Abstract
Pelvic lymph node metastases are regarded as the most important risk factor and a predictor of poor prognosis for patients with cervical cancer. Exploration of metastasis-related molecules is helpful toward improving the prognosis in cervical cancer. To identify the role of miR-375 in metastasis and progression of cervical cancer, we examined the expression of miR-375 in 170 cervical cancer tissues and 68 normal cervical tissues, using stem-loop quantitative PCR, and found that the expression of miR-375 in cervical cancer tissues was significantly decreased by 4.45-fold, compared with 68 normal tissues. A significant correlation existed between miR-375 expression and clinicopathologic parameters, including lymph node metastasis of cervical cancer. Overexpressed miR-375 suppressed cell proliferation, blocked G1-to-S cell-cycle transition, and inhibited cell migration and invasion in human cervical SiHa and CaSki cells. SP1, a potential target gene of miR-375, was inversely correlated with miR-375 expression in cervical cancer tissues. Moreover, SP1 was negatively regulated by miR-375, and knockdown of SP1 by siRNA inhibited cell malignant behaviors. Thus, our findings suggest that down-regulated miR-375 promotes cell malignant behaviors via the target gene SP1 and may consequently contribute to the progression of cervical cancer.
Cervical cancer remains the second most common female malignant disease and is still one of the leading causes of cancer-related deaths worldwide. Although early-stage cervical cancer can be treated by radical surgery with or without chemotherapy and/or radiotherapy, some patients with high risk factors have an unfavorable prognosis.1, 2 Various clinical data have shown that factors influencing prognosis in cervical cancer include tumor size, depth of stromal invasion, lymphovascular space involvement, parametrial invasion, and pelvic lymph node metastasis; of these, pelvic lymph node metastasis is regarded as the most important risk factor and predictor of poor prognosis for patients treated by surgery.3 The 5-year survival rate of patients with lymph node metastasis is diminished by 20% to 30%, compared with those without lymph node metastasis.4 Thus, to identify molecular regulators associated with metastasis and to investigate their functions would likely contribute to improving the prognosis in cervical cancer. Some molecules have been identified, but their effect and mechanism are still uncertain. The finding of miRNAs provides a new approach and insight for exploring new molecules and mechanisms of metastasis and progression in cervical cancer.
The miRNAs are a group of conserved noncoding RNAs that can bind to the 3′ untranslated region (UTR) of target mRNA and regulate stability and translation of mRNAs, resulting in either inhibition of translation or degradation of the target mRNA.5 miRNAs can be classified as oncogenes or as tumor suppressors, and by targeting various transcripts they participate in diverse processes, including proliferation, apoptosis, metabolism, and cellular differentiation.6 Accumulated evidence has revealed that miRNAs are involved in metastasis and progression of various cancers, but their functions differ. For example, the overexpression of miR-3407 and miR-218 promoted occurrence of lymph node metastasis, whereas the reduced expression of miR-10b and miR-3739 increased the risk of lymph node metastasis in breast carcinoma. To date, except for miR-127,10 few miRNAs have been reported to be associated with metastasis and progression in cervical cancer.
In a previous study, we screened the different expression of miRNAs between cervical normal and cancer tissues through miRNA array and found that miR-375 was remarkably down-regulated in cervical cancer tissues, compared with normal tissues,11 which implies that miR-375 may participate in the pathogenesis and development of cervical cancer. In the present study, we examined miR-375 expression in cervical cancer tissues and investigated the function of miR-375 in proliferation, migration, and invasion of cervical cancer cells, and we identified SP1 as a target gene for miR-375. miR-375 might act as a tumor suppressor and serve as a potential therapeutic target in cervical cancer.
Materials and Methods
Patients and Tissue Specimens
Cervical tissue samples and clinicopathologic data from 170 patients with primary cervical squamous cell carcinoma who underwent radical hysterectomy with pelvic lymph nodes dissection from July 2008 through December 2009 were collected in Women's Hospital, School of Medicine, Zhejiang University. Mean age was 44.8 years (range, 28 to 66 years). Of these 170 cervical cancer cases, 114 cases were International Federation of Gynecology and Obstetrics (FIGO) stage I and 56 were stage II; 29 cases were with lymph node metastasis and 141 without; 6 cases were well differentiated (G1), 148 were moderately differentiated (G2), and 16 were poorly differentiated (G3). No patient received chemotherapy or radiotherapy before the tissues were obtained. Normal cervical tissues with high-risk human papilloma virus (HR-HPV) infection (n = 32) or without HPV infection (n = 36) were collected as controls from women (mean age, 43.1 years; range, 27 to 64 years) who underwent hysterectomy for nonmalignant conditions. All histological diagnoses were made by two pathologists (Xiaoduan Chen and Bingjian Lv) independently. All specimens were immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. The study was approved by the Ethics Committee of the hospital.
Total RNA Extraction and Real-Time RT-PCR
RNA was isolated by the TRIzol method (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol, and the quantity and concentration of RNA were spectrophotometrically assessed by measuring absorbance at A260/280. Quantitative RT-PCR (qRT-PCR) for miRNA and mRNA was performed as described previously.11 For miRNA quantification, each reverse transcript reaction consisted of 0.5 μg of total RNA, mixed with 2.0 μL of 5× RT buffer containing dNTPs (Takara Bio, Otsu, Japan), 0.2 μL of 10 μmol/L stem-loop RT primer (Invitrogen), 0.2 μL RNase inhibitor protein (Takara Bio), and 0.8 μL reverse transcriptase (Takara Bio) in a final volume of 10 μL, and incubated at 42°C for 60 minutes and at 85°C for 5 minutes. Real-time PCR was performed with an Applied Biosystems 7900HT system (Foster City, CA) using SYBR Premix Ex Taq (Takara Bio). PCR volume was 20 μL, containing 1 μL reverse transcript product. Cycling conditions were 1 cycle of 95°C for 30 s and 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. PCR was performed in triplicate. For measurement of the SP1 transcript from total RNA, total cDNA was synthesized using a TaKaRa reverse transcription kit (Takara Bio). Real-time PCR was performed using SYBR Premix Ex Taq (Takara Bio). The U6 snRNA and EEF1A112 were used as endogenous control for miRNA and mRNA, respectively. The sequences of all primers are given in Table 1. The ΔΔCt method was used to determine relative quantitation of miRNA and mRNA expression in tissue samples, and fold change was determined as 2−ΔΔCt.
