAP-Tpt1 (Tpt1) (lanes 5–8). Input activity is shown in lanes 1–4.
As Pat1 and Tpt1 both alter telomere length (albeit in opposite directions) in a manner consistent with them somehow regulating telomerase action, we next examined whether we could detect a stable interaction between telomerase and either protein. In initial experiments, we made telomerase extracts from TAP-Pat1– or TAP-Tpt1–expressing cells by conventional approaches, affinity purified the TAP-tagged protein, and assayed for copurifying telomerase activity. We were unable to detect any activity by this approach (unpublished data). These Tetrahymena telomerase preparations, however, were made by following the standard protocol in which cells are extracted with non-ionic detergent (Igepal CA630) and 0–100 mM sodium acetate. The insoluble material is then discarded. Under these conditions, Pat1 and Tpt1 are not removed from the telomere (B. Linger, unpublished observations), so the experiments assayed only for an interaction between telomerase and soluble nontelomere-bound Pat1 or Tpt1.
Because telomerase may interact with Pat1 or Tpt1 only in the context of a whole telomere, we developed a procedure to prepare cell extracts that contained both telomerase and telomere-bound Pat1 and Tpt1. Cells were subjected to a series of 5-s sonication pulses to lyse the cells and fragment the genomic DNA into small enough pieces to remain soluble. This sonicated lysate was then centrifuged to remove the insoluble material. Lysates prepared in this way contained active telomerase (Figure 6B, lanes 1–4), and a control experiment with a cell line expressing TERT-TAP verified that that telomerase activity could be affinity purified (Figure 6B, lane 6). Affinity purification of TAP-Pat1or TAP-Tpt1 from sonicated lysates, however, still failed to recover telomerase activity (Figure 6B, lanes 6–8). To verify that this extraction procedure released soluble telomeric DNA fragments with associated telomere proteins, we isolated DNA present from TAP-Pat1 affinity-purified samples or samples precipitated with Pot1a antibody and assayed for enrichment of telomeric over nontelomeric DNA. Multiplex PCR demonstrated that the precipitated material contained DNA and that telomeric DNA was enriched over nontelomeric DNA (unpublished data). Thus our results indicate that telomerase does not form a stable complex with either soluble or telomere-bound Pat1or Tpt1. We cannot, however, rule out an interaction that is too transient or labile to be detected by an affinity purification approach.
Organization of the Pot1a-Tpt1-Pat1 complex
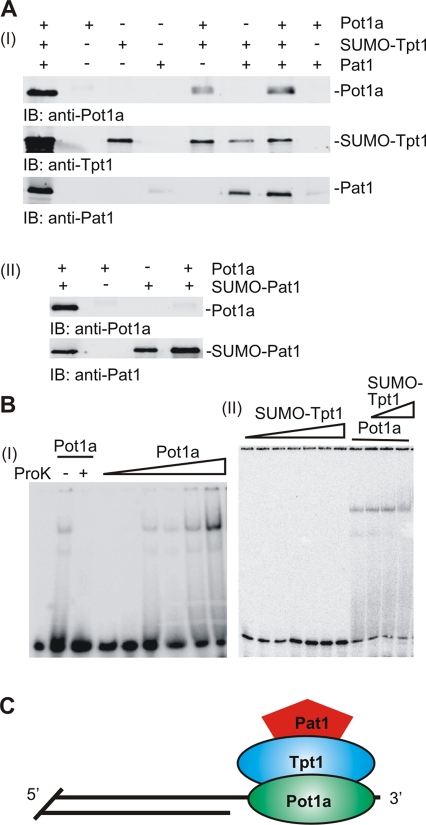
The original affinity purification used to identify Tpt1 and Pat1 indicated that they form a complex with Pot1a but did not give information about the configuration of the complex. We therefore performed a series of pull-down experiments with purified proteins to determine which proteins interact. TAP-tagged Pot1a was isolated from Tetrahymena after overexpression from an rDNA expression vector (Supplemental Figure 5A; Yao et al., 2007). Tpt1 and Pat1 were expressed in Escherichia coli with a 6His-SUMO tag (Supplemental Figure 5, B and C). Experiments were performed with both tagged and untagged forms of each protein. When Pot1a and Pat1 were incubated with SUMO-Tpt1 either individually or together, both proteins copurified with the SUMO-Tpt1 on Ni-Sepharose (Figure 7A). Similar results were seen when SUMO-Pat1 was incubated with Tpt1 plus Pot1a (unpublished