The sulfur assimilation and phospholipid biosynthesis pathways interact metabolically and transcriptionally. Genetic analysis, genome-wide sequencing, and expression microarrays show that regulators of these pathways, Met4p and Opi1p, control cellular methylation capacity that can limit the growth rate.
Abstract
A yeast strain lacking Met4p, the primary transcriptional regulator of the sulfur assimilation pathway, cannot synthesize methionine. This apparently simple auxotroph did not grow well in rich media containing excess methionine, forming small colonies on yeast extract/peptone/dextrose plates. Faster-growing large colonies were abundant when overnight cultures were plated, suggesting that spontaneous suppressors of the growth defect arise with high frequency. To identify the suppressor mutations, we used genome-wide single-nucleotide polymorphism and standard genetic analyses. The most common suppressors were loss-of-function mutations in OPI1, encoding a transcriptional repressor of phospholipid metabolism. Using a new system that allows rapid and specific degradation of Met4p, we could study the dynamic expression of all genes following loss of Met4p. Experiments using this system with and without Opi1p showed that Met4 activates and Opi1p represses genes that maintain levels of S-adenosylmethionine (SAM), the substrate for most methyltransferase reactions. Cells lacking Met4p grow normally when either SAM is added to the media or one of the SAM synthetase genes is overexpressed. SAM is used as a methyl donor in three Opi1p-regulated reactions to create the abundant membrane phospholipid, phosphatidylcholine. Our results show that rapidly growing cells require significant methylation, likely for the biosynthesis of phospholipids.
INTRODUCTION
In the yeast Saccharomyces cerevisiae, the reactions and enzyme-encoding genes of several metabolic pathways, like the sulfur and phospholipid pathways studied here, have been identified and partially characterized. Studies of synthetic interactions between genes have sought to identify higher-order, indirect interactions between biological pathways (Tong et al., 2001). Data from these studies have enabled the creation of quantitative models that integrate yeast metabolism and transcriptional regulation (Herrgard et al., 2006). However, our understanding of the yeast cell is still incomplete, as exemplified by the fact that no existing model is able to perfectly predict the in vivo response of a cell to external stimuli or genetic perturbations (Zomorrodi and Maranas, 2010). One possible reason for this failure is that many genes have multiple cellular roles and participate in indirect or condition-specific genetic interactions that are difficult to identify and understand mechanistically. Likewise, many metabolites participate in multiple reactions, leading to complex connections between metabolic pathways.
One example is the sulfur assimilation pathway, which is a critical and ancient pathway present in all microbes and eukaryotes. This pathway incorporates extracellular sulfate into several key sulfur-containing compounds, including methionine, cysteine, homocysteine, and S-adenosylmethionine (SAM). The activity of this pathway has widespread influence on other cellular pathways, largely because SAM is required for most methyl transfer reactions (Thomas and Surdin-Kerjan, 1997). Accordingly, the biosynthetic genes of the sulfur assimilation pathway are controlled by a complex regulatory system that maintains the sulfur-containing compounds at appropriate levels (Lee et al., 2010). At the center of this regulatory system is the transcriptional activator Met4p, which is recruited to specific promoters by the site-specific DNA-binding cofactors Met31, Met32, and Cbf1 (Thomas and Surdin-Kerjan, 1997; Lee et al., 2010).
Transcriptional regulation occurs primarily at the level of Met4p activity, which is negatively regulated by SAM (Thomas et al., 1988; Kuras and Thomas, 1995), creating a negative feedback loop that decreases sulfur assimilation when its products are sufficiently abundant. When SAM is abundant, Met4p is ubiquitinated, which, depending on other conditions, either leads to Met4p degradation by the proteasome or a change in Met4p activity (Kaiser et al., 2000; Rouillon et al., 2000; Kuras et al., 2002; Flick et al., 2004; Chandrasekaran et al., 2006). Besides controlling methionine metabolism, Met4p, when artificially overexpressed, causes a G1-S arrest, suggesting that Met4p mediates a cell cycle checkpoint ensuring that cells contain sufficient levels of metabolites before embarking toward cell division (Patton et al., 2000).
In this study, we found that the sulfur assimilation pathway genetically interacts with another metabolic pathway, the phospholipid biosynthesis pathway. Phospholipids, such as phosphatidylcholine (PC), make up a large fraction of membrane bilayers. The enzymatic genes of this pathway are repressed by Opi1p, a protein that directly senses the levels of phosphatidic acid (PA), a precursor of phospholipid biosynthesis. When the levels of PA are low because PA has been consumed by phospholipid biosynthesis, Opi1p represses transcription by inhibiting, through an unknown mechanism, the DNA-binding transcriptional activators Ino2p and Ino4p (White et al., 1991; Santiago and Mamoun, 2003; Loewen et al., 2004; Jesch et al., 2005). Of interest, there is a metabolic connection between sulfur assimilation and phospholipid biosynthesis; PC is biosynthesized de novo from another phospholipid, phosphatidylethanolamine, in three steps that require SAM, formed by the sulfur assimilation pathway (Chin and Bloch, 1988).
Deleting MET4 results in methionine auxotrophy (Masselot and De Robichon-Szulmajster, 1975) because Met4p is required for the expression of many of the biosynthetic MET genes (Lee et al., 2010). Although there is no doubt that Met4p is required for methionine biosynthesis, there is conflicting evidence (described in Table 1) about whether Met4p is required for growth in the presence of exogenous methionine. Here we found that a met4Δ strain does indeed have a severe growth defect, suggesting that Met4p has a function in addition to regulating MET genes. Furthermore, we found that suppressors of the growth defect of a met4Δ strain arise very frequently. To understand how met4Δ growth is impaired, we used genome-wide tiling arrays, sequencing, and standard genetic analysis to characterize the suppressor mutations. We found that the suppression was most often caused by loss-of-function mutations in OPI1. Expression microarray studies using a Met4p-degradation system showed that Met4p activates genes involved in producing SAM, whereas Opi1p represses some of these genes in addition to genes whose products catalyze methyltransferase reactions of phospholipid biosynthesis. These results suggested that a met4Δ cell does not produce sufficient SAM for growth and a subsequent opi1 mutation stimulates SAM production. Indeed, we found that adding SAM to the media or overexpressing the SAM synthetase gene allowed met4Δ cells to grow like wild-type cells. These results suggest that coordinated expression of the sulfur and phospholipid pathways contributes to optimal growth by ensuring that cells can maintain the considerable requirement for methylation in the biosynthesis of cell membrane phospholipids.
TABLE 1:
Strain backgrounds and phenotypes of met4Δ alleles.
| Strain background | met4Δ growth phenotypea | Reference |
|---|---|---|
| S288C | – | Mountain et al. (1993) |
| S288C | – | Fauchon et al. (2002)b |
| S288C | Inviable | Giaever et al. (2002) |
| S288C | Slow-growing | Snoek and Steensma (2006) |
| W303 | – | Patton et al. (2000) |
| W303 | – | Aranda and del Olmo (2004)c |
| W303 | – | Leroy et al. (2006)d |
| W303 | – | Lee et al. (2010)d |
| BF264-15D | – | Kaiser et al. (2000)e |
| CY4 | – | Wheeler et al. (2002)f |
| 4094-B | – | Masselot and De Robichon-Szulmajster (1975)g |
aPhenotypes in the presence of exogenous methionine. –,no phenotype besides methionine auxotrophy.
bStrains in this study are made from the YPH98 strain, derived from YNN216, which is “congenic with S288C” (Sikorski and Hieter, 1989).
cThis study used a met4Δ strain in the W303 background that was created and used in earlier studies (Thomas et al., 1992; Kuras et al., 1996).
dThis study used a met4Δ strain in the W303 background that was created in an earlier study (Rouillon et al., 2000).
eThis study used the BF264-15D strain from Bruce Futcher (Reed et al., 1985).
fThis study used the CY4 strain first mentioned in a previous study (Grant et al., 1996).
gThis study used a methionine auxotroph that was isolated by UV mutagenesis. The identity of the mutation is not known.