Table 1.
Sequences of Primers and siRNA
| Primers and siRNA | Sequence |
|---|---|
| miR-375 RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACGC-3′ |
| miR-375 forward | 5′-AGCCGTTTGTTCGTTCGGCT-3′ |
| miR-375 reverse | 5′-GTGCAGGGTCCGAGGT-3′ |
| U6 snRNA RT | 5′-AACGCTTCACGAATTTGCGT-3′ |
| U6 snRNA forward | 5′-CTCGCTTCGGCAGCACA-3′ |
| U6 snRNA reverse | 5′-AACGCTTCACGAATTTGCGT-3′ |
| Sp1 forward | 5′-GGCTCGGGGGATCCTGGC-3′ |
| Sp1 reverse | 5′-TATGGCCCATATGTCTCTG-3′ |
| EEF1A1 forward | 5′-TGCGGTGGGTGTCATCAAA-3′ |
| EEF1A1 reverse | 5′-AAGAGTGGGGTGGCAGGTATTG-3′ |
| siRNA targeting Sp1 | 5′-ATCACTCCATGGATGAAATGA-3′ |
| Sp1 mUTR sense | 5′-GGAATGATAGCCCAGATGTAAAAAGAAATCTTGTCTTAC-3′ |
| Sp1 mUTR antisense | 5′-GTAAGACAAGATTTCTTTTTACATCTGGGCTATCATTCC-3′ |
EEF1A1, eukaryotic translation elongation factor 1 alpha 1; mUTR: mutant untranslated region; RT, reverse transcriptase; siRNA, small interfering RNA; Sp1, Sp1 transcriptional factor.
Cell Culture and Transfection
The HPV 16-positive human cervical carcinoma cell lines SiHa and CaSki were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2 in a humidified incubator. Cells were transfected with Dharmacon miRIDIAN miR-375 mimic (miR-375) and miRIDIAN microRNA mimic negative control 1 (negative control) (Thermo Fisher Scientific, Lafayette, CO) at a final concentration of 100 nmol/L, and small interfering RNA targeting SP1 (siR-SP1) and siR-Ribo negative control (siR-Cont) (RiboBio, Guangzhou, China) at a final concentration of 30 nmol/L, using DharmaFECT 1 transfection reagent (Thermo Fisher Scientific) in accordance with the manufacturer's instructions. The sequence of siRNA targeting SP1 is given in Table 1.
Construction of Plasmids and Luciferase Activity Assay
The pmirGLO dual-luciferase miRNA target expression vector (pmirGLO vector) containing both firefly luciferase gene and Renilla luciferase gene was purchased from Promega (Madison, WI). Human SP1 3′UTR including the predicted binding site of miR-375 was amplified by RT-PCR (Table 1) and inserted into the 3′UTR region downstream of the firefly luciferase gene of pmirGLO vector (pmirGLO-UTR) using the XbaI and SacI restriction sites. A site-directed gene mutagenesis kit (Beyotime, Jiangsu, China) was used to construct the mutant type of miR-375-binding sites vector (pmirGLO-mUTR) according to the manufacturer's protocol. Both constructs were confirmed by restriction enzyme digestion and sequencing (Invitrogen). Cotransfection of miRNA mimics (50 nmol/L) and reporter vectors (0.2 μg/mL) was performed using DharmaFECT 1 transfection reagent. Luciferase activities were measured at 48 hours after transfection using a Dual-Glo luciferase assay system (Promega), and firefly luciferase activities were normalized to Renilla luciferase activities. For each comparison, values for cells with empty pmirGLO vector were set equally to 1. Experiments were performed in triplicate and repeated twice. Data are presented as means ± SD.
Western Blot Analysis
At 72 hours after transfection with miR-375 mimics or siR-SP1, total protein extracts from the cells were prepared for Western blot analysis as described previously.11 The primary antibodies used for Western blot analysis were anti-SP1 antibody (1:100) and anti-β actin antibody as an endogenous control (1:2000), both from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Proliferation Assay
To determine the effect of miR-375 or siR-SP1 on proliferation of cervical cell lines, SiHa (5 × 103) and CaSki (4 × 103) cells were suspended in Dulbecco's modified Eagle's medium (100 μL) containing 10% fetal bovine serum and cultured in 96-well plates overnight and then transfected with miR-375 mimic or siR-SP1. The cell proliferation was determined with MTT assay at 24, 48, 72, and 96 hours after transfection respectively, and the absorbance of samples was measured with a spectrophotometer reader at 490 nm. All experiments were performed in six replicates and were repeated three times independently. Data are presented as means ± SD of three separate experiments.
Wound-Healing Assay
An in vitro wound-healing assay was used to assess capacity for tumor cell motility. Briefly, SiHa cells (5 × 105/well) and CaSki cells (5 × 105/well) were seeded in 12-well plates, cultured overnight, and transfected with miR-375 or siR-SP1. When the culture had reached nearly 90% confluency, the cell layer was scratched with a sterile plastic tip and then was immediately washed with growth medium twice and cultured again in Dulbecco's modified Eagle's medium (including 1% fetal bovine serum) at 37°C in a humidified incubator with 5% CO2 for up to 48 hours. At different time points, photographic images of the plates were acquired under a microscope. Wound healing was measured at 0, 24, and 48 hours, and the data were summarized based on six assays for each experiment.
