Abstract
We have changed the potential phosphorylation site, a threonine residue at position 2 of the D2 polypeptide of the photosystem II complex of Chlamydomonas reinhardtii, to alanine, valine, aspartate, proline, glycine, or glutamate. Mutants with neutral amino acid changes did not display any phenotype with regard to photoautotrophic growth, light sensitivity, fluorescence transients, or photoinhibition. Pulse labeling of these mutants with 32P indicated that a phosphorylated protein of the same size as D2 is absent in these mutants, suggesting that threonine-2 is indeed the unique phosphorylation site of D2. In contrast, mutants in which threonine-2 has been replaced with acidic residues are deficient in photosystem II. Use of chimeric genes containing the psbD 5′-untranslated region revealed that the initiation of translation was not affected in these mutants, but the mutations interfered with a later step of D2 synthesis and accumulation.
Several polypeptides of the PSII complex can be phosphorylated in higher plants. They include the D1 and D2 proteins encoded by the psbA and psbD genes, as well as the products of the psbC and psbH genes. In spinach, the phosphorylated residue of all four proteins has been identified as an O-phosphothreonine near the N terminus, which is exposed to the stromal face of thylakoid membranes (Michel and Bennett, 1987; Michel et al., 1988).
Phosphorylation of the PSII complex has also been reported in the unicellular alga Chlamydomonas reinhardtii (de Vitry et al., 1987, 1991). The products of the psbC and psbH genes have been found to be phosphorylated (Dedner et al., 1988; de Vitry et al., 1991). Two different sites of phosphorylation exist in the 9-kD PsbH subunit and give rise to two bands, L5 and L6, as detected by PAGE. An additional small protein of approximately 5 kD is also phosphorylated and has been identified as the product of psbI or psbF (de Vitry et al., 1987). The detection of phosphorylated D1 and D2 proteins proved difficult in C. reinhardtii because of contamination of PSII particles with LHCII proteins. In this organism, LHCII proteins are difficult to resolve from D1 and D2 by PAGE and can be heavily labeled with radioactive phosphate (Owens and Ohad, 1982; Wollman and Delepelaire, 1984). For this reason, the labeled band first believed to originate from the phosphorylated form of the D1 protein was found to represent a member of the LHCII protein family (de Vitry et al., 1991). Thus, despite its sequence similarity with higher plants (Erickson et al., 1984), the N-terminal part of the D1 protein is not a target for phosphorylation in C. reinhardtii.
In contrast to the D1 protein, phosphorylation of the D2 protein does occur in C. reinhardtii. In vivo protein pulse labeling of cells with [14C]acetate revealed the presence of a new band, D2.1, corresponding to the expected size of the phosphorylated D2 protein (Delepelaire, 1984). This band disappeared after phosphatase treatment, with a concomitant increase in band D2.2, the unphosphorylated form of the D2 protein (de Vitry et al., 1987). Moreover, proteolytic treatment of the isolated D2.1 and D2.2 bands generated similar fragments, confirming the presence of two forms of the same protein (Delepelaire, 1984). The exact site of phosphorylation of the D2 protein in C. reinhardtii has not been determined. Because the N-terminal region of D2 is conserved between C. reinhardtii and higher plants, it is likely that the same Thr residue at position 2 is phosphorylated in this alga (Erickson et al., 1986). However, some doubts about the existence of this phosphorylation site were raised in a recent report (Andronis et al., 1998).
As in higher plants, the PSII complex of C. reinhardtii appears to be assembled in the unappressed region of the thylakoid membranes (Schuster et al., 1988; de Vitry et al., 1989) before it migrates to the grana region, where it accumulates (Vallon et al., 1985; de Vitry et al., 1989). Mutants of C. reinhardtii deficient in the expression of the PsbC or the D1 protein are unable to assemble the PSII complex. In these mutants, the synthesis of D2 can still be detected by [14C]acetate pulse labeling. However, the band corresponding to the phosphorylated form never appears (de Vitry et al., 1987, 1989), suggesting that phosphorylation of the D2 protein occurs only in fully assembled complexes, which is in agreement with the delayed appearance of D2.1 in [14C]acetate pulse-labeled wild-type cells (Delepelaire, 1984). Phosphorylation of PSII proteins is still observed in mutants of C. reinhardtii lacking PSI, ATPase, or the Cyt b6/f complex (de Vitry et al., 1987; Wollman and Lemaire, 1988). The independence of phosphorylation of PSII proteins from a functional Cyt b6/f complex has also been reported in maize (Bennett et al., 1988). This feature distinguishes PSII phosphorylation from the well-studied phenomenon of state transition, which relies on Cyt b6/f-dependent LHCII protein phosphorylation (Wollman and Lemaire, 1988; Allen, 1992). LHCII and PSII protein phosphorylation are dependent on the reduction of the plastoquinone pool (Allen, 1992; Aro et al., 1993; Ebbert and Godde, 1996; Rintamäki et al., 1996). However, one remarkable exception concerns the phosphorylation of D2 in C. reinhardtii, which is induced by the oxidation of the plastoquinone pool (Delepelaire, 1984; Delepelaire and Wollman, 1985).
Many different roles have been suggested for PSII phosphorylation, including spatial separation of PSII complexes between grana and stromal lamellae (Mattoo et al., 1989) and influence on the biogenesis, stability, and dimerization of the PSII complex (de Vitry et al., 1989; Kruse et al., 1997; Summer et al., 1997). However, the best-characterized function of the phosphorylation of the D1 and D2 proteins appears to be related to photoinhibition. In higher plants, phosphorylation of these proteins protects against proteolytic degradation, indicating a role in the degradation-repair cycle of D1 and D2 during photoinhibition (Schuster et al., 1988; Aro et al., 1993; Koivuniemi et al., 1995; Ebbert and Godde, 1996).
To gain new insights into the role of D2 phosphorylation in C. reinhardtii, the putative phosphorylation site was changed and the properties of the mutants were analyzed. Substitution of Thr-2 with acidic residues (either Asp or Glu) interfered drastically with the expression of the D2 protein at a step following the initiation of translation. Substitution with neutral amino acids (Ala, Val, Pro, or Gly) affected phosphorylation of the D2 protein. However, no detectable change in phenotype was observed with these mutant cells.