data). When SUMO-Pat1 was incubated with Pot1a alone, however, the proteins did not copurify (Figure 7A). This was also the case when Pat1 alone was incubated with TAP-Pot1a (data not shown). Since TAP-Pot1a is fully functional in vivo (Jacob et al., 2007), these results indicate that the lack of interaction was unlikely to be caused by the affinity tags. Thus, within the Pot1a-Tpt1-Pat1 complex, Tpt1 interacts with both Pot1a and Pat1, but Pot1a and Pat1 do not appear to interact directly with each other (Figure 7C). The direct interaction between Pot1a and Tpt1, together with the similar telomere length and growth arrest phenotypes after Pot1a and Tpt1 depletion, raised the possibility that Tpt1 is the functional homologue of mammalian TPP1. TPP1 also binds POT1, and depletion of TPP1 causes a similar phenotype to a Pot1 knockout (Wang et al., 2007; Kibe et al., 2010).
FIGURE 7:
Configuration of the Pot1a-Tpt1-Pat1 complex. (A) Protein–protein interactions within the complex. (I) His-SUMO–tagged Tpt1 was incubated with Pot1a and Pat1, complexes were isolated on Ni-Sepharose, and copurified proteins were detected by Western blotting. Lane 1 shows the input protein. (II) As for (I) but with SUMO-Pat1 and Pot1a. (B) Mobility shift gels to examine Pot1 and Tpt1 binding to telomeric DNA. (I) Addition of increasing amounts of Pot1a (∼0.25–50 nM) to 15 pM TelG20 oligonucleotide TG4(T2G4)2T2G. (II) Addition of increasing concentrations of Tpt1 to 15 pM TelG20 ± ∼25 nM Pot1a. Lanes 1–7, 50–1000 nM Tpt1; lane 8, Pot1a alone; lanes 9–11, 50–200 nM Tpt1 + Pot1a. (C) Model showing organization of the Pot1a telomeric complex.
Because dimerization with mammalian TPP1 is known to enhance binding of POT1 to telomeric G-strand DNA (Wang et al., 2007), we used gel-shift assays to examine the effect of Tpt1 on Pot1a binding. We were not able to obtain sufficient purified Pot1a to perform a full analysis of binding constants; however, preliminary experiments indicated that Pot1a bound robustly to oligonucleotides with the same sequence as the Tetrahymena telomeric G-strand overhang (Figure 7B). The minimum length needed for binding was 10-12 nt of T2G4 repeat (unpublished data). Addition of an excess of Tpt1 did not appear to alter the affinity of Pot1a for a 20 nt G-strand oligonucleotide, the length of the G-overhang on many Tetrahymena telomeres (Figure 7B; Jacob et al., 2001). Although addition of Tpt1 did not alter the mobility of the Pot1a-DNA complex, supershift experiments with either Pot1a or Tpt1 antibody confirmed that the complexes contained both Pot1a and Tpt1 (unpublished data). Titration experiments with Tpt1 or Pat1 alone indicated that neither protein bound specifically to single- or double-stranded telomeric DNA (Figure 7B and unpublished data).
DISCUSSION
Here we describe two new telomere proteins, Tpt1 and Pat1, that form a complex with the G-overhang binding protein Pot1a. Tpt1 seems to be the Tetrahymena version of the Pot1/TEBPα interaction partners from mammalian cells (TPP1), fission yeast (Taz1), and Oxytricha (TEBPβ) as Tpt1 binds directly to Pot1a, and its depletion causes the same telomere elongation and growth arrest phenotype observed previously after Pot1a depletion. In contrast, Pat1 is a novel protein that interacts with Tpt1 but not Pot1a. Removal of Pat1 causes gradual telomere shortening rather than a sudden growth arrest. Thus Pat1 appears to be required for telomerase action at the telomere rather than for telomere protection.