RESULTS
Met4p is required for normal growth, even in the presence of excess exogenous methionine
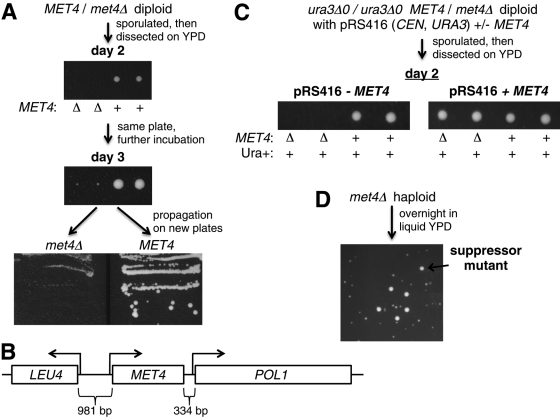
A met4Δ strain is a methionine auxotroph (Masselot and De Robichon-Szulmajster, 1975) but has been reported to have varying phenotypes in rich media, such as yeast extract/peptone/dextrose (YPD), that contain the exogenous methionine required for growth (Table 1). We noticed that fresh met4Δ haploid transformants formed very small colonies on YPD plates, even when provided with additional methionine. This phenotype was unstable: plating after overnight growth in YPD produced a mixture of small and large colonies. We decided to investigate this phenomenon by transforming a met4Δ allele into a diploid strain. The met4Δ/MET4 heterozygous diploid formed large colonies, similar to wild type, suggesting that a cell with one copy of MET4 is not haploinsufficient. To test more rigorously the growth phenotype of met4Δ haploids, we sporulated this diploid and followed growth of the haploid spore colonies on YPD plates (Figure 1A). The MET4 wild-type haploid grew as expected, but there was no met4Δ growth until day 3, suggesting that MET4 is not absolutely essential for growth but is required for the optimal rate of growth. This growth defect always segregated 2:2 with the met4Δ allele, indicating that it was caused by loss of MET4.
FIGURE 1:
Deleting MET4 causes slow growth, and this slow growth is spontaneously suppressed. (A) A met4Δ haploid grows poorly compared with MET4. A met4Δ/MET4 heterozygote (DBY12042) was sporulated, and the resulting tetrads were dissected on YPD plates. To show that the growth defect was not a simply a germination defect and persisted over many divisions, the spore colonies were restreaked onto fresh YPD plates and grown for 2 d (bottom). A MET4 wild type (FY4) is used as a control. Restreaking multiple times had the same effect. (B) The genomic locus surrounding the MET4 ORF (denoted by a box), showing that expression of the essential POL1 gene may be affected by deleting the MET4 ORF. (C) The met4Δ growth defect can be rescued by a plasmid containing MET4. A met4Δ/MET4 heterozygote containing either a MET4 plasmid (DBY12210, right) or a control plasmid (DBY12211, left) was sporulated, and the resulting tetrads were dissected on YPD plates. (D) When met4Δ haploid cells (DBY12213) are grown overnight in liquid rich medium, fast-growing cells arise in the population. This was repeated with met4Δ cells from ∼30 independent tetrads generated by sporulation of a met4Δ/MET4 heterozygote, with very similar results.
We initially tested three obvious explanations for met4Δ slow growth and found that none of them accounted for the phenotype. First, this growth defect is not the result of abnormal germination, the process by which the spore transitions to mitotic cycling, because this slow-growth phenotype is maintained even when postspore cells are streaked onto a fresh YPD plate (Figure 1A, bottom). Second, the poor growth of a met4Δ mutant was not due to insufficient import of exogenous methionine, because overexpression of the methionine transporters did not rescue this phenotype (Supplemental Figure S1). Third, the slow growth is not due to the absence of aerobic respiration, which requires sulfur metabolism (Bihlmaier et al., 2007), because met4Δ can grow on a nonfermentable carbon source (glycerol/ethanol) (Supplemental Figure S2).
The MET4 gene is only 334 base pairs upstream of the essential POL1 gene (Figure 1B), so we were concerned that the slow growth of a MET4-deletion mutant actually resulted from a perturbation of POL1. To rule out this possibility, we complemented met4Δ with a plasmid containing only the MET4 gene (Figure 1C). This plasmid did indeed allow met4Δ cells to grow like wild type, whereas a control plasmid lacking MET4 had no effect on met4Δ slow growth. These results indicated that it was the loss of MET4, and not loss of POL1 or other genes, that was responsible for the growth defect. The plasmid that we used contains the selectable gene URA3, so we attempted to select for loss of the plasmid by growing cells on 5-fluoroorotic acid (5-FOA), a drug that kills URA3-expressing cells. However, we found that met4Δ cells with a MET4 plasmid did not grow at all on 5-FOA because met4Δ cells alone do not grow on 5-FOA, even after extended incubation periods (Supplemental Figure S3).
The growth defect of met4Δ can be spontaneously suppressed
When met4Δ cells from an overnight liquid culture in YPD are plated, they give rise to a mixture of small and large colonies (Figure 1D). The size of the small colonies is similar to that of the previously observed met4Δ colonies. The size of the larger colonies is similar to that of wild-type colonies, suggesting that the met4Δ growth phenotype has been effectively suppressed. This large size is maintained when cells from a large colony are streaked onto fresh YPD plates, indicating that large colony size is mitotically stable (Supplemental Figure S4A). These cells remain methionine auxotrophs, separating the role of Met4p in methionine biosynthesis from its role in controlling cell growth. In addition, we found that this suppression phenotype is meiotically inherited by a mutation in an unlinked gene (Supplemental Figure S4B). To show this, the suppressed met4Δ strain was crossed to a MET4 strain and the resulting diploid was dissected. Half of the met4Δ spores produced small colonies and half produced wild type–sized colonies, as would be expected if the suppression were the result of a mutation in a single gene unlinked to MET4.
Identifying mutations that suppress the growth defect of met4Δ
To identify the gene(s) in which suppressor mutations arise, we isolated such suppressors from 24 independent cultures, each of which was started by a met4Δ mutant (small colony) from tetrad dissection of a heterozygous diploid, as in Figure 1D. In each case, large colonies were present at a high frequency, and we selected one of these colonies from each independent culture for further analysis. We performed initial complementation testing and found that these 24 mutants fell into several complementation groups, suggesting that suppression can be caused by a mutation in any of several different genes. To find potential causative mutations, we hybridized genomic DNA from several met4Δ suppressor mutants and their parental MET4 strains to whole-genome tiling arrays and then used a computer program (SNPScanner) (Gresham et al., 2006) to compare the genome sequence of each mutant to its parent. In this way, we found single-nucleotide polymorphisms (SNPs) in one of three genes: OPI1, TUP1, or DOT6. Then we sequenced the entire OPI1, TUP1, and DOT6 open reading frames in all 24 strains and found that nine strains contained potential mutations in OPI1, 2 had potential mutations in TUP1, and 1 had a potential mutation in DOT6 (Table 2). Two additional strains (DBY12223 and DBY12222 in Table 3 later in the paper) have genomic rearrangements at OPI1 that should produce an opi1 loss-of-function mutant phenotype. Finally, there were 10 strains in which no mutations were observed, possibly because either 1) the strains contained OPI1, TUP1, or DOT6 mutations (e.g., genomic rearrangements) that could not be detected by our analysis, or 2) the strains have mutations in other genes. Genetic analysis (see later discussion) confirmed that the OPI1 and TUP1 mutations are indeed causal; the DOT6 SNP was not studied further.
TABLE 2:
Identity of the met4Δ suppressor mutations.
| Nucleotide mutationa | Amino acid change |
|---|---|
| OPI1 | |
| G376Tb | E126amber |
| A409T | K137amber |
| G565T | E189amber |
| C614G | S205umber |
| A676G | K226E |
| T764C | L255S |
| C859T | Q287amber |
| C907T | Q303ochre |
| TUP1 | |
| C2091G | S697R |
| G2095T | D699Y |
| DOT6 | |
| C293A | S98amber |
aRelative to beginning of open reading frame.
bObserved in two suppressor mutants.
TABLE 3:
Strains used in this study.