Invasion Assay
Invasion assay of SiHa and CaSki cells was examined in Transwell chambers with members coated with Cultrex Basement Membrane Extract without Phenol Red (R&D Systems, Minneapolis, MN). For this purpose, 2 × 104 cells transfected with miR-375 mimic or siR-SP1 were suspended in 200 μL Dulbecco's modified Eagle's medium without serum and seeded on the upper chamber; the lower chamber was filled with 10% fetal bovine serum as the chemoattractant. After 48 hours, cells on the upper side of the membrane were wiped off; cells on the lower side of the membrane were fixed and stained with crystal violet solution. The cells under the microscopic fields (100× objective) in each chamber were photographed and counted; values were expressed as fold induction. All invasion assays were done in triplicate for at least three independent experiments.
Statistical Analysis
Data are presented as means ± SD from at least three independent experiments. Student's t-test was used to analyze differences in experiments with cell lines. Correlation between expression levels of miR-375 and its target genes in cervical cancer tissues was analyzed using Pearson's correlation coefficient. Association between expression level of miR-375 and each clinicopathologic parameter was evaluated using Pearson's χ2 test. All statistical tests were two-sided, and P values of <0.05 were considered statistically significant. All analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL).
Results
miR-375 Expression Correlates Negatively with Lymph Node Metastasis and Other Clinicopathologic Parameters in Early Cervical Cancer
The expression of miR-375 was detected with real-time RT-PCR; U6 snRNA served as endogenous control. The relative quantitative analysis was calculated as 2-ΔΔCt. The 75th percentile of 2-ΔΔCt was used as a cut-off point for patients with high or low levels of miR-375, respectively.13 The mean relative miR-375 expression in 170 cervical cancer tissues was significantly decreased, by 4.45-fold, compared with that of 68 normal controls (P = 5.697 × 10−20) (Figure 1). In cervical cancer tissues, relative expression of miR-375 in tissues with lymph node metastasis was significantly lower than without the metastasis (χ2 = 6.263, P = 0.012). In addition, miR-375 expression was significantly correlated with other clinicopathologic parameters, including FIGO stage (χ2 = 9.394, P = 0.002), deep stromal invasion (χ2 = 9.392, P = 0.002), lymphovascular space involvement (χ2 = 4.453, P = 0.035), and vaginal wall extension (χ2 = 5.435, P = 0.020) (Table 2).
Figure 1.
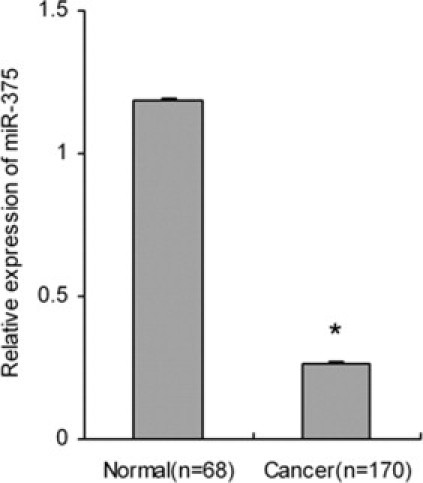
Relative expression of miR-375 in cervical cancer and normal tissues. The expression of miR-375 in cervical cancer was significantly lower than in normal tissues. Data are presented as means ± SE. *P < 0.05.
Table 2.
Relations between Expression of miR-375 and Clinicopathologic Characteristics of Cervical Cancer Tissues
| Characteristic | No. | miR-375 |
χ2 | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Patient age (years) | |||||
| ≤35 | 22 | 18 | 4 | 0.676 | 0.411 |
| >35 | 148 | 109 | 39 | ||
| FIGO stage | |||||
| IB1/IB2 | 114 | 77 | 37 | 9.394 | 0.002 |
| IIA | 56 | 50 | 6 | ||
| LNM | |||||
| Yes | 29 | 27 | 2 | 6.263 | 0.012 |
| No | 141 | 100 | 41 | ||
| Differentiation | |||||
| well | 6 | 4 | 2 | 0.213 | 0.899 |
| moderate | 148 | 111 | 37 | ||
| poor | 16 | 12 | 4 | ||
| Tumor size (cm) | |||||
| <4 | 139 | 100 | 39 | 1.592 | 0.207 |
| ≥4 | 25 | 21 | 4 | ||
| undetermined | 6 | 4 | 2 | ||
| Stromal invasion | |||||
| <2/3 | 105 | 70 | 35 | 9.392 | 0.002 |
| ≥2/3 | 65 | 57 | 8 | ||
| Vaginal wall extension | |||||
| Yes | 27 | 25 | 2 | 5.435 | 0.02 |
| No | 143 | 102 | 41 | ||
| Parametrial extension | |||||
| Yes | 14 | 12 | 2 | 0.978 | 0.323 |
| No | 156 | 115 | 41 | ||
| LVSI | |||||
| Yes | 58 | 49 | 9 | 4.453 | 0.035 |
| No | 112 | 78 | 34 | ||
FIGO, International Federation of Gynecology and Obstetrics; LNM, lymph node metastasis; LVSI, lymph vascular space invasion.