MATERIALS AND METHODS
Strains and Media
The Chlamydomonas reinhardtii mutants FuD7 and nac2-26 have been described previously (Bennoun et al., 1986; Kuchka et al., 1989; Nickelsen et al., 1994). FuD7 contains a deletion of approximately 9 kb, which removes the chloroplast psbA gene; nac2-26 is a nuclear mutant that specifically lacks the psbD mRNA. Wild-type and mutant strains were grown as described by Harris (1989). If necessary, TAP medium and HSM were solidified with 2% Bacto-agar (Difco, Detroit, MI) and supplemented with spectinomycin (Sigma).
DNA Constructs
Thr-2 Mutants
Preparation of plasmids was as described by Sambrook et al. (1989). The chloroplast EcoRI fragment R3 (Rochaix et al., 1984) was cloned into the EcoRI site of pBluescript SK− (Stratagene) in which both PvuII sites had been destroyed. The unique NsiI site was blunt ended, and a 1.8-kb EcoRV-SmaI fragment of pUC-atpX-AAD (Goldschmidt-Clermont, 1991) was inserted at this site to obtain plasmid pSK-108#14. This plasmid contains the 5′ part of psbD and the aadA-rbcL cassette (Goldschmidt-Clermont, 1991) upstream of psbD oriented in the opposite direction.
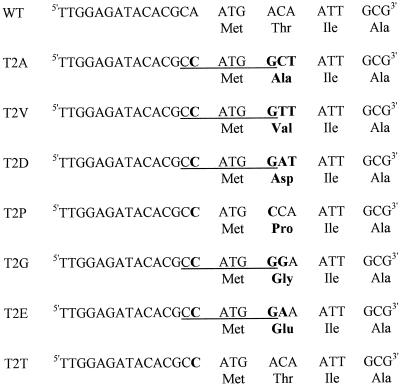
Mutagenesis of psbD was performed according to the megaprimer method (Sarkar and Sommer, 1990). A PCR reaction using the oligonucleotides 1963 (5′-AGAAACAGCTGCTGTTAA-3′) and 1965 (5′-TTTGGAGATACACGCCATGG[A/G/C/T][A/T]ATTGCGAT-3′) and pSK-108#14 as a template was performed first. The product of this reaction, the megaprimer, was used with oligonucleotide 1365 (5′-CCATCGATAAGCTTGATTTTTTATATCATAATAATAAA-3′) on the same template in a second PCR reaction. The PCR product was cloned into pBluescript SK− (Stratagene) by the T-vector method (Marchuk et al., 1990) and sequenced. Finally, fragments containing different mutations at the second codon of psbD were recloned into pSK-108#14 using the unique ClaI and PvuII sites of the vector. The resulting plasmids were called pSK-140(T2A), pSK-141(T2V), pSK-142(T2D), pSK-143(T2P), pSK-144(T2G), and pSK-145(T2E), and each contained a different substitution of the second psbD codon (Fig. 1). These plasmids were used to transform C. reinhardtii. pSK-146(T2T) was constructed similarly except that oligonucleotide 146 (5′-TTGGAGATACACGCCATGACAATTGCG-3′) was used instead of oligonucleotide 1965.
Figure 1.
Sequences of the psbD codon 2 mutations and the corresponding D2 sequences. Mutated nucleotides and amino acids are indicated in boldface. When present, the NcoI restriction site is underlined. WT, Wild type.
Fusion Proteins
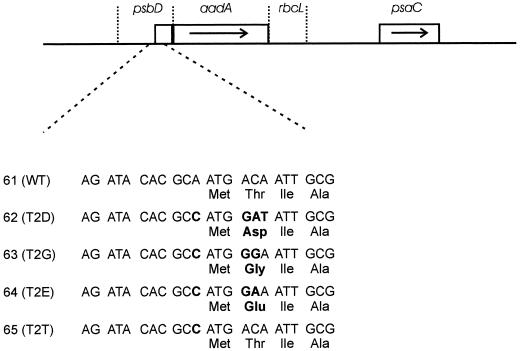
PCR fragments of 399 bp were obtained using oligonucleotides 1365 and psbD-BspHI (5′-TACTGAACGAGTCATGACAAACTGACG-3′) on plasmids pSK-108#14, pSK-142, pSK-144, pSK-145, and pSK-146. After they were cloned in pBluescript SK− and sequenced, each plasmid was cut with ClaI and BspHI and the resulting fragment was used to replace the promoter-containing atpA ClaI-NcoI fragment of plasmid pUC-atpX-AAD. The ClaI-SacI fragments containing psbD-aadA-rcbL were then used to replace the 1.8-kb ClaI-SacI fragment in pBS-5.8-aadA (Fischer et al., 1997). This yielded plasmids pBS-61(WT), pBS-62(T2D), pBS-63(T2G), pBS-64(T2E), and pBS-65(T2T) (see Fig. 6), which were used to introduce the psbD-aadA reporter genes near psaC by chloroplast transformation.
Figure 6.
Structure and sequence of the chimeric psbD-aadA genes. The chimeric psbD-aadA gene and the psaC gene are shown. psbD, Promoter, 5′-untranslated region, and coding sequence for the 35 N-terminal codons of psbD. aadA, Coding sequence for the aminoglycoside adenine transferase. rbcL, 3′-untranslated region of rbcL. The sequence around the start of translation is shown. Mutations are indicated in boldface. WT, Wild type.
Chloroplast Transformation
Chloroplast transformation was carried out as described previously (Fischer et al., 1997). The plasmids pSK-108#14, pSK-140, pSK-141, pSK-142, pSK-143, pSK-144, pSK-145, and pSK-146 were used to transform C. reinhardtii wild-type cells (mt+) by selecting for growth at low light (6 μE m−2 s−1) on TAP plates supplemented with 150 μg/mL spectinomycin. The corresponding transformants were WT-aadA, 140(T2A), 141(T2V), 142(T2D), 143(T2P), 144(T2G), 145(T2E), and 146(T2T), respectively.