The rapid telomere elongation that occurs in the absence of cell division after Pot1a depletion is likely to result from the DNA terminus becoming more accessible to telomerase after Pot1a is lost from the 3′ overhang. Our observation that Tpt1 depletion causes a similar phenotype suggests that Tpt1 functions in conjunction with Pot1a to limit telomerase access to the overhang. This hypothesis fits with our finding that Tpt1 is needed for Pot1a to localize to the nucleus. Given that Tpt1 does not increase the affinity of Pot1a for G-strand DNA, it is unlikely to directly affect telomerase access by enhancing the ability of Pot1a to compete with telomerase for binding to the 3′overhang. If the Pot1-Tpt1-Pat1 complex, however, is linked to the proteins that bind the telomeric duplex DNA, the Pot1a-Tpt1 interaction may also enhance overhang binding by serving to increase the local concentration of Pot1a.
The gradual nature of the telomere shortening seen after Pat1 depletion, together with the very minor effect on cell growth, indicates that the loss of telomeric DNA is most likely caused by a deficiency in telomerase action rather than nuclease activity following telomere deprotection. Pat1 might function to promote telomerase action in a number of ways. One possibility is that it acts like Est1 from budding yeast to form a bridge between telomerase and the DNA terminus. It could also stimulate telomerase activity (DeZwaan and Freeman, 2009; Li et al., 2009). Alternatively, Pat1 may not interact with telomerase directly but may instead help alter the conformation of the Pot1a-Tpt1 complex so that the G-overhang becomes accessible to telomerase. We favor the latter hypothesis in part because we were unable to detect a stable interaction between Pat1 and telomerase. Also the Teb1 subunit of telomerase seems to function as the Tetrahymena version of Est1 in that it has a high affinity for telomeric G-strand DNA and is known to stabilize telomerase binding to the G-overhang (Min and Collins, 2010). In Tetrahymena, most G-overhangs are 14 or 20 nt, which is too short for Pot1 and Teb1 to bind simultaneously (Jacob et al., 2001). Thus it seems likely that Pot1a binding will be subject to cell-cycle regulation, which could occur via a conformational change modulated by Pat1 and/or Tpt1.
In some ways the organization of the Tetrahymena Pot1a-Tpt1-Pat1 complex resembles the Pot1-Tpz1-Ccq1 complex found in fission yeast as, in contrast to the situation in mammalian cells, the subunit that regulates telomerase action on the chromosome terminus is not a direct Pot1 binding partner (Miyoshi et al., 2008; Latrick and Cech, 2010). The Tetrahymena telomeric dsDNA binding proteins have not yet been identified, however, and it remains to be seen whether the telomeric DNA is packaged by a single complex akin to that found in mammals or fission yeast or whether it is packaged by separate duplex DNA and G-overhang binding complexes as in budding yeast (Linger and Price, 2009).
MATERIALS AND METHODS
Tetrahymena growth and transformation
Tetrahymena thermophila cells were grown in 1.5× PPYS medium at 30°C as previously described (Jacob et al., 2001). To obtain growth curves, the culture was adjusted regularly to keep the cells in log phase (<2.0 × 105 cells/ml). For conditional TAP-Tpt1, TAP-Pat1, and conditional TERT expression, cells were grown in 2 μg/ml CdCl2. Cells expressing TAP-tagged Tpt1 or Pat1 and HA-tagged Pat1 were generated using biolistic transformation to introduce a gene replacement construct into the native TPT1 or PAT1 gene locus (Figure 1). The TAP-Tpt1 and TAP-Pat1 constructs replaced the endogenous gene promoter with MTT1 along with an upstream neomycin resistance cassette. Cells were selected in paromomycin to obtain partial gene replacement and with paromomycin in the presence of 2 μg/ml CdCl2 to obtain full replacement. Cells were checked at regular intervals to ensure they retained the full gene replacement and had not reverted to KD status. The affinity tag on TAP-Tpt1 and TAP-Pat1 contains a 6-His motif followed by two protein A motifs and a TEV cleavage site. The TAP-Pot1a cells have the affinity tag added to the N terminus of the endogenous POT1A gene locus. The tag consists of two protein A motifs, a TEV cleavage site, and one calmodulin binding domain (Jacob et al., 2007). The TERT-TAP cells have the TAP tag on the C terminus of the endogenous gene locus. The tag consists of one calmodulin binding domain followed by a TEV cleavage site and two terminally located protein A motifs (Witkin and Collins, 2004). To overexpress Pot1a, cells were transformed with an rDNA expression construct (Yao et al., 2007). The Pot1a expression cassette was regulated by the MTT1 promoter, and it encoded a Pot1a cDNA with an N-terminal TAP tag containing a 6-His motif, two protein A motifs, and a TEV cleavage site.