miR-375 Suppresses Cell Proliferation and Blocks G1-to-S Transition in Cervical Carcinoma Cells
SiHa and CaSki cells were transiently transfected with miR-375 mimic, which induced significant up-regulation of miR-375 compared with the negative control (see Supplemental Figure S1 at http://ajp.amjpathol.org). Cell proliferation was assayed by the MTT method. The proliferation rate of SiHa and CaSki cells at 96 hours after transfection was significantly reduced (P = 0.003 and P = 2.717 × 10−5, respectively), compared with that of miRNA negative control (Figure 2A). Analysis of cell cycle distribution by flow cytometry showed that the percentage of G1 phase of SiHa cells at 48 hours after transfection was significantly increased, from 63.4 ± 2.1% to 84.6 ± 2.9% (P = 6.197 × 10−4), and the percentage for S phase was significantly decreased, from 27.0 ± 1.1% to 8.6 ± 1.1% (P = 7.558 × 10−5). Similarly, the percentage of G1 of CaSki cells was significantly increased, from 66.4 ± 2.6% to 79.2 ± 4.6% (P = 1.354 × 10−5), and the percentage for S phase was significantly decreased, from 23.6 ± 1.6% to 12.0 ± 3.1% (P = 2.019 × 10−7) (Figure 2C). Thus, overexpression of miR-375 caused a significant G1 arrest, suggesting that elevated miR-375 blocks G1-to-S transition and consequently inhibits the progression of cell cycle in cervical cancer cells.
Figure 2.
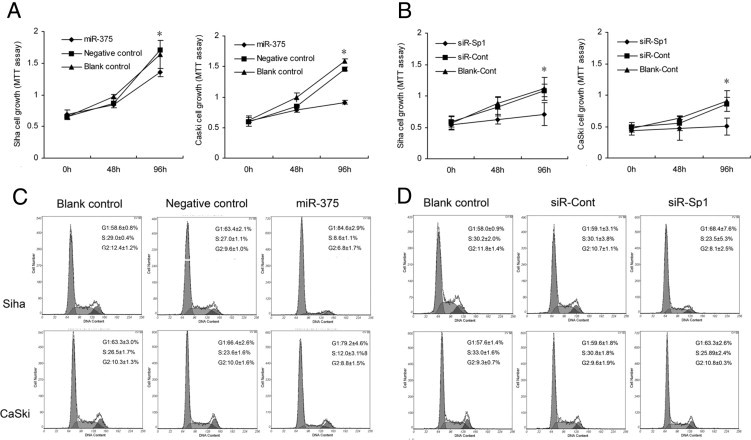
miR-375 or SP1 regulated proliferation and cycle in SiHa and CaSki human cervical carcinoma cells. A: Overexpression of miR-375 by transfection with miR-375 mimic inhibited cell growth, compared with miRNA negative control, in SiHa and CaSki cells at 96 hours after transfection. *P < 0.05. B: Knockdown of target gene SP1 using siRNA inhibited cell proliferation rate in SiHa and CaSki at 96 hours after transfection. *P < 0.05. C: Overexpression of miR-375 blocked G1-to-S transition in SiHa and CaSki cells. D: Knockdown of SP1 induced G1 arrest in SiHa and CaSki cells. Data are presented as means ± SD from three independent experiments.
miR-375 Inhibits Migration and Invasion of Cervical Carcinoma Cells
The wound-healing assay showed that SiHa and CaSki cells with miR-375 overexpression using transient transfection with miR-375 mimic presented a slower closing of scratch wound, compared with the miRNA negative controls (P = 6.535 × 10−7 and P = 2.091 × 10−8, respectively) (Figure 3A). Moreover, Transwell invasion assay revealed that the invasion potential of SiHa and CaSki cells transfected with miR-375 was significantly decreased (P = 0.001 and P = 0.011, respectively), compared with miRNA negative control (Figure 4A). Our results indicate that miR-375 served as a tumor suppressor miRNA and contributed to inhibition of migration and invasion of cervical carcinoma cells.
Figure 3.
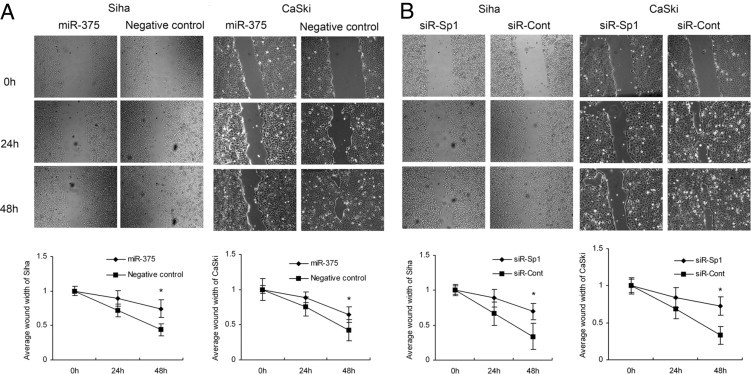
Overexpression of miR-375 or knockdown of SP1 inhibited cell migration in SiHa and CaSki. A: Overexpression of miR-375 presented a slower closing of scratch wound, compared with miRNA negative control, at 48 hours after transfection in SiHa and CaSki cells. B: Knockdown of target gene SP1 by siRNA also induced a slower wound recovery. Data are presented as means ± SD from three independent experiments. *P < 0.05. Original magnification, ×100.
Figure 4.