Plasmids pBS-61, pBS-62, pBS-63, pBS-64, and pBS-65 were used to transform the mutant psaCΔSpcs (Fischer et al., 1996) and selected on TAP plates in the presence of light (60 μE m−2 s−1) to obtain mutants 61(WT), 62(T2D), 63(T2G), 64(T2E), and 65(T2T), respectively.
Fluorescence Transients
Fluorescence transients were measured on whole cells grown on TAP plates, as described previously (Fenton and Crofts, 1990). The curves were then normalized to their maximal values using the Excel program (Microsoft, Redmond, WA).
Immunoblot Analysis
Proteins from whole cells or subcellular fractions were separated by SDS-PAGE, as described previously (Sambrook et al., 1989). For LDS-PAGE, SDS was replaced with LDS in the gel and in the buffers, and electrophoresis was performed at 4°C instead of at room temperature (Ebbert and Godde, 1996).
Immunoblotting and enhanced chemiluminescence detection were carried out according to the manufacturer's protocol (Amersham). Detection with an antibody raised against the D2 protein was modified as follows. The nitrocellulose membranes were saturated with 5% yeast extract instead of nonfat milk and incubated with the antibody overnight at 4°C. It was necessary to double the number of washings with this antibody.
35S Pulse Labeling
Approximately 107 cells in exponential growth were inoculated in 100 mL of low-S TAP medium (TAP without NH4Cl, CaCl2, or MgSO4) and grown overnight. Cells were pelleted, washed once with minus-S TAP medium (20 mm Tris-acetate-OH, pH 7.0, and 1 mm K2HPO4/KH2PO4, pH 7.0), and resuspended in 20 mL of the same medium. The cell suspension was then incubated at 25°C with agitation for 2 to 4 h, during which time the chlorophyll concentration was measured according to the method of Porra et al. (1989). Cells corresponding to 40 μg of chlorophyll were pelleted, resuspended in 1 mL of minus-S TAP medium, and incubated for 5 min at room temperature with agitation. After the addition of cycloheximide (6 μg/mL final concentration), the cell suspension was incubated for 10 min at room temperature before 100 μCi of Na235SO4 was added. Labeling was carried out at room temperature with agitation for different times. Aliquots were withdrawn, microcentrifuged for 2 min, and quickly frozen. For analysis, thawed cells were washed with TE buffer (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA) containing 1 mm PMSF, resuspended in loading buffer, and fractionated by SDS-PAGE (Sambrook et al., 1989). The gel was then stained with Coomassie blue, dried, and subjected to autoradiography (Sambrook et al., 1989).
RNA Analysis
Fifty milliliters of an exponentially growing cell culture was centrifuged at 4000g for 5 min. The pellet was washed once with 5 mL of 10 mm Tris-HCl, pH 7.5, and frozen in dry ice. Two milliliters of TEN-SDS buffer (200 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 10 mm EDTA, and 0.2% SDS) and 2 mL of phenol:chloroform (1:1, v/v) were added to the frozen pellet before it was resuspended by homogenization in a Polytron (Brinkmann). The solution was centrifuged at 8000g for 5 min at 4°C. The aqueous phase was retrieved and extracted for a second time before the nucleic acids were precipitated with 5 mL of ethanol and washed with 70% ethanol. Finally, the nucleic acids were resuspended in 400 μL of 1 mm EDTA with 0.1% diethylpyrocarbonate. RNA was quantified by spectrophotometry (Sambrook et al., 1989) and kept frozen. Northern-blot analysis was carried out as described previously (Sambrook et al., 1989; Stampacchia et al., 1997).
32P Pulse Labeling
The protocol for in vivo pulse labeling with radioactive phosphate was adapted from the method of Wollman and Delepelaire (1984). Approximately 5 × 108 cells were pelleted and resuspended in 20 mL of minus-P TAP medium (TAP without K2HPO4 or KH2PO4). The chlorophyll concentration of the cell suspension was determined according to the method of Porra et al. (1989), and the equivalent of 200 μg of chlorophyll was pelleted. The pellet was resuspended in minus-P TAP medium to a final concentration of 25 μg chlorophyll mL−1 followed by the addition of K2HPO4 and KH2PO4, pH 7.0 (5 mm final concentration), and 32Pi (1 mCi/mL final concentration). Labeling was carried out at room temperature with agitation in the presence of medium light (approximately 30 μE m−2 s−1) for 2 h, with the addition of DCMU (ICN) after 1 h. Labeled cells were pelleted and washed with 5 mL of minus-P TAP medium containing 10 mm NaF.
Thylakoid membrane isolation has been described (Chua and Bennoun, 1975). The labeled cells were resuspended in 500 μL of buffer H1 (25 mm Hepes-KOH, pH 7.5, 5 mm MgCl2, and 0.3 m Suc) with 1 mm PMSF. Cells were broken by the addition of 700 mg of glass beads and vortexing at maximum speed for 2 min. Membrane fractions were recovered in 1 mL of buffer H1 and pelleted by microcentrifugation for 3 min. The pellet was washed with 1 mL of buffer H2 (5 mm Hepes-KOH, pH 7.5, 10 mm EDTA, and 0.3 m Suc) and resuspended in 1 mL of buffer H3 (5 mm Hepes-KOH, pH 7.5, 10 mm EDTA, and 1.8 m Suc). Thylakoid membranes were then purified on a discontinuous Suc gradient consisting of 1 mL of buffer H3 (containing the membrane fraction), 1 mL of buffer H4 (5 mm Hepes-KOH, pH 7.5, 10 mm EDTA, and 1.3 m Suc), and 1 mL of buffer H5 (5 mm Hepes-KOH, pH 7.5, and 0.5 m Suc) in an ultracentrifugation tube (SW60, Beckman). The gradient was centrifuged at 24,000 rpm for 1 h at 4°C. The upper green band containing the thylakoid membranes was retrieved and washed with 1 mL of buffer H6 (5 mm Hepes-KOH, pH 7.5, and 10 mm EDTA). Finally, thylakoid membranes were resuspended in 100 μL of buffer H6 with 10% glycerol and kept at −70°C. Analysis of labeled proteins was carried out as described for 35S-labeled samples.