| Name | Genotype | Reference |
|---|---|---|
| FY4 | MATa | Winston et al. (1995) |
| FY5 | MATa | Winston et al. (1995) |
| DBY11250 | MATa/a | This study |
| DBY12000 | MATa HAP1+ | Hickman and Winston (2007) |
| DBY12001 | MATa HAP1+ | Hickman and Winston (2007) |
| DBY12007 | MATa/a HAP1+/HAP1+ | Hickman and Winston (2007) |
| DBY12042 | MATa/a HAP1+/HAP1+met4Δ::KanMX/MET4 | This study |
| DBY12043 | MATa/a HAP1+/HAP1+met4Δ::NatMX/MET4 | This study |
| DBY12210 | MATa/a HAP1+/HAP1+ura3Δ0/ura3Δ0 met4Δ::KanMX/MET4 pRS416+MET4 | This study |
| DBY12211 | MATa/a HAP1+/HAP1+ura3Δ0/ura3Δ0 met4Δ::KanMX/MET4 pRS416 | This study |
| DBY12212 | MATa/a HAP1+/HAP1+ura3Δ0/ura3Δ0 met4Δ::KanMX/MET4 | This study |
| DBY12213 | MATa HAP1+met4Δ::KanMX | This study |
| DBY12214 | MATa HAP1+met4Δ::NatMX | This study |
| DBY12215 | MATa HAP1+met4Δ::KanMX opi1-G376Ta | This study |
| DBY12216 | MATa HAP1+met4Δ::NatMX opi1-A409Ta | This study |
| DBY12217 | MATa HAP1+met4Δ::KanMX opi1-G565Ta | This study |
| DBY11402 | MAT? hap1−met4Δ::NatMX opi1-C614Ga | This study |
| DBY12218 | MATa HAP1+met4Δ::KanMX opi1-A676Ga | This study |
| DBY12219 | MATa HAP1+met4Δ::NatMX opi1-T764Ca | This study |
| DBY12220 | MATa HAP1+met4Δ::NatMX opi1-C859Ta | This study |
| DBY12221 | MATa HAP1+met4Δ::NatMX opi1-C907Ta | This study |
| DBY12222 | MAT HAP1+met4Δ::NatMX opi1b | This study |
| DBY12223 | MAT HAP1+met4Δ::KanMX opi1b | This study |
| DBY12224 | MATa HAP1+met4Δ::NatMX tup1-C2091Ga | This study |
| DBY11405 | MATa hap1−met4Δ::NatMX tup1-G2095Ta | This study |
| DBY11406 | MATa hap1−met4Δ::NatMX dot6-C293Aa | This study |
| DBY12225 | MATa/a HAP1+/HAP1+met4Δ::KanMX/MET4 opi1Δ:: NatMX/OPI1 | This study |
| DBY12226 | MATa HAP1+met4Δ:: NatMX opi1Δ::KanMX | This study |
| DBY12227 | MAT a HAP1+met4Δ:: NatMX opi1Δ::KanMX | This study |
| DBY12228 | MATa/a HAP1+/HAP1+met4Δ::NatMX/MET4 KanMX::GAL1pr-INO1/INO1 | This study |
| DBY12229 | MATa/a HAP1+/HAP1+met4Δ::KanMX/MET4 opi1Δ::NatMX/OPI1 ino1Δ::HphMX4/INO1 | This study |
| DBY12230 | MATa/a HAP1+/HAP1+met4Δ::NatMX/MET4 KanMX::GAL1pr-MUP1/MUP1 | This study |
| DBY12231 | MATa/a HAP1+/HAP1 met4Δ::NatMX/MET4 KanMX::GAL1pr-MUP3/MUP3 | This study |
| DBY12232 | MATa/a HAP1+/HAP1+met4Δ::KanMX/MET4 tup1Δ::NatMX/TUP1 | This study |
| DBY12233 | MATa/a HAP1+/HAP1 met4Δ::NatMX/MET4 KanMX::GAL1pr-SAM2/SAM2 | This study |
| DBY12049 | MATa HAP1 gal1Δ::GAL1pr-TEV::HphMX4 leu2Δ0::ACT1pr-GEV::NatMX gal4Δ::LEU2 | McIsaac et al. (2011) |
| DBY12055 | MATa HAP1 gal1Δ::GAL1pr-TEV::HphMX4 leu2Δ0::ACT1pr-GEV::NatMX gal4Δ::LEU2 NDeg-MET4-13Xmyc::KanMX | McIsaac et al. (2011) |
| DBY12074 | MATa HAP1 gal1Δ::GAL1pr-TEV::HphMX4 leu2Δ0::ACT1pr-GEV::NatMX gal4Δ::LEU2 NDeg-MET4-13Xmyc::KanMX opi1Δ::KanMX | This study |
| CC849-8A | MATa ade2-1 his3-1,15 leu2-3112 trp1-1 ura3 met4Δ::TRP1 | Rouillon et al. (2000)c,d |
| CC849-1B | MATa ade2-1 his3-1,15 leu2-3112 trp1-1 ura3 met4Δ::TRP1 | Barbey et al. (2005)c |
| yMT-234 | MATa ade2-1 can1-100 his3-1,15 leu2-3112 trp1-1 ura3 | K. Nasmythc |
aRefers to the nucleotide change relative to the ATG.
bThe exact opi1 mutation is not known, but sequencing evidence suggests that there is a genomic arrangement.
cThese strains are in the W303 genetic background.
dWe found that this strain is MATa, in contrast to the reported MAT (Rouillon et al., 2000).
Mutations in TUP1 suppress the growth defect of met4Δ
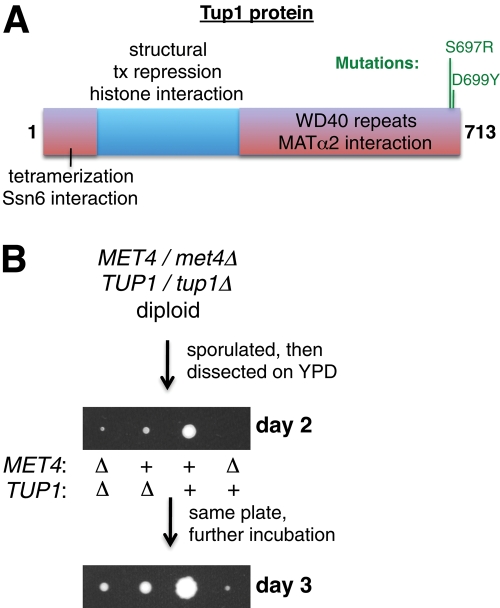
Two SNPs in the TUP1 open reading frame were found to potentially suppress the met4Δ growth defect (Table 2). These SNPs resulted in two amino acid changes that were very close together, as shown in the domain structure of the Tup1 protein (Figure 2A). Tup1 is a general transcriptional corepressor that represses many genes in yeast and is recruited to DNA by transcription factors (Malave and Dent, 2006). Of note, the mutations that we isolated specifically affect a surface of a WD40 propeller blade that interacts with MAT2 and possibly other transcription factors (Komachi and Johnson, 1997; Sprague et al., 2000), suggesting that disruption of Tup1 binding to transcription factors like Opi1p (discussed later) might be responsible for met4Δ growth suppression. To test whether deleting TUP1 suppresses the poor growth of met4Δ, we created a diploid strain that was heterozygous for both met4Δ and tup1Δ (Figure 2B). Then this diploid was sporulated and dissected on YPD plates, creating spores with all combinations of deletions. As has been widely observed, we found that a tup1Δ strain grew slower than did the wild type. Nonetheless, a met4Δ tup1Δ strain grew better than a met4Δ strain, indicating that tup1Δ suppresses the met4Δ growth defect.
FIGURE 2:
Mutations in TUP1 suppress the growth defect of met4Δ. (A) We found two met4Δ-suppressed strains with mutations in the TUP1 gene (Table 2). The mutations (S697R and D699Y) are depicted in relation to the encoded Tup1p protein and its previously determined domains (Komachi and Johnson, 1997), and they are in a region of the protein that affects Tup1p interaction with transcription factors (Sprague et al., 2000). (B) Deletion of TUP1 causes a met4Δ haploid to grow significantly better. A met4Δ/MET4 tup1Δ/TUP1 diploid (DBY12232) was sporulated, and the resulting tetrads were dissected on YPD plates. The met4Δ tup1Δ cells remained methionine auxotrophs.
Deleting OPI1 suppresses the growth defect of met4Δ
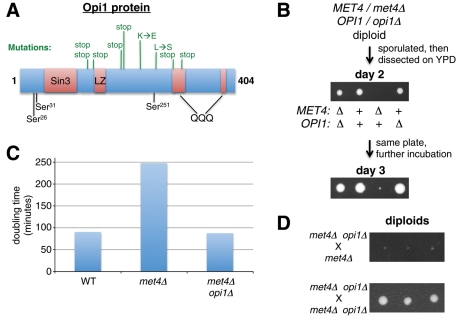
The mutations in the OPI1 gene (Table 2) were overlaid on the domain structure of the Opi1p protein (Figure 3A). This map shows that a variety of mutations, including severe mutations that prevent expression of the C-terminal two-thirds of Opi1p, potentially suppress the met4Δ growth defect. This suggested that loss of Opi1p function may suppress the met4Δ phenotype and led us to test whether an opi1Δ allele deletion also suppresses the met4Δ phenotype. First, we created a diploid strain that was heterozygous for both opi1Δ and met4Δ (Figure 3B). Then this diploid was sporulated and dissected on YPD plates, creating spores with all combinations of deletions. As expected, we observed that opi1Δ suppressed the met4Δ growth defect. As a control, opi1Δ did not affect growth of an otherwise wild-type strain. To quantify the effect of OPI1 on growth rate, we compared the doubling times of these strains in liquid YPD, which confirmed that met4Δ grows more slowly (i.e., has a greater doubling time) than either MET4 or met4Δ opi1Δ (Figure 3C). The opi1Δ allele acted recessively, as expected, because crossing a met4Δ opi1Δ strain to a met4Δ strain resulted in slow-growing diploids (Figure 3D, top). As a control, crossing a met4Δ opi1Δ strain to a met4Δ opi1Δ strain resulted in normally growing diploids (Figure 3D, bottom).