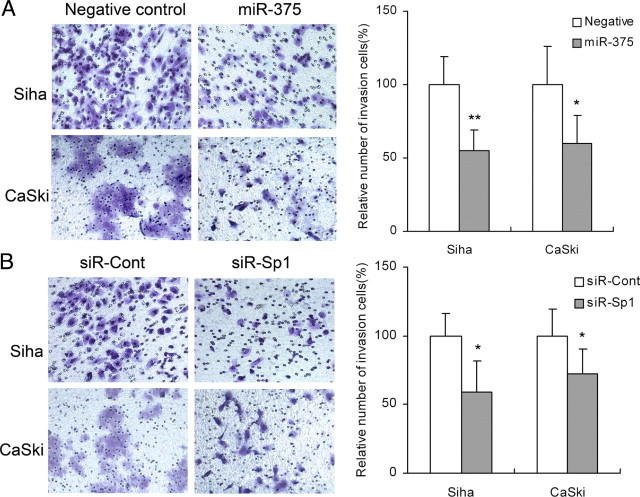
Overexpression of miR-375 or knockdown of SP1 inhibited invasion of SiHa and CaSki cells. A: Overexpression of miR-375 inhibited invasion both in SiHa and CaSki cells. B: Knockdown of SP1 by siRNA inhibited invasion both in SiHa and CaSki cells. Data are presented as means ± SD from three independent experiments. *P < 0.05; **P < 0.01.
miR-375 Negatively Regulates SP1 Gene Expression
Two public algorithms and associated databases (TargetScan version 5.1, available at http://www.targetscan.org; PicTar, available at http://pictar.mdc-berlin.de) were used to find putative target genes for miR-375 that mediate cell growth, migration, and invasion. Gene Ontology analysis using the tools of AmiGO and CateGOrizer (GO; available at http://www.geneontology.org) was used to predict the genes common to both public databases (see Supplemental Figure S2 at http://ajp.amjpathol.org). The potential binding capability of SP1 3′UTR with miR-375 was predicted by both of the public algorithms (see Supplemental Figure S3 at http://ajp.amjpathol.org). SP1 was also an important gene with effective transcriptional regulation on various downstream genes involved in cell cycle, apoptosis, proliferation, and invasion. To confirm further that SP1 is a target gene for miR-375, RT-PCR and Western blot analysis were used to detect the expression of SP1 regulated by miR-375 in SiHa and CaSki. The expression of SP1 was significantly down-regulated after forced expression of miR-375: 37.6 ± 9.5% (P = 0.035) and 52.3 ± 5.9% (P = 0.010) at the mRNA level (Figure 5A) and 43.2 ± 9.1% (P = 0.023) and 49.2 ± 6.8% (P = 0.017) at the protein level (Figure 5, B and C), compared with miRNA negative control in SiHa and CaSki, respectively. Moreover, the mRNA levels of SP1 and miR-375 expression in one set of cervical cancer tissue were assessed by qRT-PCR. There was an inverse correlation between miR-375 and SP1 mRNA expression was found (Spearman's ρ = −0.437, P = 4.409 × 10−9). Taken together, our results together suggest that miR-375 negatively regulates SP1 gene expression and that SP1 is a potential target gene.
Figure 5.
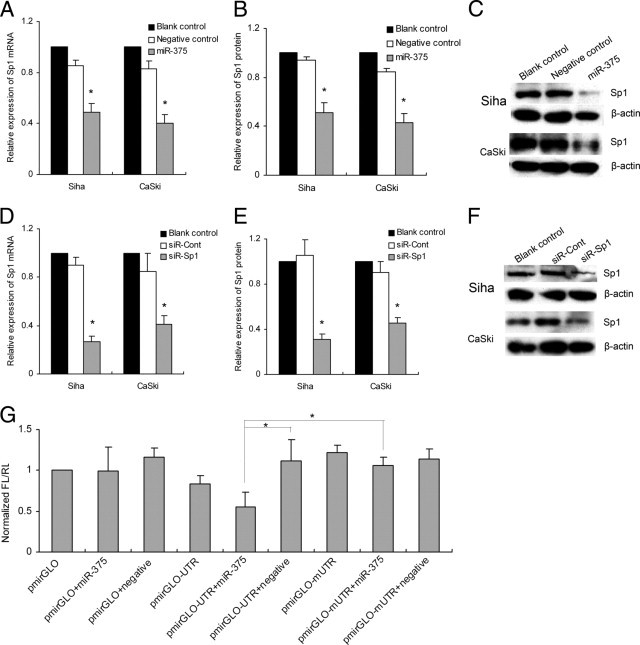
Forced expression of miR-375 negatively regulated SP1 expression. MiR-375 inhibited the expression of SP1 at the mRNA level (A) and the protein level (B and C) in SiHa and CaSki cells. siRNA targeting SP1 suppressed the expression of SP1 at the mRNA level (D) and the protein level (E and F). The expression of SP1 protein detected by Western blot was normalized to β-actin (C and F). G: Luciferase assay. SiHa cells were transiently transfected with pmirGLO, pmirGLO-UTR, or pmirGLO-mUTR vector, each with or without cotransfection with 50 nmol/L of miR-375. After 48 hours, reduced luciferase activity was observed after cotransfection of pmirGLO-UTR vector with miR-375, but not pmirGLO-mUTR or empty pmirGLO vector. Data are presented as means ± SD from three independent experiments. *P < 0.05.
miR-375 Directly Targets the SP1 3′UTR
To confirm that SP1 was a direct target of miR-375, pmirGLO-UTR containing the miR-375 binding sites was constructed to perform the reporter assay, and pmirGLO-mUTR containing mutant binding sites was used as a control. The luciferase activities of the reporter vectors were assayed at 48 hours after cotransfection with miR-375 mimic or negative control in SiHa cells. When cotransfected with miR-375, the relative luciferase activity of pmirGLO-UTR was significantly suppressed by 50% (P = 0.001), compared with cotransfection with negative control. Also, the relative luciferase activity was not altered when cotransfection was done with miR-375 and pmirGLO-mUTR containing a mutant binding site or empty pmirGLO (Figure 5G), indicating that SP1 was a direct target for miR-375.