Photoinhibition Measurement
Exponentially growing cells were diluted to a final concentration of 0.5 μg chlorophyll mL−1 in TAP medium. For photoinhibition, cell cultures were exposed to high light (1500 μE m−2 s−1) at room temperature with agitation. After 1.5 h, light was reduced to approximately 5 μE m−2 s−1 to allow for recovery of PSII activity. Fluorescence transients of the cell cultures were measured at different times using the Plant Efficiency Analyser (Hansatech, King's Lynn, UK), and the ratio of variable to maximal chlorophyll fluorescence was used as an indicator of the efficiency of light capture by PSII (Kyle et al., 1984; Schuster et al., 1988; Krause and Weis, 1991).
RESULTS
The second codon of the psbD gene of C. reinhardtii, coding for Thr, was changed to Ala, Val, Asp, Pro, Gly, or Glu by site-directed PCR mutagenesis (Fig. 1). For practical reasons, these mutations also convert the A preceding the AUG into a C to create a new NcoI site. Mutant T2T, which contains only this last substitution, was generated as a control (Fig. 1). The psbD mutations were introduced in a transformation vector carrying the 2.7-kb EcoRI chloroplast fragment R3 containing psbD (Rochaix et al., 1984), thus replacing the endogenous psbD gene. As a selectable marker, the aadA cassette (Goldschmidt-Clermont, 1991) was inserted in the opposite direction 261 bp upstream of the translation start site of psbD. The constructs were introduced into C. reinhardtii wild-type (mt+) cells by biolistic transformation, and transformants were selected on spectinomycin-containing plates. Correct integration of the transforming DNA and the homoplasmicity of the transformants were confirmed by Southern blotting, and the DNA regions that had been amplified by PCR for mutagenesis were verified by sequencing (data not shown).
Table I summarizes the growth properties of the transformants on TAP medium and HSM under different light intensities. Mutants T2A, T2V, T2P, T2G, T2T, and WT-aadA were indistinguishable from the wild type under all conditions tested. Mutant T2D did not grow on minimal medium, indicating that it is deficient in photosynthetic activity. Reduced photosynthetic activity probably explains the slow growth of mutant T2E on minimal medium. In addition, mutants T2D and T2E were sensitive to high light (1000 μE m−2 s−1) on TAP medium or HSM (Table I). This was also observed with nac2-26, a nuclear mutant that does not express the D2 protein (Kuchka et al., 1989).
Table I.
Growth properties of the psbD mutants
| Medium | Light | T2A | T2V | T2D | T2P | T2G | T2E | T2T | WT-aadAa | nac2-26 | WT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| μE m−2 s−1 | |||||||||||
| TAP | 6 | +b | + | + | + | + | + | + | + | + | + |
| 60 | ++c | ++ | + | ++ | ++ | ++ | ++ | ++ | + | ++ | |
| 1000 | ++ | ++ | −d | ++ | ++ | − | ++ | ++ | − | ++ | |
| HSM | 60 | + | + | − | + | + | +/− | + | + | − | + |
| 1000 | + | + | − | + | + | − | + | + | − | + |
WT-aadA is a wild-type strain with the aadA cassette inserted at the same site of the chloroplast genome as in the other psbD mutants.
+, Medium growth.
++, Fast growth.
−, No growth.
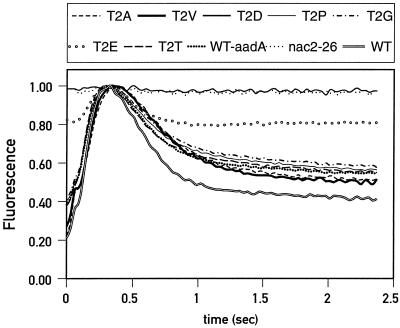
As expected for psbD mutants, the PSII activity of T2D and T2E was affected. Fluorescence transients of T2D cells displayed a flat curve characteristic of PSII-deficient mutants (Fig. 2). The fluorescence transient of mutant T2E was intermediate between that of the wild type and nac2-26, indicating partial PSII activity. The fluorescence transients of the other transformants were indistinguishable from that of the wild-type control. Therefore, despite its strong conservation across species, Thr-2 can be replaced by Ala, Val, Pro, or Gly without any apparent effect on PSII activity. We noticed, however, a small but significant difference in the stationary fluorescence level, which was consistently higher in the strains containing the aadA cassette than in the wild-type strain. The significance of this difference is not clear.
Figure 2.
Normalized fluorescence transients of wild-type (WT) and mutant cells.
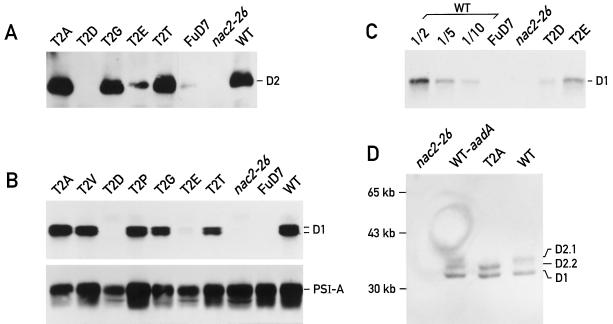
The reduction of PSII activity in the mutants T2D and T2E can be attributed to a low level of PSII complex, because less D2 protein accumulated in these mutants than in the wild type (Fig. 3A). No D2 protein was seen in mutant T2D, and it was barely detectable in mutant T2E (Fig. 3A). The accumulation of the D2 and D1 proteins followed the same pattern in all mutants examined, i.e. the amount of D1 in mutants T2D and T2E was reduced to 10% and 30%, respectively, of wild-type levels (Fig. 3C), and D1 and D2 accumulated to wild-type levels in the other mutants (Fig. 3B). It is well established that these two proteins can accumulate only in the presence of each other. In the absence of D1 in FuD7, the D2 protein is synthesized but rapidly degraded (deVitry et al., 1989), and in the absence of D2 in the nac2-26 mutant, the D1 protein is synthesized and turns over rapidly (Kuchka et al., 1989). Overexposure of the immunoblot in Figure 3A reveals trace amounts of D2 in FuD7. However, under the same conditions D2 was undetectable in the T2D and nac2-26 mutants. Taken together, these results suggest that the presence of Asp at position 2 affects a step of the synthesis or assembly process of the D1 and D2 proteins. The accumulation of the PsaA reaction center subunit of PSI was not affected in these mutants, indicating that the mutations act specifically on PSII (Fig. 3B).