FIGURE 3:
Opi1p is required for the slow growth of met4Δ cells. (A) The mutations in the OPI1 gene are depicted in relation to the encoded Opi1p protein and its previously determined domains (Sreenivas and Carman, 2003). The following structural and functional domains are depicted: Sin3 (Sin3-interacting), LZ (leucine zipper), and QQQ (glutamine rich). Protein kinase A phosphorylates residues Ser-31 and Ser-251, whereas protein kinase C phosphorylates Ser-26. (B) Deletion of OPI1 causes a met4Δ haploid to grow like wild type. A met4Δ/MET4 opi1Δ/OPI1 diploid (DBY12225) was sporulated, and the resulting tetrads were dissected on YPD plates. The tetrads showed the 2:2, 3:1, and 4:0 large:small patterns as expected. The met4Δ opi1Δ cells remained methionine auxotrophs. (C) The doubling time of met4Δ is higher than that for MET4 or met4Δ opi1Δ. The MET4 (DBY12000), met4Δ (DBY12214), and met4Δ opi1Δ (DBY12226) strains were inoculated into YPD at low density and the cell concentration was followed until they reached saturation. The exponential phase of growth was used to calculate the doubling time only for cultures that did not develop growth suppression. A representative graph from many experiments is shown. (D) The opi1Δ allele acts recessively to suppress the growth defect of a met4Δ strain. Top, a met4Δ haploid (DBY12214) was crossed to a met4Δ opi1Δ haploid (DBY12227), and the resulting met4Δ/met4Δ opi1Δ/OPI1 diploids (three are shown) were grown for 2 d. Bottom, a met4Δ opi1Δ haploid (DBY12227) was crossed to a met4Δ opi1Δ haploid (DBY12226), and the resulting met4Δ/met4Δ opi1Δ/opi1Δ diploids (three are shown) were grown for 2 d.
Simultaneously studying transcriptional regulation by Met4p and Opi1p
We further investigated the genetic interaction between Opi1p and Met4p because, in addition to the results presented earlier, we previously found that the expression of two Opi1p targets (OPI3 and CKI1) depends specifically on methionine and sulfur abundance (Petti et al., 2011). Given that both Opi1p and Met4p are transcriptional regulators, we sought to understand their genetic interaction from a transcriptional perspective. Because a met4Δ strain readily develops growth suppressor mutations, we studied Met4p transcriptional regulation using a system in which Met4p can be quickly degraded by an estradiol-inducible TEV protease (McIsaac et al., 2011). Such a system is indispensable for studying unstable deletion phenotypes like that of met4Δ, because the target protein (in this case, Met4p) is degraded with a half-life of ∼15 min, well before suppressor mutations can arise. We used the NDeg-MET4 strain containing the machinery necessary for estradiol induction, an inducible TEV protease gene, and the MET4 gene N-terminally tagged with a TEV protease site. To simultaneously study Opi1p transcriptional regulation, we used a pair of strains: 1) NDeg-MET4, in which OPI1 is wild type, and 2) opi1Δ NDeg-MET4. We also included a control strain that contains the estradiol-induction machinery and inducible TEV protease gene but wild-type MET4 and OPI1 genes.
To compare gene expression across these strains, we grew each to midexponential phase in minimal medium, added estradiol, and measured mRNA abundance at several time points up to 6 h after estradiol addition (Supplemental Data Set S1). We focused our analysis on the immediate response observed within 30 min after estradiol addition (Materials and Methods). Confirming that Met4p is degraded, we found that the canonical Met4p targets (Lee et al., 2010) are indeed turned off after estradiol addition (Figure 4). Many of these targets are slightly turned off in the control strain, but they are significantly more turned off in the NDeg-MET4 strain (see Figure 5 and Supplemental Data Set S1). Furthermore, many of the targets known to be repressed by Opi1p (Santiago and Mamoun, 2003) are derepressed in opi1Δ NDeg-MET4 compared with NDeg-MET4 (Figure 4).
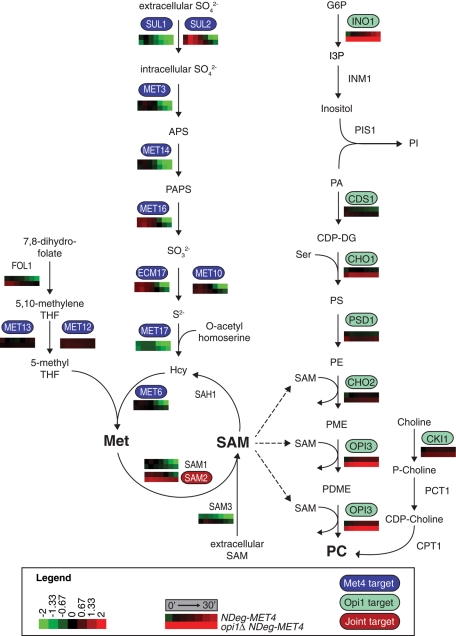
FIGURE 4:
Role of Met4p and Opi1p in regulating sulfur assimilation and PC biosynthesis pathways. Gene expression data for the biosynthesis genes were overlaid on the pathways. Gene expression time courses for a given gene are shown beneath that gene. Top and bottom, NDeg-MET4 and opi1Δ NDeg-MET4, respectively, with log2 fold change as indicated in the legend. Gene expression time courses are displayed as in the legend: from the instant before 1 μM estradiol treatment (0′) until 30 min after treatment (30′). Values at 0, 5, 15, and 30 min represent actual experimental measurements (indicated with black boxes above the time course), whereas values at 10, 20, and 25 min represent interpolated values (indicated with gray boxes above the time course) (see Materials and Methods). Genes directly regulated by Met4p (Lee et al., 2010) are shown in blue; genes directly regulated by Opi1p (Santiago and Mamoun, 2003) are shown in green; joint targets are shown in red. CDP-choline, cytidine diphosphate choline; CDP-DG, cytidine diphosphate diacylglycerol; APS, 5′-adenylylsulfate; G6P, glucose 6-phosphate; Hcy, homocysteine; I3P, inositol 3-phosphate; Met, methionine; P-choline, phosphocholine; PA, phosphatidic acid; PAPS, 3′-phospho-5′adenylylsulfate; PC, phosphatidylcholine; PDME, phosphatidyl-dimethylethanolamine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PME, phosphatidyl-monomethyl-ethanolamine; PS, phosphatidylserine; S2−, sulfide; SAM, S-adenosylmethionine; SO32−, sulfite; SO42−, sulfate; THF, tetrahydrofolate.
FIGURE 5:
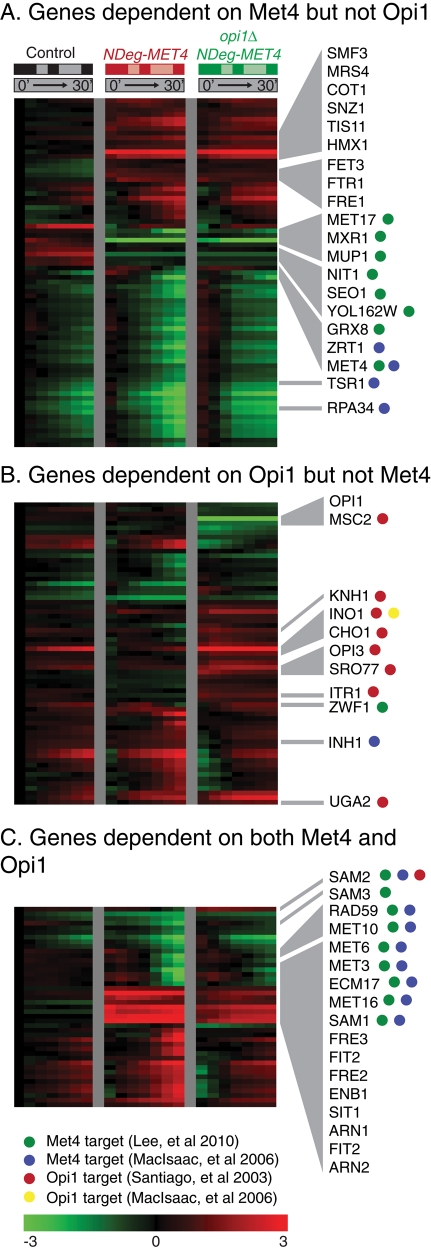
Genome-wide expression analysis of Met4p and Opi1p regulation. Multiple regression was used to identify genes whose expression is primarily dependent on Met4p abundance (A), Opi1p abundance (B), or both (C). Three strains (the control strain, NDeg-MET4, and opi1Δ NDeg-MET4) were treated with 1 μM estradiol. Gene expression is shown from the instant before estradiol treatment (0′) until 30 min after treatment (30′), with log2 fold change as indicated in the legend. Values at 0, 5, 15, and 30 min represent actual experimental measurements (indicated with black, red, or dark green boxes above the time courses, respectively), whereas values at 10, 20, and 25 min represent interpolated values (indicated with gray, pink, or light green boxes above the time courses, respectively; see Materials and Methods). Only genes that change by a factor of at least 1.5 in at least one time course are shown. Gene names discussed in the text are shown in the figure. Previously identified targets of Met4p are indicated with green (Lee et al., 2010) or blue (MacIsaac et al., 2006). Previously identified targets of Opi1p are indicated with red (Santiago and Mamoun, 2003) or yellow (MacIsaac et al., 2006) dots.