Knockdown of SP1 Reduces Proliferation, Migration, and Invasion in Cervical Carcinoma Cells
To investigate the role of SP1 in cell proliferation, migration, and invasion, siRNA targeting SP1 (siR-SP1) was used. Expression of SP1 was inhibited by 69.9 ± 7.2% (P = 0.008) and 52.4 ± 12.4% (P = 0.019) at the mRNA level (Figure 5D) and 70.2 ± 3.2% (P = 0.045) and 50.9 ± 8.4% (P = 0.017) at the protein level after knockdown of the SP1 gene in SiHa and CaSki cells (Figure 5, E and F). Consistently, the knockdown of SP1 markedly induced G1 arrest (P = 0.017 and P = 0.028) (Figure 2D) and inhibited cell proliferation (P = 9.315 × 10−7 and P = 3.905 × 10−4) (Figure 2B), migration (P = 3.738 × 10−4 and P = 0.002) (Figure 3B), and cell invasion (P = 0.013 and P = 0.027) (Figure 4B) in SiHa and CaSki, respectively, suggesting that up-regulated SP1 (induced by suppression of miR-375) participates in proliferation, migration, and invasion of cervical cancer cells and that knockdown of SP1 can reduce the malignant behaviors induced by diminished miR-375.
HR-HPV16 E6/E7 Does Not Directly Regulate miR-375 Expression
To explore whether decreased miR-375 expression was induced by HPV oncoproteins, the expression of miR-375 was detected and compared in 32 normal cervical tissues with (n = 32) and without (n = 36) high-risk HPV infection. No difference of miR-375 relative expression was found between the two groups (P = 0.461). Furthermore, neither overexpression of HPV16 E6 by transfection with HPV-16 E6 expression vector14 into C33A cells nor down-regulated expression of HPV16 E6/E7 using siRNAs15 in SiHa cells significantly altered miR-375 transcripts (P = 0.277 and P = 0.583), suggesting that suppressed miR-375 expression in cervical cancer cells and tissue was not directly induced by HR-HPV16 E6/E7.
Discussion
miRNAs have been identified as important regulators in tumorigenesis and cancer progression through various molecular pathways.16 miR-375 was initially found to be expressed in pancreatic islet, where it regulates the secretion of insulin and metabolism of glucose17 and influences pancreatic α- and β-cell mass and pancreatic development.17, 18, 19 Recently, miR-375 was reported to be associated with cancer development; this miRNA was down-regulated in some malignancies, including liver cancer, gastric cancer,20, 21, 22 head and neck squamous cell carcinoma,23, 24 and pancreatic adenocarcinomas,25 and was up-regulated in an ERα+ breast cancer cell line.26 In the present study, we found as low as 4.45-fold reduced expression of miR-375 in cervical cancer tissues, compared with that in normal cervical tissues. We also found that lower expression of miR-375 was closely correlated with lymph node metastasis, advanced clinical stage, deeper stromal invasion, lymphovascular space involvement, and vaginal wall extension. Considering the association of all these clinicopathological parameters with poor prognosis for cervical cancer patients, our results imply that miR-375 may participate in the progression and influence the prognosis in cervical cancer. To explore the effect of suppressed miR-375 expression on metastasis and progression of cervical cancer, we examined the alteration of cell growth and behavior by miR-375 expression in cervical cancer cells. Elevated expression of miR-375 suppressed proliferation, blocked G1-to-S cell cycle transition, and inhibited cell migration and invasion in cervical cancer cells. Taken together, our results suggest that miR-375 as a tumor suppressor plays a role in the metastasis and progression of cervical cancer.
Some genes, such as those encoding myotrophin,27 PDK1,20 14-3-3ζ,20 JAK2,22 and RASD1,26 are targets for miR-375. We used a public algorithm and found that SP1 is a putative target gene for miR-375, mediating cell growth, migration, and invasion. Previous studies have shown that that SP1 is a key transcriptional regulator that exerts a variety of biological functions.28, 29 SP1 also participates in cancer development and progression.30, 31, 32 SP1 expression is increased in gastric cancer, pancreatic carcinoma, and breast carcinoma.33, 34, 35 In the present study, we found an inverse correlation between miR-375 and SP1 expression in the cervical cancer tissues. Furthermore, expression of SP1 at both mRNA and protein levels could be down-regulated significantly by overexpressed miR-375 using miR-375 mimic transfection, and knockdown of SP1 using specific siRNA in turn inhibited proliferation, migration, and invasion of SiHa and CaSki cells. Additionally, we confirmed the existence of a binding site for miR-375 at SP1 3′UTR by luciferase activity assay. Thus, we have verified that SP1 is a direct target for miR-375 related to progression of cervical cancer. Taken together, our results suggest that elevated SP1, induced by suppressed miR-375, participates in progression of cervical cancer. Both miR-375 and its target gene SP1 might be therapeutic targets, and either restoring miR-375 expression or abolishing SP1 expression could diminish cell malignant behaviors and consequently block the progression of cervical cancer.
HR-HPV infection is a necessary factor in the development of cervical cancer, of which HPV16 type accounts for >60%. Viral oncoprotein E6 or E7 can induce alteration of cellular miRNA expression in different ways during the development and progression of cervical cancer. HPV 16 infection reduced miR-218 expression, resulting in increased expression of LAMB3 (which encodes laminin-5 β3),36 and HPV 16 E6 induced a reduction of miR-34a14, 37 and miR-23b,38 due to the degradation of p53 in cervical cancer cell lines. To determine whether the reduction of miR-375 expression is regulated by HR-HPV oncoproteins, we examined the expression of miR-375 in normal cervical tissues with or without HR-HPV infection. We observed no difference between the two groups. Moreover, we observed no change in miR-375 expression after overexpression of HPV 16 E6 in C33A cells or after siRNA-mediated reduced expression of HPV16 E6/E7 in SiHa cells. Our results suggest that HPV 16 E6/E7 does not directly regulate the expression of miR-375. An analysis with the online miRBase database (Release 17) (available at http://www.mirbase.org) showed that miR-375 maps to chromosome 2q35, a genomic region frequently deleted in early cervical cancer.39 Thus, it is possible that the diminished expression of miR-375 arises from deletion of the 2q35 genomic region. An alternative mechanism may involve the change of epigenetic marks and local DNA methylation states.26 Nevertheless, further study is needed to determine the exact mechanism of decreased miR-375 expression during the progression of cervical cancer.