Figure 3.
Accumulation of the PSII proteins. A, The equivalent of 2.5 μg of chlorophyll from each sample was analyzed by immunoblotting using antibodies directed against the D2 protein. B, The equivalent of 2 μg of chlorophyll from each sample was analyzed by immunoblotting using antibodies directed against the D1 or PsaA proteins. C, Thirty micrograms of protein extract from each sample was analyzed by immunoblotting using antibodies directed against the D1 protein. Dilutions of the wild type (WT) were done in FuD7 extracts to keep total protein concentration constant. D, The equivalent of 1.5 μg of chlorophyll from each sample was analyzed by 12.5% LDS-PAGE at 4°C. Immunoblotting was performed with antibodies directed against the D1 and D2 proteins.
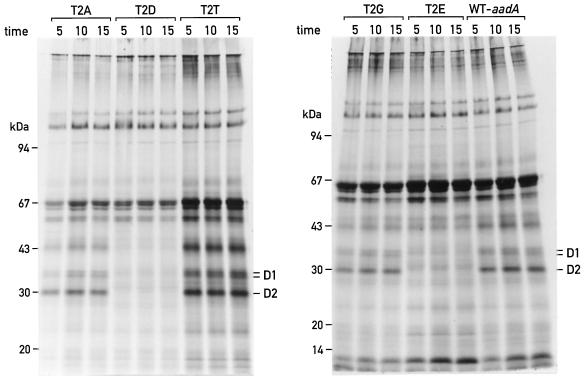
To examine the synthesis of the D2 protein in mutants T2D and T2E, the chloroplast proteins of these cells were pulse labeled with 35S (Fig. 4). It can be seen that the synthesis of D2 was strongly diminished in mutant T2D compared with the other mutants or the control strains. D2 protein synthesis was also affected in mutant T2E, but to a lesser extent. Synthesis of the D2 protein in mutant T2E was estimated at approximately 30% of the wild-type level by quantification of Figure 4 (not shown).
Figure 4.
35S pulse labeling of wild-type (WT) and mutant cells. Different strains were labeled in vivo with 35S in the presence of cycloheximide for 5, 10, or 15 min. Labeled chloroplast proteins were analyzed on a 7.5% to 15% PAGE gradient gel and subjected to autoradiography.
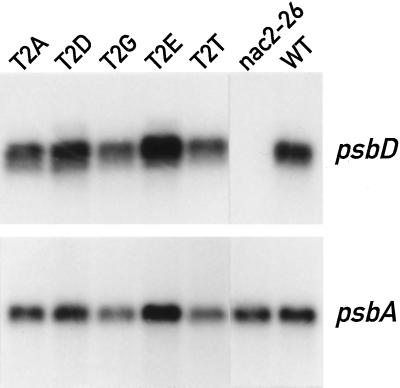
Reduction of D2 protein synthesis in mutants T2D and T2E cannot be explained by an effect on the transcription of the psbD gene or on stabilization of its mRNA, because the psbD mRNA accumulated normally in these mutants (Fig. 5). Nor can reduction of the D2 protein be explained by an indirect effect of low PSII activity in these mutants, because D2 protein synthesis is unaffected in mutants lacking D1 (de Vitry et al., 1989). This implies that the mutations in strains T2D and T2E diminish the expression of psbD at the level of mRNA translation or protein stability.
Figure 5.
Accumulation of psbD mRNA in wild-type (WT) and mutant cells. Total RNA of the different strains was analyzed by northern blotting. PsbD probe, 917-bp DpnI-EcoRI fragment of psbD (nucleotides 13 to 930 relative to the translation start site). PsbA probe, 1.8-kb BamHI-XbaI fragment of psbA (nucleotides −50 to1750 relative to the translation start site).
To determine whether translation initiation was affected, chimeric genes were constructed between psbD and aadA (Fig. 6). These genes encode proteins containing the N-terminal 35 amino acids of the D2 protein fused to the aadA reporter protein and are driven by the promotor and the 5′-untranslated region of psbD. These chimeric genes were introduced into the chloroplast genome near the psaC gene (see Methods). Correct insertion and homoplasmicity of these constructs in the transformants were confirmed by Southern blotting (data not shown).
Expression of the fusion protein was monitored by growth of the transformants with increasing amounts of spectinomycin (Table II). All transformants tested were resistant to a low concentration of spectinomycin (50 μg/mL), in contrast to the wild type or psaCΔSpcs, the recipient strain used for transformation. No correlation could be found between the negative charge on residue 2 of D2 and the reduction in expression of the chimeric gene. Instead, a clear increase in spectinomycin resistance was observed with the T2D mutant. This increase must have been caused by enhanced translation or stabilization of the fusion protein, because the level of the chimeric mRNA was the same in all transformants (data not shown). Comparison of the spectinomycin-resistance levels showed that the negative charge at the N terminus did not interfere with the initiation of translation. Therefore, the reduced levels of the D2 protein in mutants T2D and T2E must be attributed to a step subsequent to translation initiation.
Table II.
Growth properties of the transformants containing the psbD-aadA mutant genes
| Medium | Light | Spectinomycin | 61 (WT)a | 62 (T2D) | 63 (T2G) | 64 (T2E) | 65 (T2T) | nac2-26 | T2D | WT | psa CΔSpcs b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| μE m−2 s−1 | μg mL−1 | ||||||||||
| TAP | 6 | 500 | −c | − | − | − | − | − | + | − | − |
| 6 | 300 | − | +d | − | − | − | − | + | − | − | |
| 6 | 200 | − | + | − | − | − | − | + | − | − | |
| 6 | 100 | + | + | − | − | − | − | + | − | − | |
| 6 | 50 | + | + | + | + | + | − | + | − | − | |
| 6 | 0 | + | + | + | + | + | + | + | + | + | |
| HSM | 60 | 0 | + | + | + | + | + | − | − | + | − |
WT, Wild type.