In an initial, semiquantitative comparison of NDeg-MET4 and opi1Δ NDeg-MET4, we found that INO1 exhibited the most striking difference between the strains (Figures 4 and 5B). INO1, a gene required for inositol biosynthesis, is the best-described target of Opi1p (White et al., 1991). It is also a target of Cbf1, a DNA-binding partner of Met4p (Shetty and Lopes, 2010). Even before estradiol addition, the INO1 transcript is ∼32 times more abundant in opi1Δ NDeg-MET4 than in the other strains. INO1 expression increases over time in all three strains (including the control), reaching a final abundance in opi1Δ NDeg-MET4 that is 23 times greater than in NDeg-MET4 and 9 times greater than in the control.
Deleting or manipulating expression of INO1 has no effect on the growth of either met4Δ or met4Δ opi1Δ strains
To test whether the slow growth phenotype of met4Δ is due to INO1 misregulation, we made a met4Δ/MET4 GAL1pr-INO1/INO1 diploid strain, sporulated it and dissected the spores on yeast extract/peptone (YP) media either with glucose or galactose as a carbon source (to turn off or on INO1 expression, respectively). We found that repression or overexpression of INO1 had no effect on met4Δ growth (Supplemental Figure S5A). Next we wanted to know whether the normal growth of met4Δ opi1Δ was due to a change in INO1 expression. We therefore made a met4Δ/MET4 opi1Δ/OPI1 ino1Δ/INO1 diploid strain, sporulated it, and dissected the spores on YPD media. Deleting INO1 had no effect on the growth of any strains, including a met4Δ mutant and a met4Δ opi1Δ mutant (Supplemental Figure S5B). These data show that INO1 is not involved in the regulation of growth by Met4p and Opi1p.
Gene expression analysis suggests that Met4p and Opi1p both regulate SAM levels
Next we searched for other gene expression differences that might explain the role of Met4p and Opi1p in growth regulation. We began by using multiple regression to quantify the dependence of each gene in the original data set on Met4p and Opi1p (see Materials and Methods for data processing and filtering). This analysis identified most of the known Met4p and Opi1p target genes (Santiago and Mamoun, 2003; Lee et al., 2010), as well as other genes that may either be indirectly regulated or newly identified targets (Figure 5). All of these genes can be grouped into three categories: 1) 75 genes affected only by Met4p abundance (Figure 5A), 2) 65 genes affected only by Opi1p abundance (Figure 5B), and 3) 43 genes affected by both Met4p and Opi1p (Figure 5C). This coregulation by Met4p and Opi1p is statistically significant (p < 0.0001), suggesting that the biochemical processes regulated by Met4p (sulfur assimilation) and Opi1p (phospholipid biosynthesis) are interdependent. However, almost all coregulation by Met4p and Opi1p is carried out by indirect or nontranscriptional mechanisms, because only one gene, SAM2, has been shown to be regulated by both Met4p and Opi1p (Santiago and Mamoun, 2003; Lee et al., 2010). The SAM2 gene will be discussed later.
To further understand how Opi1p specifically affects growth of a met4Δ strain, we analyzed the 108 genes that are Opi1p regulated (i.e., differentially expressed between opi1Δ NDeg-MET4 and NDeg-MET4; false-discovery rate [FDR] < 0.1; Figure 5, B and C). This set is highly enriched for genes whose products 1) associate with the cellular membrane and 2) function in cation homeostasis, oxidation–reduction, sulfur assimilation, the trichloracetic acid cycle, carbohydrate metabolism, and transmembrane transport (Supplemental Data Set S2). Notably, this set also contains three of the five genes that regulate intracellular SAM abundance: the two SAM synthetase genes (SAM1 and SAM2) and the SAM transporter gene (SAM3; p = 0.0016 by hypergeometric distribution). From this analysis, we speculated that deleting OPI1 suppresses the growth defect of a met4Δ strain by affecting one or more of these Opi1p-regulated processes.
Inspection of the metabolic pathways governing phospholipid biosynthesis reveals that PC biosynthesis, which is repressed by Opi1p, requires SAM at three steps (Figure 4; Chin and Bloch, 1988). We overlaid gene expression data on the metabolic pathways regulated by Met4p and Opi1p and found that several SAM biosynthesis genes are activated by Met4p and repressed by Opi1p. Most notably, SAM2 is weakly activated by Met4 and strongly repressed by Opi1 (p = 9.4 × 10−3 based on use of the F test to compare opi1Δ NDeg-MET4 and NDeg-MET4; see Materials and Methods). Average SAM2 abundance, which changes little in response to estradiol, is 5.5 times higher in opi1Δ NDeg-MET4 than in NDeg-MET4 (p = 0.038 by Student's t test). SAM3 behaves similarly (p = 0.003 by the F test); on average, SAM3 abundance is 12 times higher in opi1Δ NDeg-MET4 than in NDeg-MET4 (p = 0.078 by Student's t test). Although SAM1 follows a similar pattern, it is only weakly repressed by OPI1: during the first 30 min after estradiol addition, SAM1 abundance falls nearly fourfold in NDeg-MET4 and 2.1-fold in opi1Δ NDeg-MET4. Thus the interdependence of Met4p- and Opi1p-regulated pathways may be based on SAM metabolism. In previous work, we found that SAM is rapidly and dramatically depleted in methionine auxotrophs (Petti et al., 2011). We therefore hypothesized that met4Δ mutants grow slowly because they lack sufficient SAM for PC biosynthesis.
Increased amounts of SAM suppress the growth defect of met4Δ
SAM is required for growth and can be either synthesized endogenously by Sam1p or Sam2p or added exogenously to a SAM auxotroph. However, SAM is not present in YPD because it is a highly unstable metabolite and thus likely does not survive the autoclaving process (Kuras et al., 2002). Therefore we predicted that met4Δ cells grow slowly because they are unable to express the Met4p-regulated SAM2 gene and thus synthesize SAM. To test this, we first sporulated a met4Δ/MET4 heterozygote and dissected the spores on YPD with freshly added SAM (Figure 6A). Indeed, we found that a met4Δ spore grew almost as well as a wild-type spore under these conditions. Next we created a met4Δ/MET4 GAL1pr-SAM2/SAM2 diploid strain, sporulated it, and dissected spores on YP media either with glucose or galactose as a carbon source (to turn off or on SAM2 expression, respectively) (Figure 6B). As expected, a met4Δ GAL1pr-SAM2 spore grew like wild type on galactose (when overexpressing SAM2) and poorly on glucose (with SAM2 turned off). Thus the poor growth of met4Δ cells can be suppressed by increasing the intracellular concentration of SAM.