Affinity purification and immunoprecipitation
Nuclei (chromatin) were isolated as previously described (Blackburn and Chiou, 1981). To prepare nuclear extract, nuclei were resuspended in 20 mM Tris, pH 7.5, 200 mM NaCl, and 1.5 mM MgCl2 plus protease inhibitor cocktail (5 μg/ml antipain, 2 μg/ml aprotinin, 16 μg/ml benzamidine, 6 μg/ml chymostatin, 1 μg/ml E64, 5 μg/ml leupeptin, and 1 μg/ml pepstatin A) and then incubated for 1 h at 4°C and centrifuged at 16,000 × g. To immunoprecipitate endogenous Pot1a, 500 μl of supernatant was incubated with Pot1a antibody for 1 h at 4°C, and immune complexes were precipitated with protein A Sepharose for 1 h at 4°C. TAP-tagged proteins were precipitated with IgG Sepharose for 1 h at 4°C. Beads were washed three times with five volumes of 20 mM Tris, pH 7.5, 200 mM NaCl, and 1.5 mM MgCl2 plus protease inhibitor cocktail. For Ni pull-down, nuclei were extracted as described earlier in the text but with 300 mM NaCl. 6-His tagged proteins were precipitated with chelating Sepharose charged with Ni. Beads were washed twice with 8 mM imidazole in 20 mM Tris, pH 7.5, 300 mM NaCl, and 1.5 mM MgCl2 plus protease inhibitor cocktail and twice with 40 mM imidazole in 20 mM Tris, pH 7.5, 300 mM NaCl, and 1.5 mM MgCl2 plus protease inhibitor cocktail.
Mass spectrometry
TAP-tagged samples were eluted from IgG Sepharose with TEV cleavage and precipitated with trichloroacetic acid. Two experimental and two control samples (mock purification from a Tetrahymena cell line not expressing TAP-Pot1a) were separated by one-dimensional SDS–PAGE and visualized by colloidal Coomassie. The entire lane was excised, divided into 16 pieces, and prepared for MS analysis by in-gel reduction, alkylation, and trypsin digestion. The eluted samples were then analyzed by reverse-phase nano-electrospray tandem MS as described previously (Mead et al., 2010). The MS/MS spectra emanating from the gel slices for each lane were concatenated and searched against tryptic peptides predicted from the Tetrahymena genome (TTA1_08302006.pep; ftp://ftp.ciliate.org/Tetrahymena/sequence/) using the X!Tandem (www.thegpm.org/TANDEM) and Mascot (Matrix Science, Boston, MA) search engines. Putative interacting proteins were those identified in both experimental samples and not in either control sample.
Protein purification
Tpt1 and Pat1 were expressed in E. coli from cDNAs that had the codon usage altered to remove stop codons. Both proteins were expressed with a 6-His-SUMO tag. The soluble protein was affinity purified on Ni-Sepharose. The tag was removed by ULP1 cleavage (Mossessova and Lima, 2000).
Tpt1 and Pat1 antibody
Pat1 antibody was made by giving injections to rabbits of native-folded, full-length recombinant Pat1, and Tpt1 antibody was made with denatured full-length recombinant protein. Both antibodies were affinity purified using purified protein. Pot1a antibody was previously described (Jacob et al., 2007). HA antibody (mouse monoclonal 12CA5) was a gift from William Miller, University of Cincinnati.