The psaCΔSpcs strain lacks psaC (Fischer et al., 1996) and was used as the recipient strain for transformation with the chimeric psbD-aadA genes.
−, No growth.
+, Growth.
The protein pulse-labeling patterns in Figure 4 show that, in addition to the reduction in the rate of synthesis of D2, D1 synthesis was also considerably diminished in mutants T2D and T2E. To determine whether this occurred at the level of initiation of translation, a chimeric gene consisting of the psbA 5′-untranslated region fused to aadA was inserted into the chloroplast genome. However, the expression of this chimeric gene could not be tested properly because its transcript was present in considerably lower amounts in the transformed wild-type strain than in the transformed nac2-26 and FuD7 strains, for reasons that are not clear (data not shown).
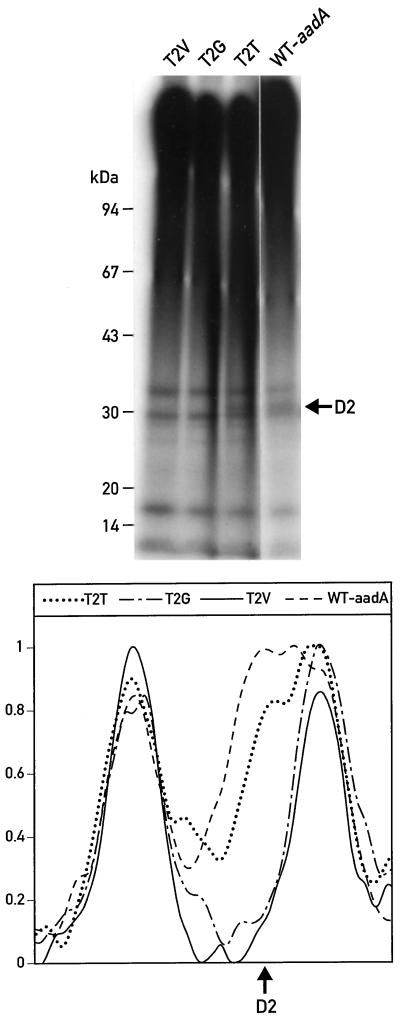
Figure 3A reveals small differences in the size of the protein detected by the D2 antibody. In samples from mutants T2A and T2G, the D2 protein had a slightly smaller apparent molecular mass than in samples from T2T and the wild type. An improved gel resolution was obtained with LDS-PAGE at 4°C (Fig. 3D). Under these conditions, the D2 band of lower mobility, labeled D2.1, represents the phosphorylated form of D2, whereas the faster-migrating band, D2.2, represents the unphosphorylated form (Delepelaire, 1984). Although D2.1 was absent in T2A, this band could be detected in both the wild type and WT-aadA (Fig. 3D), suggesting that Thr-2 is the unique phosphorylation site of D2.
To test directly whether phosphorylation is impeded by substitutions of Thr-2, mutant cells were pulse labeled in vivo with 32Pi (Fig. 7). The labeling was performed under conditions that lead to the oxidation of the plastoquinone pool, and hence favor phosphorylation of the D2 protein, and that reduce phosphorylation of the LHCII proteins (Delepelaire and Wollman, 1985). Analysis of purified thylakoid membranes from these cells indicated the presence of a phosphorylated protein the size of D2 in both the wild type and T2T. This band was not detectable in the mutants T2V (Fig. 7), T2G (Fig. 7), or T2A (data not shown). Quantification of the radioactive signals on this gel excluded the possibility that in mutants T2V and T2G the D2 signal was shifted and hidden by one of the LHCII protein bands (Fig. 7). These observations are compatible with the idea that the D2 protein is phosphorylated on Thr-2 in C. reinhardtii, as it is in spinach (Michel et al., 1988).
Figure 7.
32P pulse labeling. Mutant strains were labeled in vivo with 32Pi for 2 h under oxidizing conditions. Purified thylakoid membranes were analyzed on a 7.5% to 15% PAGE gradient gel and subjected to autoradiography. A scan of the autoradiogram in the region of D2 is shown in the lower part of the figure. WT, Wild type.
Because strains T2V and T2G accumulate PSII, in which D2 does not appear to be phosphorylated, we tested whether phenotypic differences could be detected in these cells. Strains T2V and T2G grew as well as the wild type on minimal medium (Table I), indicating that photosynthesis was not appreciably affected. In higher plants, phosphorylation of the D1 and D2 proteins has been shown to play a role in the turnover of the proteins under high light (Koivuniemi et al., 1995; Ebbert and Godde, 1996). In C. reinhardtii, the mutant cells T2V and T2G grew normally at light intensities as high as 1000 μE m−2 s−1 (Table I), indicating that phosphorylation of the D2 protein has no obvious role in the protection against photoinhibition.
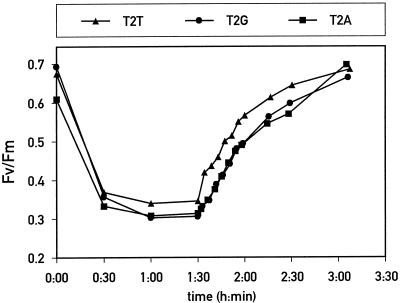
To measure photoinhibition more quantitatively, the ratio of variable to maximal chlorophyll fluorescence was determined for mutant and control cells during photoinhibition and the subsequent recovery period (Fig. 8). The overall kinetics of photoinhibition and recovery were identical for mutants and control cells. The small differences in absolute values were not reproducible from one experiment to another, so they are not significant. Because mutants T2A and T2G prevented phosphorylation of D2, these data show that this posttranslational modification is not important for the protection of PSII during photoinhibition and the recovery of PSII activity. Therefore, the function ascribed to D2 protein phosphorylation in higher plants is not conserved in C. reinhardtii.
Figure 8.
Kinetics of photoinhibition and recovery. Cell suspensions at a concentration of 2 μg chlorophyll mL−1 were subjected to high light for 90 min, followed by an incubation in low light for 2 h. The ratio of variable to maximal chlorophyll fluorescence (Fv/Fm), calculated from fluorescence measurements, is plotted versus time for each mutant.