FIGURE 6:
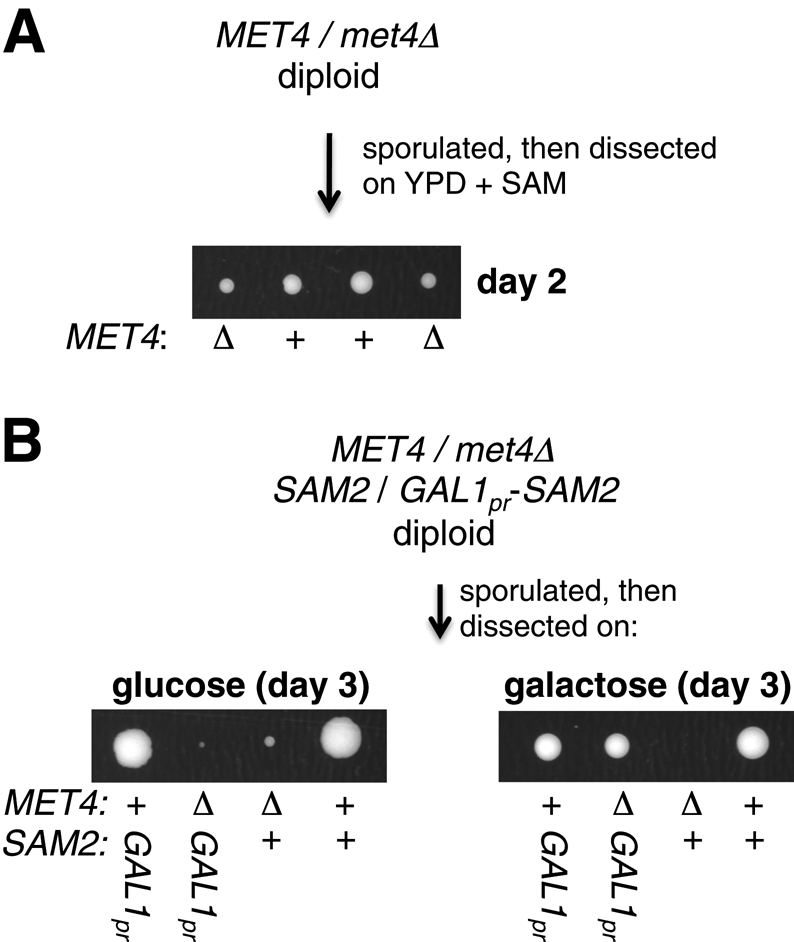
Increasing intracellular levels of SAM suppresses the growth defect of met4Δ. (A) Adding SAM to YPD rescues growth of a met4Δ haploid. A met4Δ/MET4 heterozygote (DBY12043) was sporulated, and the resulting tetrads were dissected on YPD plates with 0.2 mM SAM. Many SAM concentrations (0.05–0.5 mM) were tested, with 0.2 mM supporting the optimal met4Δ growth. The met4Δ cells remained, as expected, methionine auxotrophs. (B) Overexpression of the SAM2 gene rescues growth of a met4Δ haploid. A met4Δ/MET4 GAL1pr-SAM2/SAM2 heterozygote (DBY12233) was sporulated, and the resulting tetrads were dissected on YPD or YPGal plates.
Expression data suggest that Met4p and Opi1p both indirectly affect membrane function
The Met4p-dependent genes that we identified earlier have a wide variety of cellular functions (Supplemental Data Set S2). One group of genes, those repressed by Met4p (i.e., induced during Met4p degradation), is enriched for iron homeostasis genes (FTR1, SMF3, COT1, MRS4, FET3, FRE1; Figure 5A; FDR < 0.1). These genes were derepressed even before estradiol was added to the NDeg-MET4 strain, suggesting that Met4p may be slightly unstable in this strain due to modification and that these genes are very sensitive to the levels of Met4p. Consistent with these data, we previously observed dramatic induction of the same iron homeostasis genes upon deletion of MET31 and MET32 (Petti et al., 2011).
We found that these and additional iron homeostasis genes (FRE3, FRE2, FIT2, SIT1, ARN1, FIT3, MSC2, ARN2, ENB1) are also activated by Opi1p (i.e., less strongly induced by Met4p degradation in opi1Δ NDeg-MET4 than in NDeg-MET4; Figure 5C). Surprisingly, none of these genes is a known direct target of Met4p or Opi1p (or Met31/Met32/Cbf1; Figure 5, A–C), indicating that Met4p and Opi1p affect iron homeostasis by indirect regulation of expression. Of note, all of the proteins encoded by these genes are physically located in or at the cellular membrane, suggesting the following hypothetical chain of regulatory events: Met4p degradation leads to a drop in SAM levels and a subsequent decrease in PC biosynthesis. This PC decrease perturbs a wide variety of processes, such as iron homeostasis, that depend on cellular membranes for transport and intracellular homeostasis. Deleting OPI1 bypasses the decrease in PC biosynthesis, alleviating the perturbation in iron homeostasis.
The met4Δ slow-growth phenotype occurs in the W303 strain background
We found that deleting MET4 in the S288C genetic background causes slow growth. We wanted to test whether this is the case in another background, W303, which has been used extensively in studying Met4p (Table 1). We found that two previously generated met4Δ strains have frameshift opi1 mutations that allow them to grow normally. On crossing these strains to a wild-type strain and sporulating the diploid, we found that half of the resulting met4Δ spores grew slowly (no suppression) and the other half grew normally (suppressed). The met4Δ slow growth can be rescued by adding SAM to the media (Supplemental Figure S6).
DISCUSSION
Met4p has long been known to regulate the genes of the sulfur assimilation pathway, which is responsible for the incorporation of extracellular sulfate into methionine, cysteine, homocysteine, and SAM. Indeed, Met4p was previously shown to be required for methionine biosynthesis (Masselot and De Robichon-Szulmajster, 1975). We show here that Met4p also contributes to the biosynthesis of SAM, an essential metabolite that serves as the methyl donor in most methyltransferase reactions (Thomas and Surdin-Kerjan, 1997). Specifically, we found that SAM must be added to YPD in order for met4Δ cells to grow at wild-type rates, indicating that met4Δ is not only a methionine auxotroph but also a partial SAM auxotroph even in the presence of excess methionine, either provided in the medium or by overexpression of a methionine transporter. However, met4Δ cells are able to grow, albeit slowly, on YPD, indicating that met4Δ cells either carry out limited SAM biosynthesis from methionine or import some SAM from rich media.
The increased SAM requirement of met4Δ is consistent with the known role of Met4p in regulating the expression of genes required for SAM biosynthesis, specifically the biosynthetic MET and SAM genes (Thomas and Surdin-Kerjan, 1987; Thomas et al., 1988; Rouillon et al., 2000; Lee et al., 2010). These genes are directly induced by Met4p (Lee et al., 2010), consistent with our microarray analysis showing that expression of these genes decreases upon Met4p degradation (Figures 4 and 5 and Supplemental Data Set S1). In addition, Met4p degradation decreases the expression of FOL1, encoding an enzyme in the folate metabolism pathway that contributes a methyl group in the conversion of homocysteine to methionine.
The met4Δ growth defect can be suppressed by increasing endogenous SAM levels, as described earlier, or by spontaneous loss-of-function mutations in a variety of genes, including OPI1 and TUP1. Independently deleting OPI1 or TUP1 in a met4Δ strain recapitulated the effects of these mutations. Our expression microarray analysis showed that Opi1p represses SAM2 expression, whereas Met4p induces SAM2, consistent with previous results (Kodaki et al., 2003; Santiago and Mamoun, 2003; Jesch et al., 2005). This suggests that opi1Δ suppresses met4Δ slow growth by derepressing SAM2 and thereby increasing SAM levels. Indeed, overexpression of SAM2 is sufficient to suppress the growth defect of a met4Δ strain. The ability of a tup1Δ allele to suppress the met4Δ growth defect may also lie in its ability to compensate for the SAM deficiency of met4Δ. This is because Tup1, like Opi1p, represses SAM2, as Tup1 is required for repression of Opi1p-regulated genes (Wagner et al., 2001).
Our experiments suggest that derepression of SAM2 is the main mechanism of suppressing the met4Δ growth defect, but Opi1p inactivation might compensate for the SAM deficiency through four alternative mechanisms. First, an opi1 mutation derepresses all of the PC biosynthetic genes (CDS1, CHO1, PSD1, CHO2, and OPI3), thereby increasing flux through the pathway. If PC is the major consumer of SAM, as we argue later, this may allow cells to produce sufficient PC when SAM levels are low. Second, an opi1 mutation derepresses expression of CKI1, required for the Kennedy pathway that produces PC without SAM. However, this pathway uses choline obtained from the breakdown of PC and thus cannot be used for de novo PC biosynthesis. Third, an opi1 mutation derepresses methionine biosynthetic genes, presumably leading to increased production of methionine and SAM. Fourth, an opi1 mutation derepresses the FOL1 gene, a component of the folate cycle that donates a methyl group for methionine biosynthesis. The result could be increased levels of methionine and subsequently SAM.