Chromatin immunoprecipitation (ChIP)
Cells were fixed with formaldehyde and DNA sheared, and supernatant of each extract was prepared as previously described (Jacob et al., 2007). Precipitation was performed for 1 h at 4°C with IgG Sepharose for TAP-tagged proteins or with antibody and protein A Sepharose for endogenous proteins. Precipitates were washed, cross-linking reversed, and DNA purified as previously described (Jacob et al., 2001). The DNA was analyzed by multiplex PCR using the Int and Tel primers.
Telomere length analysis
Telomere length was determined by Southern hybridization to restriction-digested genomic DNA using a subtelomeric probe to the rDNA telomere (Jacob et al., 2004).
Telomerase activity assay
Cultures were grown to <5.0 × 105 cells/ml, lysed in T2MG (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol), and the extract was sonicated for 3 × 5 s pulses. The extract was then centrifuged at 13,000 × g for 15 min. Telomerase was batch purified by incubating the supernatant with an equal volume of DEAE Sepharose Fast Flow beads (Amersham, Buckinghamshire, UK) in T2MG with 100 mM sodium acetate, followed by elution in four volumes of T2MG with 350 mM sodium acetate. Activity assays were performed as described previously (Miller et al., 2000).
Supplementary Material
Acknowledgments
We thank Doug Chalker and Kathy Collins for reagents, Serena Heyse for helpful discussions, Michael Kuzyk for MS analysis, Christina Wicker for help with Pot1a purification and the DNA binding assays, and Wanda Manieri for antibody production. This work was supported by National Institutes of Health (NIH) grant GM088728 (to C.M.P.) and by the BC Cancer Agency (to G.B.M.). B.R.L. was supported by NIH grant T32 CA117846.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- dsDNA
double-stranded DNA
- MS
mass spectrometry
- MTT1
metallothionein
- rDNA
ribosomal DNA
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0551) on September 7, 2011.
REFERENCES
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Gooding AR, Cech TR. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol. 2010;30:4965–4976. doi: 10.1128/MCB.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Chiou SS. Non-nucleosomal packaging of a tandemly repeated DNA sequence at termini of extrachromosomal DNA coding for rRNA in Tetrahymena. Proc Natl Acad Sci USA. 1981;78:2263–2267. doi: 10.1073/pnas.78.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Blackburn EH. Two types of telomeric chromatin in Tetrahymena thermophila. J Mol Biol. 1998;280:327–344. doi: 10.1006/jmbi.1998.1867. [DOI] [PubMed] [Google Scholar]
- Coyne RS, et al. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics. 2008;9:562. doi: 10.1186/1471-2164-9-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- DeZwaan DC, Freeman BC. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci USA. 2009;106:17337–17342. doi: 10.1073/pnas.0905703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Kirk KE, Price CM. Generation of telomeric g strand overhangs involves both g and C strand cleavage. Mol Cell. 2003;11:1021–1032. doi: 10.1016/s1097-2765(03)00131-x. [DOI] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 2001;20:4299–4308. doi: 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Stout AR, Price CM. Modulation of telomere length dynamics by the subtelomeric region of tetrahymena telomeres. Mol Biol Cell. 2004;15:3719–3728. doi: 10.1091/mbc.E04-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T. Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol. 2010;30:1059–1066. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Mead CL, Kuzyk MA, Moradian A, Wilson GM, Holt RA, Morin GB. Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J Neurochem. 2010;113:1491–1503. doi: 10.1111/j.1471-4159.2010.06690.x. [DOI] [PubMed] [Google Scholar]
- Miller MC, Liu JK, Collins K. Template definition by tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- O'sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Price CM, editor. Ciliate Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. [Google Scholar]
- Robart AR, O'Connor CM, Collins K. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA. 2010;16:563–571. doi: 10.1261/rna.1936410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res. 2011 doi: 10.1016/j.mrfmmm.2011.08.011. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkewitz AP, Orias E, Kapler G. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 2002;18:35–40. doi: 10.1016/s0168-9525(01)02560-4. [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J Cell Sci. 2007;120:1978–1989. doi: 10.1242/jcs.006502. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.