DISCUSSION
The Phosphorylation Site of D2 Is Not Essential for PSII Function
The N-terminal region of the D2 protein is highly conserved among different photosynthetic organisms. In particular, Thr-2, which has been shown to be phosphorylated in spinach (Michel et al., 1988), is present in all D2 sequences that have been determined. It is surprising, therefore, that this residue can be changed to Ala, Val, Pro, or Gly without any apparent effect on PSII activity. The same observation was reported recently with Ala and Ser substitutions at the same site (Andronis et al., 1998). In that report, photoautotrophic growth rates, fluorescence induction and decay, thermoluminescence, and photoinhibition were not significantly different in the D2 mutants compared with the wild type. Although Andronis et al. (1998) noticed differences in the migration of the D2 protein, they favored the idea that this protein is not phosphorylated in C. reinhardtii. Our data indicate that mutations of Thr-2 of D2 abolish specifically the phosphorylation of a polypeptide of the same size as D2 (Fig. 7). This implies that the mutations of Thr-2 affect the only phosphorylation site of the protein. It could still be argued that the modifications introduced into the D2 protein affect phosphorylation indirectly on other residues. However, because these modifications have no apparent effect on PSII activity, any structural perturbation of the protein seems unlikely. Nevertheless, unequivocal identification of the phosphorylation site on the D2 protein still awaits MS analysis as performed for higher plants (Michel et al., 1988).
It appears that phosphorylation of the PSII proteins differs between C. reinhardtii and higher plants. The D1 protein, which is most prominently phosphorylated in higher plants, is not phosphorylated in this unicellular alga (de Vitry et al., 1991). Phosphorylation of the D2 protein is regulated in an opposite manner than phosphorylation of LHCII with respect to the plastoquinone redox state (Delepelaire, 1984; Delepelaire and Wollman 1985). Phosphorylation of D2 in C. reinhardtii was also demonstrated by analysis of purified PSII complexes after labeling of cells in vivo or thylakoids in vitro (Ikeuchi et al., 1987). However, we and others (Andronis et al., 1998) were not able to detect phosphorylation of the D2 protein by in vitro phosphorylation of thylakoid membranes (data not shown). One possibility is that the kinase associated with this process is only loosely associated with the thylakoid membranes, in contrast to the situation in higher plants (Koivuniemi et al., 1995). Finally, the relationship between PSII phosphorylation and photoinhibition does not seem to hold for C. reinhardtii (Andronis et al., 1998; this work).
The D2 Polypeptide Is Destabilized by Thr-2 Asp/Glu Changes
Substitution of Thr-2 of the D2 protein with acidic residues had a drastic effect on its synthesis and, consequently, on PSII stability. By using chimeric genes containing the first 35 residues of the D2 protein fused to the aadA sequence, no reduction in expression of the chimeric genes carrying the T2D and T2E mutations was observed compared with genes containing the wild-type residue. It is highly unlikely, therefore, that the loss of D2 in these mutants is attributable to an inhibition of initiation of translation. In these mutants, the effect of the mutations could occur at the level of translation elongation or at the level of stability of D2. Acidic residues near the N-terminal end of D2 could induce a conformational change that makes the protein very susceptible to proteases. One possibility is that the negative charge at the N terminus interferes with the insertion of the protein into the thylakoid membranes. Because the D2 protein inserts into the membranes before it interacts with the D1 protein (Jensen et al., 1986; de Vitry et al., 1989), this last model would explain the more pronounced destabilization of D2 in the T2D mutant compared with FuD7, which lacks D1 but is still able to synthesize D2. The fact that the presence of Asp and Glu reduced D2 to different levels indicates that, in addition to its negative charge, the nature of the residue is also important.
The Absence of D2 Synthesis Leads to the Absence of D1 Synthesis
Our protein pulse-labeling studies have shown that substitution of Thr-2 with neutral amino acids has no effect on the rate of synthesis and accumulation of D2 and D1. In contrast, in the T2D and T2E mutants, in addition to D2, D1 was also strongly destabilized. Similar effects on D1 have been observed in other mutants in which D2 is no longer synthesized or in which D2 is rapidly degraded (Erickson et al., 1986; de Vitry et al., 1989). It is not yet known through which step of gene expression this effect is mediated.
ACKNOWLEDGMENTS
We thank P. Nixon for antibodies against D2, A. Auchincloss, M. Goldschmidt-Clermont, and M. Hippler for helpful comments, and N. Roggli for drawings and photography.
Abbreviations:
- HSM
high-salt minimal medium
- LDS
lithium dodecyl sulfate
- LHCII
light-harvesting antenna of PSII
- TAP
Tris-acetate-phosphate
Footnotes
This work was supported by grant no. 3100-050895.97 from the Swiss National Science Foundation.