Our results point to transcriptional and metabolic coordination between the sulfur assimilation pathway, regulated by Met4p, and the PC biosynthetic pathway, regulated by Opi1p. Previous work implied that these pathways are connected through SAM-dependent methylation (Santiago and Mamoun, 2003; Malanovic et al., 2008; Petti et al., 2011). Here we show that Opi1p and Met4p genetically interact because they both contribute to regulating SAM levels. The sulfur assimilation pathway is required for the biosynthesis of SAM, which provides a methyl group during three steps in PC biosynthesis (Figure 4). Opi1p directly represses SAM2 expression as well as PC biosynthetic genes, likely to maintain coordination of SAM levels with the SAM requirement of PC biosynthesis. Our search of the SAM2 literature and for potential binding sites in the SAM2 promoter shows that, besides the Met4p transcriptional complex, Opi1p is the only known direct regulator of SAM2. From this finding, we hypothesize that PC biosynthesis accounts for the majority of SAM-dependent methylation reactions, although this remains to be tested. Phospholipids represent a major portion of the dry weight of a yeast cell (Nurminen et al., 1976; Strathern et al., 1982), with PC making up at least 30% of phospholipids (Strathern et al., 1982). As shown in Figure 4, PC is biosynthesized de novo from another phospholipid, phosphatidylethanolamine (PE), in three SAM-consuming methyltransferase reactions catalyzed by Opi3 and Cho2 (Chin and Bloch, 1988; Summers et al., 1988; McGraw and Henry, 1989; Kodaki et al., 2003). In contrast, ergosterol, another membrane lipid synthesized by SAM-dependent methylation, may be as abundant as PC (Nurminen et al., 1976), but the synthesis of one ergosterol molecule only consumes one SAM (McCammon et al., 1984). Thus the SAM requirement is three times higher to make PC than to make ergosterol. Although SAM is used in several other reactions, such as during the biosynthesis of biotin and polyamine and in the modification of RNA and proteins (Thomas and Surdin-Kerjan, 1997), these reactions are likely not significant consumers of SAM because the products are at relatively low levels in comparison to the membrane lipids.
OPI1 inactivation was the most common spontaneous mutation causing met4Δ suppression. We found that the mechanism of suppression is consistent with an increase in SAM levels, and OPI1 inactivation is likely the simplest way to achieve this. Opi1p represses transcription by inhibiting the transcriptional activators Ino2 and Ino4, and therefore an opi1Δ mutant causes relative activation of Ino2/Ino4–dependent genes like SAM2 (Santiago and Mamoun, 2003; Jesch et al., 2005; Chen et al., 2007). Because it is more likely that a mutation causes a loss of function (LOF) than a gain of function (GOF), increased SAM2 expression is more likely to be caused by LOF opi1 mutations than GOF mutations in INO2, INO4, the SAM2 promoter, or other positive regulatory genes. There are other SAM2 negative regulators, such as TUP1, that could be the target of LOF mutations. However, since TUP1 itself is required for normal growth, few mutations in this gene will cause suppression without affecting growth. Indeed, the two tup1 SNPs that we identified likely were this type of mutation because they were close together on the TUP1 gene and resulted in growth that was better than that with tup1Δ.
Finally, our findings may be of clinical importance. It was previously shown that inhibiting the methylation steps of de novo PC biosynthesis, through deletion of SAH1, OPI3, or CHO2, leads to triacylglycerol accumulation, an indicator of several diseases, including heart disease (Malanovic et al., 2008). We found that SAM deficiency caused by met4Δ may also inhibit the PC pathway. Our work shows that inactivation of Opi1p might prevent this inhibition and the consequent accumulation of triacylglycerol. Although humans do not have an Opi1p orthologue, they may have a similar mechanism for sensing phospholipid levels (Loewen and Levine, 2005). Thus targeting this mechanism could be a way to treat diseases of lipid accumulation. More generally, a more complete understanding of the regulatory relationships between metabolic pathways will be useful in understanding the etiology of metabolic diseases.
MATERIALS AND METHODS
Strains
All S. cerevisiae strains are listed in Table 3, and almost all are isogenic with a GAL2+ derivative of S288C containing a repaired HAP1 allele (Winston et al., 1995; Hickman and Winston, 2007). The rest of the strains are in the W303 background. Strains were constructed by standard methods, either by crosses or by transformation (Ausubel, 2001). The deletion alleles were created by replacing the respective open reading frame (ORF) with the KanMX or NatMX (Brachmann et al., 1998) or HphMX4 markers (Carter and Delneri, 2010). The KanMX::GAL1pr-ORF alleles (where ORF = SAM2, MUP1, MUP3, or INO1) was constructed by placing the KanMX marker and the GAL1 promoter upstream of the respective ORF (Longtine et al., 1998). The selection drugs used were ClonNat (100 μg/ml; Werner BioAgents, Jena, Germany), G418 (100 μg/ml; Cellgro, Manassas, VA), and hygromycin (2900 U/ml; Calbiochem, La Jolla, CA).
Media and growth conditions
Cells were grown at 30°C in 1% yeast extract, 2% peptone, 2% glucose (YPD), unless otherwise noted. Yeast extract/peptone/galactose (YPGal) and YP + glycerol/ethanol contained 2% galactose and 2% of a 50/50 glycerol/ethanol mixture, respectively, in place of glucose. SAM (A7007; Sigma-Aldrich, St. Louis, MO) was made up in water at 20 mg/ml, filter sterilized, and added to YPD plates 1 d before plating cells. The met4Δ allele was maintained in a met4Δ/MET4 heterozygous diploid to prevent suppressor formation; there was no haploinsufficiency or suppressor formation seen in this diploid. To create a met4Δ haploid with or without other mutations, the relevant diploid was sporulated by growing it to mid-log in YPD, washed in water, and grown in sporulation media (1% potassium acetate) for 3 d at room temperature. The resulting tetrads were dissected by dissection microscopy. As denoted in the figures, pictures were taken after 2 or more days of growth. In the figures, a representative tetrad may be shown, but at least 10 complete tetrads were analyzed for each diploid. The resulting spores were tested for all of the relevant marker phenotypes. All experiments in liquid media were plated upon completion to check for growth suppression of the met4Δ growth defect.
Mutation analysis
Suppressor strains were colony purified by plating and then grown overnight in liquid YPD. Genomic DNA was isolated from saturated cultures using a Qiagen (Valencia, CA) Genomic DNA kit and prepared for hybridization to high-density, whole-genome tiling arrays (GeneChip S. cerevisiae Tiling 1.0R; Affymetrix, Santa Clara, CA), as described previously (Gresham et al., 2006). The hybridization data were processed by the SNP Scanner program, which calculated the probability of a mutation at each nucleotide in the genome. These data were visualized using Integrated Genome Browser (http://bioviz.org/igb/) to determine mutations that were specific to met4Δ suppressor strains compared with a reference (FY4) and parental strains (DBY12000 and DBY12001).
Estradiol-induction mRNA abundance–time courses
A single colony of NDeg-MET4, opi1Δ NDeg-MET4, or the control strain was grown to mid-log phase. Estradiol was added to a final concentration of 1 μM or 10 nM, and 5-ml aliquots of culture were filtered and flash-frozen in liquid nitrogen at 0 (immediately before estradiol addition), 5, 15, 30, 60, 120, 180, and 360 min after estradiol addition. RNA was isolated using standard phenol:chloroform extraction, purified using an RNeasy RNA purification kit (Qiagen), labeled using the QuickAmp labeling kit (Agilent, Santa Clara, CA), hybridized to 8 × 15k Yeast Aligent Oligo V2 microarrays using Agilent reagents and protocols, washed, and scanned as described (Brauer et al., 2005).
Expression data analysis
All expression data (for both 1 μM and 10 nM estradiol) are provided in Supplemental Data Set S1 and are available in the Princeton University MicroArray database (http://puma.princeton.edu). Expression data were processed as described (Petti et al., 2011) in order to select genes whose time dependence is statistically significant (p ≤ 0.05) and whose maximum fold change is at least 1.5 in any of the three strains. We also included genes with no time dependence if they showed constitutive differences in expression among the strains (analysis of variance q value ≤ 0.1; Storey and Tibshirani, 2003). To focus our analysis on the most direct effects of Met4p degradation, we used data from the first 30 min (during which the known Met4p targets reached their maximum level of repression) in subsequent processing steps. We also measured gene expression in the same three strains using 10 nM estradiol. These data were used to check each gene for qualitative consistency across different doses of estradiol but were not used in quantitative analyses because other work in our lab showed that 1 µM estradiol is saturating for the estradiol-induction machinery (McIsaac et al., 2011). Genes meeting the foregoing criteria were hierarchically clustered using the MultiExperiment Viewer (Saeed et al., 2006; Supplemental Figure S7) and used as a starting point for the analyses to be described.
Classification of transcription factor dependence
Multiple regression was used to determine whether each gene depends on Met4p, Opi1p, or both. NDeg-MET4 was compared with the control using the regression model in Eq. 1A, and opi1Δ NDeg-MET4 was compared with NDeg-MET4 using the regression model in Eq. 1B. In Eq. 1A, the dummy variable DM classifies the strain as an NDeg-MET4 strain or a control strain (DM = 1 for NDeg-MET4; DM = 0 for control). In Eq. 1B, the dummy variable Do classifies the strain as an opi1Δ NDeg-MET4 strain or an NDeg-MET4 strain (Do = 1 for opi1Δ NDeg-MET4; Do = 0 for NDeg-MET4). Regression significance was calculated from the F statistic comparing the fit of the full model (Eq. 1A or Eq. 1B) to the fit of the reduced model (Eq. 1C).