In conclusion, miR-375 expression is down-regulated in cervical cancer tissues and is correlated with pelvic lymph node metastasis, FIGO stage, and other poor-prognosis clinicopathological variables. The loss of miR-375 promotes cell proliferation, migration, and invasion of cervical cancer cells in vitro. SP1 contains a binding site for miR-375 at 3′UTR and was negatively regulated by miR-375. Down-regulation of SP1 inhibits cell malignant behavior. Taken together, our results suggest that the suppression of miR-375 expression induces the elevation of target gene SP1 expression, and consequently promotes cell malignant behaviors and probably contributes to the progression of cervical cancer. Thus, miR-375 and its target gene SP1 have potential as biomarkers and as therapeutic targets for cervical cancer.
Acknowledgments
We thank Dr. Yifan Cheng for sample collection and technical assistance. We also thank Drs. Xiaoduan Chen and Bingjian Lv for their histological diagnoses of all samples.
Footnotes
Supported by grants from the National Natural Science Foundation of China (no. 30872752 and no. 30973173) and Zhejiang Provincial Natural Science Foundation of China (no. Y2090355 and no. Z2110056).
F.W. and Y.L. contributed equally to the present work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.07.037.
Supplementary Data
Relative expression of miR-375 in SiHa and CaSki human cervical cancer cells transfected with miR-375 mimic and negative control. The expression of miR-375 was significantly up-regulated in SiHa and CaSki transfected with miR-375 mimic, compared with negative control. Data are expressed as means ± SD from three independent experiments. *P < 0.05.
The target genes of miR-375 as predicted by two online databases (TargetScan, available at http://www.targetscan.org, and PicTar, available at http://www.pictar.mdc-berlin.de). Gene Ontology analysis (GO; available at http://www.geneontology.org) was used to analyze the target genes predicted by both databases and to form the gene network shown.
SP1 was a potential target gene predicted by both TargetScan and PicTar, and bioinformatics analysis showed the predicted binding site of miR-375 and SP1 3′UTR. The reporter pmirGLO-mUTR was constructed by site-directed mutagenesis.
References
- 1.Landoni F., Maneo A., Colombo A., Placa F., Milani R., Perego P., Favini G., Ferri L., Mangioni C. Randomised study of radical surgery versus radiotherapy for stage Ib-IIa cervical cancer. Lancet. 1997;350:535–540. doi: 10.1016/S0140-6736(97)02250-2. [DOI] [PubMed] [Google Scholar]
- 2.Biewenga P., van der Velden J., Mol B.W., Stalpers L.J., Schilthuis M.S., van der Steeg J.W., Burger M.P., Buist M.R. Prognostic model for survival in patients with early stage cervical cancer. Cancer. 2011;117:768–776. doi: 10.1002/cncr.25658. [DOI] [PubMed] [Google Scholar]
- 3.Potter M.E., Alvarez R.D., Shingleton H.M., Soong S.J., Hatch K.D. Early invasive cervical cancer with pelvic lymph node involvement: to complete or not to complete radical hysterectomy? Gynecol Oncol. 1990;37:78–81. doi: 10.1016/0090-8258(90)90312-9. [DOI] [PubMed] [Google Scholar]
- 4.Shingleton H.M., Jones W.B., Russell A., Fremgen A., Chmiel J.S., Ocwieja K., Winchester D.P., Clive R. Hysterectomy in invasive cervical cancer: a national patterns of care study of the American College of Surgeons. J Am Coll Surg. 1996;183:393–400. [PubMed] [Google Scholar]
- 5.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z.S., Wu Q., Wang C.Q., Wang X.N., Huang J., Zhao J.J., Mao S.S., Zhang G.H., Xu X.C., Zhang N. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer. 2011;117:2842–2852. doi: 10.1002/cncr.25860. [DOI] [PubMed] [Google Scholar]
- 8.Yan L.X., Huang X.F., Shao Q., Huang M.Y., Deng L., Wu Q.L., Zeng Y.X., Shao J.Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negrini M., Calin G.A. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.W., Choi C.H., Choi J.J., Park Y.A., Kim S.J., Hwang S.Y., Kim W.Y., Kim T.J., Lee J.H., Kim B.G., Bae D.S. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Wang F., Xu J., Ye F., Shen Y., Zhou J., Lu W., Wan X., Ma D., Xie X. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224:484–495. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y., Li Y., Ye F., Wang F., Lu W., Xie X. Identification of suitable reference genes for measurement of gene expression in human cervical tissues. Anal Biochem. 2010;405:224–229. doi: 10.1016/j.ab.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z.L., Zhao X.H., Wang J.W., Li B.Z., Wang Z., Sun J., Tan F.W., Ding D.P., Xu X.H., Zhou F., Tan X.G., Hang J., Shi S.S., Feng X.L., He J. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725–10734. doi: 10.1074/jbc.M110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B., Hu Y., Ye F., Li Y., Lv W., Xie X. Reduced miR-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection. Int J Gynecol Cancer. 2010;20:597–604. doi: 10.1111/IGC.0b013e3181d63170. [DOI] [PubMed] [Google Scholar]