LITERATURE CITED
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Andronis C, Kruse O, Deák Z, Vass I, Diner B, Nixon PJ. Mutation of residue threonine-2 of the D2 polypeptide and its effect on photosystem II function in Chlamydomonas reinhardtii. Plant Physiol. 1998;117:515–524. doi: 10.1104/pp.117.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Bennett J, Shaw EK, Michel H. Cytochrome b6/f complex is required for phosphorylation of light-harvesting chlorophyll a/b complex II in chloroplast photosynthetic membranes. Eur J Biochem. 1988;171:95–100. doi: 10.1111/j.1432-1033.1988.tb13763.x. [DOI] [PubMed] [Google Scholar]
- Bennoun P, Spierer-Herz M, Erickson J, Girard-Bascou J, Pierre Y, Delosme M, Rochaix J-D. Characterization of photosystem II mutants of Chlamydomonas reinhardtii lacking the psbA gene. Plant Mol Biol. 1986;6:151–160. doi: 10.1007/BF00021484. [DOI] [PubMed] [Google Scholar]
- Chua N-H, Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedner N, Meyer HE, Ashton C, Wildner GF. N-terminal sequence analysis of the 8 kDa protein in Chlamydomonas reinhardtii: localization of the phosphothreonine. FEBS Lett. 1988;26:77–82. [Google Scholar]
- Delepelaire P. Partial characterization of the biosynthesis and integration of the photosystem II reaction centers in the thylakoid membrane of Chlamydomonas reinhardtii. EMBO J. 1984;3:701–706. doi: 10.1002/j.1460-2075.1984.tb01872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P, Wollman F-A. Correlations between fluorescence and phosphorylation changes in thylakoid membranes of Chlamydomonas reinhardtii in vivo: a kinetic analysis. Biochim Biophys Acta. 1985;809:277–283. [Google Scholar]
- de Vitry C, Diner BA, Lemoine Y. Chemical composition of photosystem II reaction centers (PSII): phosphorylation of PSII polypeptides. In: Biggins J, editor. Progress in Photosynthesis Research, Vol II. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987. pp. 105–108. [Google Scholar]
- de Vitry C, Diner BA, Popot J-L. Photosystem II particles from Chlamydomonas reinhardtii: purification, molecular weight, small subunit composition and protein phosphorylation. J Biol Chem. 1991;266:16614–16621. [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman F-A. Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol. 1989;109:991–1006. doi: 10.1083/jcb.109.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbert V, Godde D. Phosphorylation of PSII polypeptides inhibits D1 protein degradation and increases PSII stability. Photosynth Res. 1996;50:257–269. doi: 10.1007/BF00033124. [DOI] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Malnoë P, Rochaix J-D. Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J. 1986;5:1745–1754. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Rochaix J-D. Chlamydomonas reinhardtii gene for the 32000 mol wt protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984;3:2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton JM, Crofts AR. Computer-aided fluorescence imaging of photosynthetic systems: application of video imaging to the study of fluorescence induction in green plants and photosynthetic bacteria. Photosynth Res. 1990;26:59–66. doi: 10.1007/BF00048977. [DOI] [PubMed] [Google Scholar]
- Fischer N, Sétif P, Rochaix J-D. Targeted mutations in the psaC gene of Chlamydomonas reinhardtii: preferential reduction of FB at low temperature is not accompanied by altered electron flow from photosystem I to ferredoxin. Biochemistry. 1997;36:93–102. doi: 10.1021/bi962244v. [DOI] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix J-D. Selectable marker recycling in the chloroplast. Mol Gen Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Plumley F, Schmidt GW. Identification of phosphorylated reaction center polypeptides in thylakoids of Chlamydomonas reinhardtii and Pisum sativum. In: Biggins J, editor. Progress in Photosynthesis Research, Vol II. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987. pp. 805–808. [Google Scholar]
- Jensen KH, Herrin DL, Plumley PL, Schmidt GW. Biogenesis of photosystem II complexes: transcriptional, translational and post-translational regulation. J Cell Biol. 1986;103:1315–1325. doi: 10.1083/jcb.103.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A, Aro E-M, Andersson B. Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry. 1995;34:16022–16029. doi: 10.1021/bi00049a016. [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E. Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:313–349. [Google Scholar]
- Kruse O, Zheleva D, Barber J. Stabilization of photosystem two dimers by phosphorylation: implication for the regulation of the turnover of D1 protein. FEBS Lett. 1997;408:276–280. doi: 10.1016/s0014-5793(97)00439-0. [DOI] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, Van Dillewijn J, Rochaix J-D. Mutation at the Chlamydomonas nuclear NAC-2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of photosystem II. Cell. 1989;58:869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Kyle DJ, Ohad I, Arntzen CJ. Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc Natl Acad Sci USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vector: a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1990;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Marder JB, Edelman M. Dynamics of the photosystem II reaction center. Cell. 1989;56:241–246. doi: 10.1016/0092-8674(89)90897-0. [DOI] [PubMed] [Google Scholar]
- Michel H, Hunt DF, Shabanowitz J, Bennett J. Tandem mass spectrometry reveals that three photosystem II proteins of spinach chloroplast contain N-acetyl-O-phosphothreonine at their N-termini. J Biol Chem. 1988;263:1123–1130. [PubMed] [Google Scholar]
- Michel HP, Bennett J. Identification of the phosphorylation site of an 83 kDa protein from photosystem II of spinach. FEBS Lett. 1987;212:103–108. [Google Scholar]
- Nickelsen J, van Dillewijn J, Rahire M, Rochaix J-D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GC, Ohad I. Phosphorylation of Chlamydomonas reinhardtii chloroplast membrane proteins in vivo and in vitro. J Cell Biol. 1982;93:712–718. doi: 10.1083/jcb.93.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Rintamäki E, Kettunen R, Aro E-M. Differential D1 dephosphorylation in functional and photodamaged photosystem II centers. J Biol Chem. 1996;271:14870–14875. doi: 10.1074/jbc.271.25.14870. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D, Dron M, Rahire M, Malnoe PM. Sequence homology between the 32K dalton and the D2 chloroplast membrane polypeptides of Chlamydomonas reinhardtii. Plant Mol Biol. 1984;3:363–370. doi: 10.1007/BF00033383. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarkar G, Sommer SS. The ‘megaprimer’ method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Schuster G, Timberg R, Ohad I. Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem. 1988;177:403–410. doi: 10.1111/j.1432-1033.1988.tb14389.x. [DOI] [PubMed] [Google Scholar]
- Stampacchia O, Girard-Bascou J, Zanasco J-L, Zerges W, Bennoun P, Rochaix J-D. A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer EJ, Schmid VHR, Bruns BU, Schmidt GW. Requirement for the phosphoprotein H in photosystem II of Chlamydomonas reinhardtii. Plant Physiol. 1997;113:1359–1368. doi: 10.1104/pp.113.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O, Wollman FA, Olive J. Distribution of intrinsic and extrinsic subunits of the PSII protein complex between appressed and non-appressed regions of the thylakoid membrane: an immunocytochemical study. FEBS Lett. 1985;183:245–250. [Google Scholar]
- Wollman F-A, Delepelaire P. Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J Cell Biol. 1984;97:1–7. doi: 10.1083/jcb.98.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F-A, Lemaire C. Studies on kinase-controlled state transitions in photosystem II and b6/f mutants from Chlamydomonas reinhardtii which lack quinone-binding proteins. Biochim Biophys Acta. 1988;993:85–94. [Google Scholar]