Regression model for dependence on Met4p:
Regression model for dependence on Opi1p:
Reduced regression model:
A gene was defined as uniquely Met4p dependent (or uniquely Opi1p dependent) if it 1) exhibited statistically significant strain specificity in 1A but not 1B (1B but not 1A), using the F-statistic q value as a measure of significance (q ≤ 0.1, p ≤ 0.033), or 2) exhibited constitutively different expression from the comparison strain, using the t-test q value as a measure of significance (q ≤ 0.1, p ≤ 0.0014). Because the expression profiles of some genes did not fit well to any quadratic, we also included a small number of genes whose expression differed between the strains by a factor of at least two at one or more time points, as long as expression pattern was consistent in both the 1 μM and 10 nM data sets. A gene was defined as jointly dependent on both Met4p and Opi1p if it exhibited strain specificity in both comparisons. We also analyzed genes that were Opi1p regulated but not uniquely so. (These exhibited statistically significant strain specificity in comparison 1B, without regard to comparison 1A.)
Accurately representing time-dependent data using heat maps
We introduce here a new method for representing unevenly sampled gene expression time-course data using heat maps. As in this study, it is common practice to sample biological time courses unevenly, such that measurements are taken more frequently when the variable of interest is changing most quickly. Although heat maps represent high-throughput expression data with unparalleled convenience and conciseness, they misrepresent the time dependence of unevenly sampled data. As a result, visual inspection of heat maps often leads to incorrect conclusions about expression dynamics. Scatterplots, on the other hand, represent time dependence accurately but are not amenable to high-throughput analysis and concise representation (e.g., of hierarchically clustered genes). We combined the best features of both graphical methods by linearly interpolating our unevenly sampled gene expression data and representing the interpolated data using a heat map. As a result, each pair of data points (either real or interpolated) is separated by the same time interval, and the dynamic changes in gene expression are displayed accurately. These interpolated values are only for display in Figures 4 and 5 and are not included in any of our statistical analyses.
Functional enrichment analysis
The functional enrichment of all gene clusters was measured with respect to the functional categorization of yeast genes in the Gene Ontology (http://www.geneontology.org; Ashburner et al., 2000). Enrichment for Gene Ontology terms was measured using the Generic Gene Ontology Term Finder (Boyle et al., 2004) available at http://go.princeton.edu/cgi-bin/GOTermFinder. Here we used the FDR option for multiple hypothesis correction and only report enrichments with FDR ≤ 0.1. The results of these analyses are shown in Supplemental Data Set S2.
Supplementary Material
Acknowledgments
We thank Ryan Briehof for technical help. This work was supported by National Institutes of Health grants GM046406 to D.B. and P50GM071508 to the Center for Quantitative Biology at Princeton University and by the National Science Foundation Research Fellowship Program to R.S.M.
Abbreviations used:
- PC
phosphatidylcholine
- SAM
S-adenosylmethionine
- SNP
single-nucleotide polymorphism
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0467) on September 7, 2011.
REFERENCES
- Aranda A, del Olmo ML. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol. 2004;70:1913–1922. doi: 10.1128/AEM.70.4.1913-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 2001. [Google Scholar]
- Barbey R, Baudouin-Cornu P, Lee TA, Rouillon A, Zarzov P, Tyers M, Thomas D. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. EMBO J. 2005;24:521–532. doi: 10.1038/sj.emboj.7600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter Z, Delneri D. New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast. 2010;27:765–775. doi: 10.1002/yea.1774. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran S, Deffenbaugh AE, Ford DA, Bailly E, Mathias N, Skowyra D. Destabilization of binding to cofactors and SCFMet30 is the rate-limiting regulatory step in degradation of polyubiquitinated Met4. Mol Cell. 2006;24:689–699. doi: 10.1016/j.molcel.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Chen M, Hancock LC, Lopes JM. Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim Biophys Acta. 2007;1771:310–321. doi: 10.1016/j.bbalip.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Chin J, Bloch K. Phosphatidylcholine synthesis in yeast. J Lipid Res. 1988;29:9–14. [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- Flick K, Ouni I, Wohlschlegel JA, Capati C, McDonald WH, Yates JR, Kaiser P. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol. 2004;6:634–641. doi: 10.1038/ncb1143. [DOI] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FH, Dawes IW. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Herrgard MJ, Lee BS, Portnoy V, Palsson BO. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 2006;16:627–635. doi: 10.1101/gr.4083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MJ, Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol. 2007;27:7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Tsuji S, Otani N, Yamamoto D, Rao KS, Watanabe S, Tsukatsune M, Makino K. Differential transcriptional regulation of two distinct S-adenosylmethionine synthetase genes (SAM1 and SAM2) of Saccharomyces cerevisiae. Nucleic Acids Res Suppl. 2003:303–304. doi: 10.1093/nass/3.1.303. [DOI] [PubMed] [Google Scholar]
- Komachi K, Johnson AD. Residues in the WD repeats of Tup1 required for interaction with alpha2. Mol Cell Biol. 1997;17:6023–6028. doi: 10.1128/mcb.17.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Rouillon A, Lee T, Barbey R, Tyers M, Thomas D. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol Cell. 2002;10:69–80. doi: 10.1016/s1097-2765(02)00561-0. [DOI] [PubMed] [Google Scholar]
- Kuras L, Thomas D. Identification of the yeast methionine biosynthetic genes that require the centromere binding factor 1 for their transcriptional activation. FEBS Lett. 1995;367:15–18. doi: 10.1016/0014-5793(95)00528-h. [DOI] [PubMed] [Google Scholar]
- Lee TA, Jorgensen P, Bognar AL, Peyraud C, Thomas D, Tyers M. Dissection of combinatorial control by the Met4 transcriptional complex. Mol Biol Cell. 2010;21:456–469. doi: 10.1091/mbc.E09-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C, Cormier L, Kuras L. Independent recruitment of mediator and SAGA by the activator Met4. Mol Cell Biol. 2006;26:3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Levine TP. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem. 2008;283:23989–23999. doi: 10.1074/jbc.M800830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malave TM, Dent SY. Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol. 2006;84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- Masselot M, De Robichon-Szulmajster H. Methionine biosynthesis in Saccharomyces cerevisiae. I. Genetical analysis of auxotrophic mutants. Mol Gen Genet. 1975;139:121–132. doi: 10.1007/BF00264692. [DOI] [PubMed] [Google Scholar]
- McCammon MT, Hartmann MA, Bottema CD, Parks LW. Sterol methylation in Saccharomyces cerevisiae. J Bacteriol. 1984;157:475–483. doi: 10.1128/jb.157.2.475-483.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P, Henry SA. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti A, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-05-0466. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain HA, Bystrom AS, Korch C. The general amino acid control regulates MET4, which encodes a methionine-pathway-specific transcriptional activator of Saccharomyces cerevisiae. Mol Microbiol. 1993;7:215–228. doi: 10.1111/j.1365-2958.1993.tb01113.x. [DOI] [PubMed] [Google Scholar]
- Nurminen T, Taskinen L, Suomalainen H. Distribution of membranes, especially of plasma-membrane fragments, during zonal centrifugations of homogenates from glucose-repressed Saccharomyces cerevisiae. Biochem J. 1976;154:751–763. doi: 10.1042/bj1540751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D. SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 2000;19:1613–1624. doi: 10.1093/emboj/19.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti AA, Crutchfield CA, Rabinowitz JD, Botstein D. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1101494108. 2011 Jul 6 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI, Hadwiger JA, Lorincz AT. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci USA. 1985;82:4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Santiago TC, Mamoun CB. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Shetty A, Lopes JM. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot Cell. 2010;9:1845–1855. doi: 10.1128/EC.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek IS, Steensma HY. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res. 2006;6:393–403. doi: 10.1111/j.1567-1364.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- Sprague ER, Redd MJ, Johnson AD, Wolberger C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000;19:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A, Carman GM. Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase A. J Biol Chem. 2003;278:20673–20680. doi: 10.1074/jbc.M300132200. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Jones EW, Broach JR. The Molecular Biology of the Yeast Saccharomyces. Metabolism and Gene Expression. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Jacquemin I, Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol. 1988;8:5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. SAM1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J Biol Chem. 1987;262:16704–16709. [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol Microbiol. 2001;41:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Quinn KA, Perrone G, Dawes IW, Grant CM. Glutathione regulates the expression of gamma-glutamylcysteine synthetase via the Met4 transcription factor. Mol Microbiol. 2002;46:545–556. doi: 10.1046/j.1365-2958.2002.03174.x. [DOI] [PubMed] [Google Scholar]
- White MJ, Hirsch JP, Henry SA. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Zomorrodi AR, Maranas CD. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data. BMC Syst Biol. 2010;4:178. doi: 10.1186/1752-0509-4-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.