- 15.Hong D., Lu W., Ye F., Hu Y., Xie X. Gene silencing of HPV16 E6/E7 induced by promoter-targeting siRNA in SiHa cells. Br J Cancer. 2009;101:1798–1804. doi: 10.1038/sj.bjc.6605344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avnit-Sagi T., Kantorovich L., Kredo-Russo S., Hornstein E., Walker M.D. The promoter of the pri-miR-375 gene directs expression selectively to the endocrine pancreas. PLoS One. 2009;4:e5033. doi: 10.1371/journal.pone.0005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukamoto Y., Nakada C., Noguchi T., Tanigawa M., Nguyen L.T., Uchida T., Hijiya N., Matsuura K., Fujioka T., Seto M., Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 21.Liu A.M., Poon R.T., Luk J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 22.Ding L., Xu Y., Zhang W., Deng Y., Si M., Du Y., Yao H., Liu X., Ke Y., Si J., Zhou T. miR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 23.Hui A.B., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O'Sullivan B., Waldron J., Gullane P., Cummings B., Liu F.F. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 24.Avissar M., Christensen B.C., Kelsey K.T., Marsit C.J. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloomston M., Frankel W.L., Petrocca F., Volinia S., Alder H., Hagan J.P., Liu C.G., Bhatt D., Taccioli C., Croce C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 26.de Souza Rocha Simonini P., Breiling A., Gupta N., Malekpour M., Youns M., Omranipour R., Malekpour F., Volinia S., Croce C.M., Najmabadi H., Diederichs S., Sahin O., Mayer D., Lyko F., Hoheisel J.D., Riazalhosseini Y. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Xu X., Liang Y., Liu S., Xiao H., Li F., Cheng H., Fu Z. miR-375 enhances palmitate-induced lipoapoptosis in insulin-secreting NIT-1 cells by repressing myotrophin (V1) protein expression. Int J Clin Exp Pathol. 2010;3:254–264. [PMC free article] [PubMed] [Google Scholar]
- 28.Lu S., Archer M.C. Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int J Cancer. 2009;126:416–425. doi: 10.1002/ijc.24761. [DOI] [PubMed] [Google Scholar]
- 29.Waby J.S., Chirakkal H., Yu C., Griffiths G.J., Benson R.S., Bingle C.D., Corfe B.M. Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol Cancer. 2010;9:275. doi: 10.1186/1476-4598-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Wang L., Gong W., Wei D., Le X., Yao J., Ajani J., Abbruzzese J.L., Huang S., Xie K. A high expression level of insulin-like growth factor I receptor is associated with increased expression of transcription factor Sp1 and regional lymph node metastasis of human gastric cancer. Clin Exp Metastasis. 2004;21:755–764. doi: 10.1007/s10585-005-1198-2. [DOI] [PubMed] [Google Scholar]
- 31.Yao J.C., Wang L., Wei D., Gong W., Hassan M., Wu T.T., Mansfield P., Ajani J., Xie K. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109–4117. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 32.Yuan P., Wang L., Wei D., Zhang J., Jia Z., Li Q., Le X., Wang H., Yao J., Xie K. Therapeutic inhibition of Sp1 expression in growing tumors by mithramycin a correlates directly with potent antiangiogenic effects on human pancreatic cancer. Cancer. 2007;110:2682–2690. doi: 10.1002/cncr.23092. [DOI] [PubMed] [Google Scholar]
- 33.Song J., Ugai H., Ogawa K., Wang Y., Sarai A., Obata Y., Kanazawa I., Sun K., Itakura K., Yokoyama K.K. Two consecutive zinc fingers in Sp1 and in MAZ are essential for interactions with cis-elements. J Biol Chem. 2001;276:30429–30434. doi: 10.1074/jbc.M103968200. [DOI] [PubMed] [Google Scholar]
- 34.Wang L., Wei D., Huang S., Peng Z., Le X., Wu T.T., Yao J., Ajani J., Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 35.Zannetti A., Del Vecchio S., Carriero M.V., Fonti R., Franco P., Botti G., D'Aiuto G., Stoppelli M.P., Salvatore M. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 2000;60:1546–1551. [PubMed] [Google Scholar]
- 36.Martinez I., Gardiner A.S., Board K.F., Monzon F.A., Edwards R.P., Khan S.A. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Wang H.K., McCoy J.P., Banerjee N.S., Rader J.S., Broker T.R., Meyers C., Chow L.T., Zheng Z.M. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Au Yeung C.L., Tsang T.Y., Yau P.L., Kwok T.T. Human papillomavirus type 16 E6 induces cervical cancer cell migration through the p53/microRNA-23b/urokinase-type plasminogen activator pathway. Oncogene. 2011;30:2401–2410. doi: 10.1038/onc.2010.613. [DOI] [PubMed] [Google Scholar]
- 39.Narayan G., Pulido H.A., Koul S., Lu X.Y., Harris C.P., Yeh Y.A., Vargas H., Posso H., Terry M.B., Gissmann L., Schneider A., Mansukhani M., Rao P.H., Murty V.V. Genetic analysis identifies putative tumor suppressor sites at 2q35-q36.1 and 2q36.3-q37.1 involved in cervical cancer progression. Oncogene. 2003;22:3489–3499. doi: 10.1038/sj.onc.1206432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative expression of miR-375 in SiHa and CaSki human cervical cancer cells transfected with miR-375 mimic and negative control. The expression of miR-375 was significantly up-regulated in SiHa and CaSki transfected with miR-375 mimic, compared with negative control. Data are expressed as means ± SD from three independent experiments. *P < 0.05.
The target genes of miR-375 as predicted by two online databases (TargetScan, available at http://www.targetscan.org, and PicTar, available at http://www.pictar.mdc-berlin.de). Gene Ontology analysis (GO; available at http://www.geneontology.org) was used to analyze the target genes predicted by both databases and to form the gene network shown.
SP1 was a potential target gene predicted by both TargetScan and PicTar, and bioinformatics analysis showed the predicted binding site of miR-375 and SP1 3′UTR. The reporter pmirGLO-mUTR was constructed by site-directed mutagenesis.


